It has been reported that the activation of multiple myeloma (MM) cells by CD40 induces proliferation, growth arrest, and apoptosis. To determine whether the biologic sequelae of CD40 activation in MM cells depends on p53 function, we identified temperature-sensitive p53 mutations in the RPMI 8226 (tsp53E285K) and the HS Sultan (tsp53Y163H) MM cell lines. These cells were then used as a model system of inducible wtp53-like function because wild-type-like p53 is induced at permissive (30°C) but not at restrictive (37°C) temperatures. Using p21-luciferase reporter assays, we confirmed that CD40 induces p53 transactivation in RPMI 8226 and HS Sultan cells cultured under permissive, but not restrictive, conditions. Furthermore, CD40 activation of these MM cells under permissive, but not restrictive, temperatures increased the expression of p53 and p21 mRNA and protein. Importantly, CD40 activation induced the proliferation of RPMI 8226 and HS Sultan cells at restrictive temperatures and growth arrest and increased subG1 phase cells at permissive temperatures. These data confirmed that CD40 activation might have distinct biologic sequelae in MM cells, depending on their p53 status.
Others1-6 and we have demonstrated the effect of CD40 activation on the proliferation and differentiation of B lymphocytes. Triggering of multiple myeloma (MM) cells by cell surface CD40 has been shown to induce the proliferation of tumor cells in some studies7-12 and to trigger growth arrest13 and apoptosis14 in others. Ligation of CD40 on MM cells also induces the secretion of transforming growth factor-β1 (TGF-β1)12 and increases the expression of adhesion molecules9 and the Ku86 autoantigen15 on the MM cell surface, thereby augmenting homotypic tumor cell adhesion and heterotypic binding of tumor cells to bone marrow stromal cells (BMSCs) and fibronectin. Up-regulation of MM cell adhesion to BMSCs in turn triggers the transcription and secretion of interleukin (IL)-6 mRNA expression and protein secretion in BMSCs.9 15-20
Earlier studies characterize the signaling cascades mediating IL-6-triggered growth21,22 and survival23-25 in MM cells and demonstrate that apoptosis of MM cells, induced by wild-type p53 (wtp53)26,27 and p21,28 can be abrogated by treatment with exogenous IL-6. These studies establish IL-6 as a major autocrine and paracrine growth factor, especially in CD40-activated MM cells. Cell cycle regulatory proteins, including wtp53, directly regulate IL-6 secretion; wtp53 represses the IL-6 promoter, whereas IL-6 promoter activity is not down-regulated by mtp53.29-31 These data suggest that the CD40 activation of MM cells may induce growth and survival rather than growth arrest and apoptosis, depending on their p53 status. In addition, they suggest that the activation of CD40 can alter the expression of p53 and consequently of p21 proteins. However, the direct role of CD40 activation in regulating p53 and p21 expression and p53-dependent cell cycle control has not been studied.
In this study, we first identified temperature-sensitive missense mutations in the p53 gene in 2 human MM-derived cell lines: codon 285, Glu [GAG] to Lys [AAG] in RPMI 8226, and codon 163, Tyr [TAC] to His [CAC] in HS Sultan MM cells. These cell lines were then used as a model system of inducible wtp53-like function because wt-like p53 conformation is induced in these cells at permissive, but not at restrictive, temperatures. Using p21-luciferase reporter assays, we confirmed that CD40 activation of these MM cell lines at permissive, but not at restrictive, temperatures increased p53 transactivation activity. We then showed that p21 and p53 mRNA and protein expression were increased in CD40-activated RPMI 8226 and HS Sultan cells cultured under permissive, but not restrictive, conditions. Finally, CD40 activation of RPMI 8226 and HS Sultan cells at restrictive temperatures increased G1/S transition and tumor cell proliferation; in contrast, CD40 activation of these cells at permissive temperatures increased growth arrest and subG1 phase cells. These data demonstrated that CD40 activation mediated p53-dependent cell cycle regulation in human MM cell lines.
Methods
Cell lines and transfectants
RPMI 8226 (CCL-155) and HS Sultan (CRL-1484) human MM cells were obtained from the American Type Culture Collection (ATCC, Rockville, MD) and were cultured in 90% RPMI-1640 medium with 25 IU/mL penicillin, 25 μg/mL streptomycin, and 5 mmol/L L-glutamine (all from Gibco BRL, Gaithersburg, MD), and 10% FBS (PAA Laboratories, Newport Beach, CA). Wild-type NIH3T3 murine fibroblasts (NIH3T3/wt), NIH3T3 cells transfected with the human CD40 ligand(CD40L) cDNA (NIH3T3/CD40LT), and NIH3T3 cells transfected with the vector plasmid (NIH3T3/vt) were provided by Dr Gordon Freeman (Dana-Farber Cancer Institute, Boston, MA) and were cultured in F12/Dulbecco's modified Eagle's medium.9 15 NIH3T3/CD40LT and NIH3T3/vt were further selected with 400 μg/mL Geneticin (G418) (Gibco BRL). All cells were grown at 30°C or 37°C in a humidified 5% CO2 atmosphere. For CD40 activation of MM cell lines, NIH3T3/CD40LT was fixed in 1% formalin (Sigma Diagnostics, St. Louis, MO) and cultured with MM cells (100 MM cells:1 NIH3T3 cell) for up to 144 hours. Induction of wtp53 function was attempted using γ-irradiation (2 cGy), followed by culture in complete medium for 2 hours at 30°C or 37°C.
Cell proliferation and apoptosis assays
RPMI 8226 and HS Sultan cells (0.5-1.0 × 106initiating cells/mL) were cultured in media with formalin-fixed NIH3T3/CD40LT, NIH3T3/wt, or NIH3T3/vt for up to 144 hours. Viable cell density was determined by trypan blue (Gibco BRL) exclusion. DNA synthesis was assayed using tritiated thymidine (3H-TdR) (Dupont NEN, Boston, MA) incorporation, as previously described15 32 using 0.5 μCi 3H-TdR for each sample (0.1 × 106 initiating cells/well) 6 hours before harvesting onto glass filters using the Harvestar 96 Mach II harvester (Tomtec, Orange, CT) and analysis on the 1205 Betaplate counter (Wallac, Gaithersburg, MD). Experiments were performed in triplicate, and results were expressed as mean ± SEM.
Cell cycle distribution analysis
RPMI 8226 and HS Sultan cells (0.5 × 106cells/mL) were cultured in media with formalin-fixed NIH3T3/CD40LT, NIH3T3/wt, or NIH3T3/vt for up to 144 hours. Cells were fixed and permeabilized with 80% ethanol on ice for 1 hour and resuspended in phosphate-buffered saline (PBS) with DNase-free RNase (10 μg/mL; Boehringer Mannheim, Indianapolis, IN) and stained with propidium iodide (PI) (15 μg/mL, Sigma Diagnostics). Cell cycle distribution was assessed by flow cytometric analysis on the Coulter Epics XL flow cytometer (Coulter, Hialeah, FL).
Antibodies for immunoprecipitation and Western blotting
The following antibodies (Abs) were used: Ab-5 anti-wtp53 (clone PAb 1620) monoclonal Ab (mAb), which recognizes wtp53 conformation; Ab-3 anti-mtp53 (clone PAb 240) mAb, which recognizes mtp53 conformation; and Ab-1 anti-actin mAb (all from Oncogene Science, Cambridge, MA); DO-1 horseradish peroxidase (HRP)-conjugated anti-pantropic (ptp53) mAb, C-19 goat anti-p21 polyclonal Ab (pAb), HRP-conjugated anti-mouse immunoglobulin (Ig), and HRP-conjugated donkey anti-goat IgG mAb (all from Santa Cruz Biotechnology, Santa Cruz, CA); and DM 1A antitubulin mAb (Sigma Diagnostics).
Immunoprecipitation and Western blotting
Total cell lysates were obtained as previously described15 32 and quantified by Bradford's method (Bio-Rad, Hercules, CA). For immunoprecipitation, 1 mL cell lysate (1 mg/mL) was incubated with 10 μg of the relevant Ab and 100 μL of 10% Protein-A Sepharose CL-4B (Pharmacia Biotech, Uppsala, Sweden); for Western blotting, total protein (30 μg/sample) was used. The samples were resolved by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE), transferred to polyvinylidene difluoride membranes (Millipore, Bedford, MA), blocked with 5% skim milk (Saco Foods, Middleton, WI) in PBS, 0.2% Tween-20, and 0.02% NaN3 (both from Sigma Diagnostics); and immunoblotted with 10 μg relevant Ab, followed by appropriate HRP-conjugated secondary Ab. Detection was performed by HRP chemiluminescence (ECL; Amersham Life Science) autoradiography (Biomax MR; Eastman Kodak, Rochester, NY). The relative intensity of expression was assessed using imaging densitometry (AlphaImager 2000; Alpha Innotech, San Leandro, CA).
Direct DNA sequencing of human p53 gene exons IV to IX
Genomic DNA was extracted from both MM cell lines (2 × 106/sample) using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI) and was quantified (260 nm) on the DU 640 Spectrophotometer (Beckman Instruments, Columbia, MD). In the first-step polymerase chain reaction (PCR), a 2.8-kb fragment of DNA that encodes exons IV through IX was obtained using the LA-PCR Kit (Takara, Tokyo, Japan). The PCR conditions were as follows: forward primer, 5′-AGGACCTGGTCCTCTGACTG-3′; reverse primer, 5′-TAGACTGGAAACTTCCACTTG-3′; 94°C at 30 seconds (denaturation); 58°C at 60 seconds (annealing); and 72°C at 60 seconds (extension) for 35 cycles. Single-strand DNA33 was produced by asymmetrical PCR using the deoxy chain termination method with end-labeled sequencing primers for exon IV (forward primer, 5′-TTTCACCCATCTACAGTCCCC-3′; reverse primer, 5′-GAAGTCTCATGGAAGCCAGCC-3′); exons V and VI (forward primer, 5′-TTCCTCTTCTACAGTACTCC-3′; reverse primer, 5′-AGTTGCAAACCAGACCTCAGG-3′); exon VII (forward primer, 5′-CCAAGGCGCACTGGCCTCATC-3′; reverse primer, 5′-TCAGCGGCAAGCAGAGGCTGG-3′); and exons VIII and IX (forward primer, 5′-CCTATCCTGAGTAGTGGTAAT-3′; reverse primer, 5′-TAAGAGGTCCCAAGACTTAGT-3′) of the human p53 gene (PRISM DyeDeoxy Terminator Sequencing Kit FS; Applied Biosystems, Foster City, CA). DNA sequencing was performed on the ABIPRISM 310 Genetic Analyzer (Applied Biosystems). Sequencing of the antisense DNA was used to confirm mutations.
p21-Luciferase reporter assay
p21-Luciferase reporter plasmid—either PG13-luc, which contains 13 copies of a normal p53-binding site, or MG15-luc, which contains 15 copies of a subtly mutated p53-binding site (both kind gifts of Dr Bert Vogelstein, Johns Hopkins University, Baltimore, MD)34—was transfected (10 μg/sample) into RPMI 8226 and HS Sultan MM cell lines (10 × 106/sample) using electroporation (Bio-Rad Gene Pulser; Bio-Rad). Tumor cells were incubated at 37°C for 14 hours to 16 hours and then cultured in media or with formalin-fixed NIH3T3/CD40LT for up to 40 hours. Next MM cells were harvested, and luciferase activity was expressed as relative light units (Luciferase Assay System; Promega) measured using the Monolight 2010 Luminometer (Analytical Luminescence Laboratory, Frederick, MD). Total protein content was used for the normalization of luciferase activity. Experiments were performed in triplicate, and results were expressed as mean ± SEM normalized luciferase activity.
Ribonuclease protection assay
RPMI 8226 or HS Sultan MM cells (10 × 106/sample) were cultured at restrictive (37°C) and permissive (30°C) temperatures in media and with formalin-fixed NIH3T3/CD40LT, NIH3T3/wt, or NIH3T3/vt for up to 72 hours. Total cellular RNA was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA) and quantified (260 nm) on the DU 640 Spectrophotometer (Beckman Instruments). The following cDNA templates were used: p21 cDNA cloned to pcDNA3 plasmid (Invitrogen, Carlsbad, CA) and linearized at the Kpn I1164 restriction site (Gibco BRL); p53 cDNA cloned to pcDNA3 plasmid (Invitrogen) and linearized at the BssS I1019 restriction site (New England Biolabs, Beverly, MA); and pTri-actin control cDNA (Ambion, Austin, TX). Next α-32P-UTP (Dupont NEN)-labeled antisense riboprobes were transcribed using the SP6 RNA polymerase promoter from these cDNA templates (MAXIscript T7/SP6 in vitro transcription kit; Ambion) and purified on a 5% polyacrylamide/8 mol/L urea sequencing gel. Ribonuclease protection assay (RPA) (10 μg total RNA/sample) was performed using the RPA III kit (Ambion), resolved on a denaturing 5% polyacrylamide/8 mol/L urea sequencing gel, and detected by autoradiography (Eastman Kodak). The relative intensity of expression was assessed by imaging densitometry (AlphaImager 2000; Alpha Innotech).
Results
Characterization of p53 mutations in RPMI 8226 and HS Sultan MM cell lines
To determine the molecular mechanisms for the contrasting biologic sequelae of CD40 activation in MM cells, we first characterized the p53 gene in 2 human MM cell lines, RPMI 8226 and HS Sultan. Polymerase chain reaction and direct DNA sequencing identified a previously described temperature-sensitive missense mutation (tsp53E285K, codon 285 (Glu [GAG] to Lys [AAG])) capable of producing p53 transactivation function at 30°C in RPMI 8226 cells 35and another missense mutation (mtp53Y163H, codon 163 (Tyr [TAC] to His [CAC])) in HS Sultan cells. Sequencing of the antisense DNA was used to confirm mutations.
Characterization of temperature-dependent p53 protein conformation in RPMI 8226 and HS Sultan MM cell lines
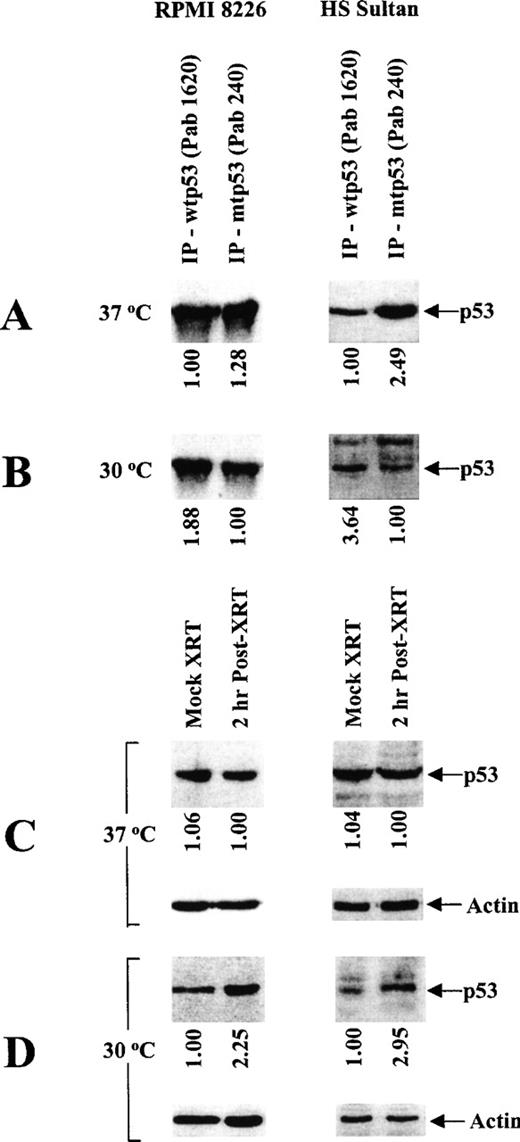
Because missense mutations in the central region (codons 101-318) of the human p53 gene often confer temperature sensitivity (tsp53),36 we next characterized the effect of temperature on the expression of p53 protein using conformation-specific mAbs directed against wtp53 (PAb 1620) and mtp53 (PAb 240) for immunoprecipitation, followed by immunoblotting with a nonconformation-specific pantropic p53 (ptp53) mAb (DO-1) (Figure1). Specifically, the effect of temperature on wtp53 expression and mtp53 conformation was assessed by culturing tumor cells at restrictive (37°C) rather than permissive (30°C) temperatures. RPMI 8226 and HS Sultan cells predominantly express mtp53 when cultured at restrictive temperatures (Figure 1A), and they express wtp53 when cultured at permissive temperatures (Figure 1B). To confirm the above pattern of wtp53 expression in these MM cells, we also carried out mock- and γ-irradiated cultures of these tumor cells at 37°C or 30°C and assayed for p53 protein expression as described above. Expression of wtp53 was increased by γ-irradiation in RPMI 8226 and HS Sultan cells cultured at permissive (Figure 1D), but not at restrictive (Figure 1C) temperatures. These data confirm the differences in wt-like p53 expression in RPMI 8226 and HS Sultan cells cultured at permissive rather than restrictive temperatures.
Characterization of temperature-dependent p53 protein conformation in RPMI 8226 and HS Sultan MM cell lines.
RPMI 8226 and HS Sultan cells were cultured at 37°C (A, C) or 30°C (B, D). Total tumor cell lysates (1 mg/sample) were immunoprecipitated (10 μg) with either Ab-5 anti-wtp53 (Pab 1620) mAb or Ab-3 anti-mtp53 (Pab 240) mAb and immunoblotted (10 μg) with DO-1 HRP-conjugated anti-ptp53 mAb (A, B). Tumor cells were also mock- or γ-irradiated (2 cGy) and cultured at 37°C (C) or at 30°C (D) for 2 hours to induce wtp53 expression. Total cell lysates (30 μg/sample) of mock- or γ-irradiated tumor cells were resolved by SDS-PAGE and immunoblotted (10 μg) with DO-1 HRP-conjugated anti-ptp53 mAb or with Ab-1 anti-actin mAb.
Characterization of temperature-dependent p53 protein conformation in RPMI 8226 and HS Sultan MM cell lines.
RPMI 8226 and HS Sultan cells were cultured at 37°C (A, C) or 30°C (B, D). Total tumor cell lysates (1 mg/sample) were immunoprecipitated (10 μg) with either Ab-5 anti-wtp53 (Pab 1620) mAb or Ab-3 anti-mtp53 (Pab 240) mAb and immunoblotted (10 μg) with DO-1 HRP-conjugated anti-ptp53 mAb (A, B). Tumor cells were also mock- or γ-irradiated (2 cGy) and cultured at 37°C (C) or at 30°C (D) for 2 hours to induce wtp53 expression. Total cell lysates (30 μg/sample) of mock- or γ-irradiated tumor cells were resolved by SDS-PAGE and immunoblotted (10 μg) with DO-1 HRP-conjugated anti-ptp53 mAb or with Ab-1 anti-actin mAb.
Effect of CD40 activation on p53 transactivation in RPMI 8226 and HS Sultan MM cell lines
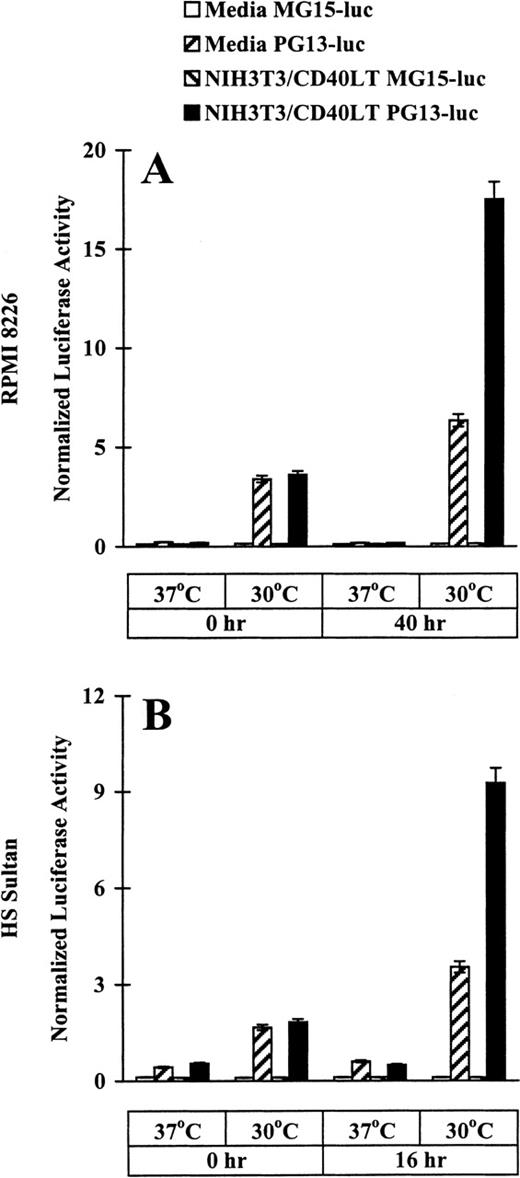
After demonstrating the tsp53 mutations in RPMI 8226 and HS Sultan cell lines and the differences in wtp53 protein expression at permissive rather than restrictive conditions, we used p21-luciferase reporter assays to assess directly the temperature-dependent specific p53 transactivation function. RPMI 8226 and HS Sultan cells were transfected with either PG13-luc, plasmid that contains 13 copies of a normal p53-binding site, or with MG15-luc, plasmid that contains 15 copies of a mutated p53-binding site. Luciferase assays were performed in cells cultured in media or with NIH3T3/CD40LT for up to 40 hours at restrictive and permissive temperatures. Baseline (time 0 hour) normalized luciferase activity was increased in RPMI 8226 (Figure2A) and HS Sultan (Figure 2B) cells transfected with PG13-luc, but not with MG15-luc, at permissive rather than restrictive temperatures. In cultures with NIH3T3/CD40LT at permissive (but not restrictive) temperatures, marked increases in normalized luciferase activity were observed in PG13-luc-transfected RPMI 8226 at 40 hours (Figure 2A) and HS Sultan cells at 16 hours (Figure 2B), but not in cells transfected with MG15-luc. These data provide strong evidence that CD40 activation induces functional wtp53 in a temperature-dependent fashion in these tumor cells.
Effect of CD40 activation on p53 transactivation in RPMI 8226 and HS Sultan MM cell lines.
RPMI 8226 (A) and HS Sultan (B) cells (10 × 106/sample) were transfected with either PG13-luc, a plasmid containing 13 copies of normal p53-binding site, or with MG15-luc, a plasmid containing 15 copies of mutated p53-binding site, using electroporation. Tumor cells were incubated at 37°C for 14 to 16 hours and cultured in media and with formalin-fixed NIH3T3/CD40LT at 37°C or 30°C for up to 40 hours. Cells were harvested, and luciferase activity was normalized to total cellular protein content. Experiments were performed in triplicate, and results are expressed as mean ± SEM normalized luciferase activity.
Effect of CD40 activation on p53 transactivation in RPMI 8226 and HS Sultan MM cell lines.
RPMI 8226 (A) and HS Sultan (B) cells (10 × 106/sample) were transfected with either PG13-luc, a plasmid containing 13 copies of normal p53-binding site, or with MG15-luc, a plasmid containing 15 copies of mutated p53-binding site, using electroporation. Tumor cells were incubated at 37°C for 14 to 16 hours and cultured in media and with formalin-fixed NIH3T3/CD40LT at 37°C or 30°C for up to 40 hours. Cells were harvested, and luciferase activity was normalized to total cellular protein content. Experiments were performed in triplicate, and results are expressed as mean ± SEM normalized luciferase activity.
Induction of p53 and p21 transcription and expression by CD40 activation of RPMI 8226 and HS Sultan MM cell lines
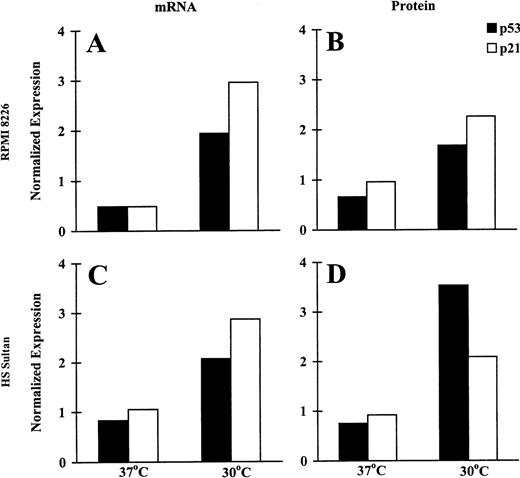
To assess the biologic significance of wtp53 activation induced by CD40 activation in RPMI 8226 and HS Sultan cells at permissive temperatures, we assayed for the CD40L-triggered induction of p53 and p21 mRNA with RPA and protein expression using Western blot analysis. Probing with β-actin probe and DM 1A antitubulin mAb served as a control for RPA and Western blot analysis, respectively. The relative intensity of expression was assessed using imaging densitometry and was normalized against the expression in control cells cultured in media without NIH3T3/CD40LT. Relevant data of CD40-triggered tumor cells are presented as normalized mRNA or protein expression in Figure3. CD40L triggered increased p53 and p21 mRNA (Figures 3A, 3C) and protein (Figures 3B, 3D) expression in both MM cell lines when cultured at permissive, but not at restrictive, temperatures. Specifically, in cultures at 30°C, CD40 activation triggered 1.94- and 2.07-fold increases in p53 mRNA in RPMI 8226 (Figure 3A) and HS Sultan (Figure 3C) cells. CD40 activation also induced 2.96- and 2.87-fold increments in p21 mRNA in RPMI 8226 (Figure3A) and HS Sultan (Figure 3C) cells, respectively, in cultures at 30°C but not at 37°C. Similar trends were noted in p21 and p53 protein expression. Specifically, after culturing with CD40L, p53 protein expression was increased 1.68- and 3.53-fold in RPMI 8226 (Figure 3B) and HS Sultan (Figure 3D) cells. Similarly, p21 protein expression was increased 2.26- and 2.09-fold in RPMI 8226 (Figure 3B) and HS Sultan (Figure 3D) cells, respectively, by CD40 activation at 30°C, but not at 37°C.
Induction of p53 and p21 transcription and expression by CD40 activation of RPMI 8226 and HS Sultan MM cell lines.
RPMI 8226 (A, B) and HS Sultan (C, D) cells were cultured in media and with formalin-fixed NIH3T3/CD40LT, NIH3T3/wt, or NIH3T3/vt at 37°C or 30°C for 72 hours. p53 and p21 mRNA expression (A, C) was assayed using RPA; p53 and p21 protein expression (B, D) was assayed using Western blotting. β-Actin probe and DM 1A anti-tubulin mAb served as controls for RPA and Western blotting, respectively. The relative intensity of expression was assessed using imaging densitometry and was normalized against expression in control cells cultured in media without NIH3T3/CD40LT. Relevant data of CD40-triggered tumor cells are presented as normalized mRNA or protein expression.
Induction of p53 and p21 transcription and expression by CD40 activation of RPMI 8226 and HS Sultan MM cell lines.
RPMI 8226 (A, B) and HS Sultan (C, D) cells were cultured in media and with formalin-fixed NIH3T3/CD40LT, NIH3T3/wt, or NIH3T3/vt at 37°C or 30°C for 72 hours. p53 and p21 mRNA expression (A, C) was assayed using RPA; p53 and p21 protein expression (B, D) was assayed using Western blotting. β-Actin probe and DM 1A anti-tubulin mAb served as controls for RPA and Western blotting, respectively. The relative intensity of expression was assessed using imaging densitometry and was normalized against expression in control cells cultured in media without NIH3T3/CD40LT. Relevant data of CD40-triggered tumor cells are presented as normalized mRNA or protein expression.
Effect of temperature on viability of CD40-activated RPMI 8226 and HS Sultan MM cell lines
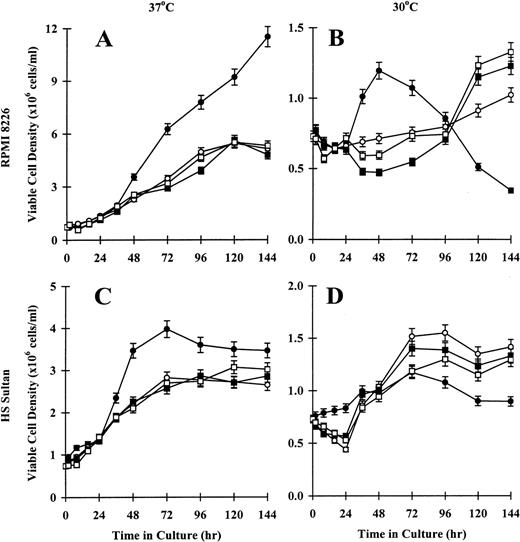
Because RPMI 8226 and HS Sultan cells underwent tsp53 mutations with temperature-dependent differences in p53 transactivation and p53 and p21 mRNA and protein expression, we studied the effect of temperature on the CD40 activation of these tumor cells. First we examined viable cell density, assessed by trypan blue exclusion. As seen in Figure4A, the density of viable CD40-activated RPMI 8226 cells cultured at 37°C increased as much as 11-12 × 106 cells/mL at 144 hours. Tumor cells cultured in media, with NIH3T3/wt, or with NIH3T3/vt showed maximal growth densities between 5-6 × 106 cells/mL at 96 hours to 144 hours. In contrast, CD40-activated RPMI 8226 cells cultured at 30°C exhibited only modest growth to 1-1.5 × 106 cells/mL at 48 hours, with the number of viable cells decreasing to below baseline at 144 hours. Tumor cells cultured in media, with NIH3T3/wt, or with NIH3T3/vt showed slow but sustained growth to approximately 1-1.5 × 106cells/mL continuing to 144 hours (Figure 4B). Similar growth patterns were observed in HS Sultan cells. Specifically, the density of viable CD40-activated HS Sultan cells cultured at 37°C increased rapidly to 3-4 × 106 cells/mL at 48 hours. Cells cultured in media, with NIH3T3/wt, or with NIH3T3/vt demonstrated slower growth, with a maximum viable cell density of 3 × 106cells/mL at 96 hours to 144 hours (Figure 4C). As seen in Figure 4D, growth of CD40-activated HS Sultan cells cultured at 30°C was reduced to 0.5-1.0 × 106 cells/mL at 144 hours, whereas cells cultured in media, with NIH3T3/wt, or with NIH3T3/vt demonstrated sustained growth to approximately 1.0-1.5 × 106 cells/mL at 144 hours. These data confirm that CD40 activation results in differences in viable cell numbers of RPMI 8226 and HS Sultan cells, which are temperature dependent.
Effect of temperature on viability of CD40-activated RPMI 8226 and HS Sultan MM cell lines.
RPMI 8226 (A, B) and HS Sultan (C, D) cells (0.7 × 106initiating cells/well) were cultured in media (○) and with formalin-fixed NIH3T3/CD40LT (•), NIH3T3/wt (□), or NIH3T3/vt (▪) for up to 144 hours. Tumor cells were cultured at 37°C (A, C) or 30°C (B, D), and viable cell density was assessed at 0, 2, 8, 16, 24, 36, 48, 72, 96, 120, and 144 hours using trypan blue exclusion. Experiments were performed in triplicate, and results are expressed as mean ± SEM.
Effect of temperature on viability of CD40-activated RPMI 8226 and HS Sultan MM cell lines.
RPMI 8226 (A, B) and HS Sultan (C, D) cells (0.7 × 106initiating cells/well) were cultured in media (○) and with formalin-fixed NIH3T3/CD40LT (•), NIH3T3/wt (□), or NIH3T3/vt (▪) for up to 144 hours. Tumor cells were cultured at 37°C (A, C) or 30°C (B, D), and viable cell density was assessed at 0, 2, 8, 16, 24, 36, 48, 72, 96, 120, and 144 hours using trypan blue exclusion. Experiments were performed in triplicate, and results are expressed as mean ± SEM.
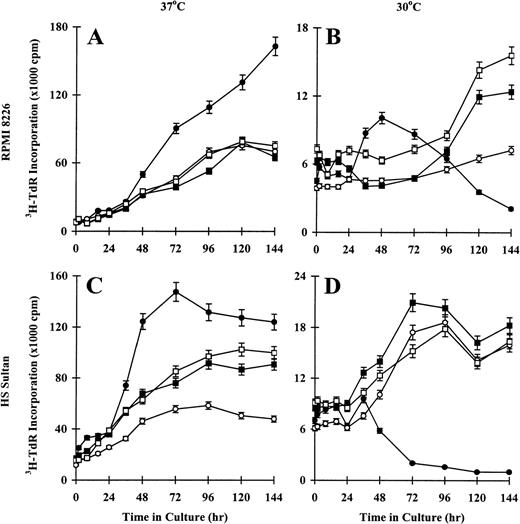
Effect of temperature on proliferation of CD40-activated RPMI 8226 and HS Sultan MM cell lines
To confirm the effect of temperature on CD40-induced proliferation of RPMI 8226 and HS Sultan cells, we performed 3H-TdR incorporation assays on tumor cells cultured at restrictive temperatures (37°C) and at permissive temperatures (30°C). Sustained increases in DNA synthesis were observed in CD40-activated RPMI 8226 cells cultured at 37°C, whereas tumor cells cultured in media, with NIH3T3/wt, or with NIH3T3/vt showed maximal3H-TdR incorporation at 96 hours to 144 hours (Figure5A). In contrast, proliferation of CD40-activated RPMI 8226 cells cultured at 30°C peaked at 48 hours and decreased to 144 hours, whereas tumor cells cultured in media, with NIH3T3/wt, or with NIH3T3/vt demonstrated slow, sustained growth to 144 hours (Figure 5B). Similar patterns of DNA synthesis were observed in HS Sultan cells. For example, maximum proliferation of CD40-activated HS Sultan cells cultured at 37°C was observed at 48 hours to 72 hours (Figure 5C). Tumor cells cultured in media, with NIH3T3/wt, or with NIH3T3/vt showed slow, sustained increases in 3H-TdR incorporation, which peaked at 96 hours to 144 hours. In contrast, DNA synthesis of CD40-activated HS Sultan cells cultured at 30°C peaked at 48 hours and decreased to 144 hours, whereas these cells cultured in media, with NIH3T3/wt, or with NIH3T3/vt demonstrated sustained increases in DNA synthesis up to 144 hours (Figure 5D). These data further support the functional significance of the observed tsp53 in RPMI 8226 and HS Sultan cells.
Effect of temperature on proliferation of CD40-activated RPMI 8226 and HS Sultan MM cell lines.
RPMI 8226 (A, B) and HS Sultan (C, D) cells (0.7 × 106 initiating cells/well) were cultured in media (○) and with formalin-fixed NIH3T3/CD40LT (•), NIH3T3/wt (□), or NIH3T3/vt (▪) for up to 144 hours. Tumor cells were cultured at 37°C (A, C) or 30°C (B, D), and DNA synthesis was assessed at 0, 2, 8, 16, 24, 36, 48, 72, 96, 120, and 144 hours using3H-TdR incorporation. Experiments were performed in triplicate, and results are expressed as mean ± SEM.
Effect of temperature on proliferation of CD40-activated RPMI 8226 and HS Sultan MM cell lines.
RPMI 8226 (A, B) and HS Sultan (C, D) cells (0.7 × 106 initiating cells/well) were cultured in media (○) and with formalin-fixed NIH3T3/CD40LT (•), NIH3T3/wt (□), or NIH3T3/vt (▪) for up to 144 hours. Tumor cells were cultured at 37°C (A, C) or 30°C (B, D), and DNA synthesis was assessed at 0, 2, 8, 16, 24, 36, 48, 72, 96, 120, and 144 hours using3H-TdR incorporation. Experiments were performed in triplicate, and results are expressed as mean ± SEM.
Effect of temperature on cell cycle profile of CD40-activated RPMI 8226 and HS Sultan MM cell lines
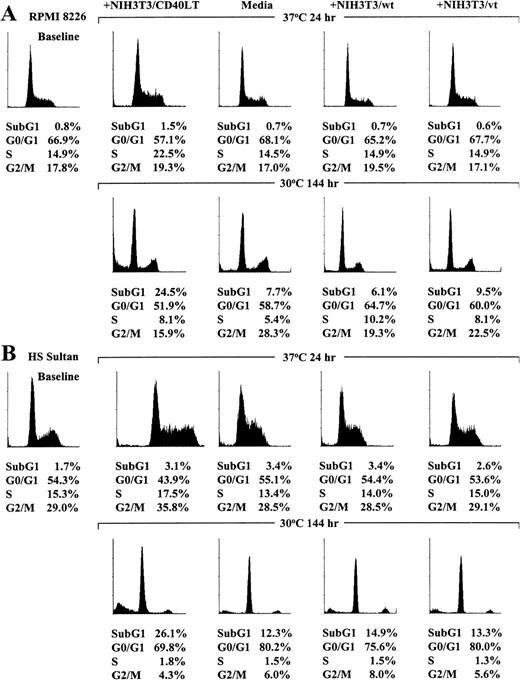
Because cell proliferation was induced by CD40 activation in RPMI 8226 and HS Sultan cells cultured at restrictive temperatures (37°C) but growth arrest was observed in these tumor cell lines cultured at permissive temperatures (30°C), we next studied the effect of temperature on the cell cycle profile, assessed using PI staining, after CD40 activation of each MM cell line. As can be seen in Figure 6A, the profile of RPMI 8226 cells cultured at 37°C for 24 hours in media, with NIH3T3/wt, or with NIH3T3/vt was similar, whereas RPMI 8226 cells cultured with NIH3T3/CD40LT demonstrated G1/S transition. In contrast, CD40-activated RPMI 8226 cells cultured at permissive temperatures (30°C) for 144 hours showed increased subG1 phase cells compared with cells cultured in media, with NIH3T3/wt, or with NIH3T3/vt.
Effect of temperature on cell cycle profile of CD40-activated RPMI 8226 and HS Sultan MM cell lines.
RPMI 8226 (A) and HS Sultan (B) cells were cultured in media and with formalin-fixed NIH3T3/CD40LT, NIH3T3/wt, or NIH3T3/vt at 37°C or 30°C for up to 144 hours. The cell cycle profile was assessed using PI staining and flow cytometric analysis (0.5 × 106cells/sample) at 0 hour and at culture conditions in which maximum cell cycle effects were noted at 24 hours for cultures at 37°C and at 144 hours for cultures at 30°C.
Effect of temperature on cell cycle profile of CD40-activated RPMI 8226 and HS Sultan MM cell lines.
RPMI 8226 (A) and HS Sultan (B) cells were cultured in media and with formalin-fixed NIH3T3/CD40LT, NIH3T3/wt, or NIH3T3/vt at 37°C or 30°C for up to 144 hours. The cell cycle profile was assessed using PI staining and flow cytometric analysis (0.5 × 106cells/sample) at 0 hour and at culture conditions in which maximum cell cycle effects were noted at 24 hours for cultures at 37°C and at 144 hours for cultures at 30°C.
The effect of temperature on the cell cycle profile in CD40-activated HS Sultan cells was similar to that observed in RPMI 8226 cells. As can be seen in Figure 6B, the cell cycle profile of HS Sultan cells cultured at 37°C for 24 hours in media, with NIH3T3/wt, or with NIH3T3/vt was similar, but HS Sultan cells cultured with NIH3T3/CD40LT demonstrated G1/S transition. In contrast, increased subG1 phase cells were observed in CD40-activated HS Sultan cells cultured at permissive temperature for 144 hours compared with cultures of cells in media, with NIH3T3/wt, or with NIH3T3/vt.
Discussion
In this study we report that CD40 activation of human MM-derived cell lines can induce either proliferation or growth arrest, depending on their p53 status. We characterized 2 MM cell lines that had naturally occurring tsp53 mutations, and we used p21-luciferase reporter assays in these 2 cell lines to demonstrate that CD40 activation increased p53 transactivation function at permissive, but not at restrictive, temperatures. Moreover, p53 and p21 mRNA expression and protein expression was increased by CD40 activation of these MM cells cultured at permissive, but not at restrictive, temperatures. Of importance, we demonstrated that CD40 activation of these tumor cells cultured at restrictive temperatures was associated with increased numbers of viable tumor cells, tumor cell proliferation, and G1/S transition but that CD40 activation of MM cells cultured at permissive temperatures was associated with growth arrest and increased subG1 phase cells. These data confirmed that CD40 activation mediated cell cycle regulation in a p53-dependent mechanism.
Earlier reports demonstrate that the ligation of CD40 induces the proliferation of MM cells,7-12 growth arrest,13and apoptosis.14 However, the mechanisms underlying these contrasting sequelae of CD40 activation have not been characterized. Furthermore, the panoply of ways to activate MM cells by their cell surface CD40, including the use of mAbs against CD40, soluble CD40L, or cells transfected with the human CD40L cDNA, has confounded the interpretation of results. For example, it appeared that growth arrest and apoptosis were associated with triggering by anti-CD40 mAbs, either in soluble form or immobilized on the surface of L cells.13 14 The current study clearly showed that both these effects, as well as proliferation or growth arrest, can be observed after CD40 activation of MM cells. These data suggested that the differences in CD40 activation responses observed in earlier studies did not result from the manner in which CD40 activation was achieved but that they may be related to inherent differences in cell cycle regulatory proteins (ie, p53) in tumor cells.
To test this hypothesis, we used 2 naturally occurring tsp53 mutant MM cell lines. We specifically studied RPMI 8226 cells, which were previously described as having a missense mutation at codon 285 (Glu [GAG] to Lys [AAG]) (tsp53E285K), and we confirmed that they were capable of transactivation at 30°C but not at 37°C.35 To strengthen our model system, we identified another p53 mutation in a second (HS Sultan) MM cell line, a missense mutation in codon 163 (Tyr [TAC] to His [CAC]; tsp53Y163H), which also conferred temperature-dependent wtp53 function. Both MM cell lines behaved similarly when activated by CD40. At restrictive temperatures, p53 transactivation function was greatly diminished, and sustained proliferation of these tumor cell lines was observed in long-term cultures. At permissive temperatures, sustained proliferation of both cell lines was observed, but at reduced rates, compared with cultures at restrictive conditions. It is significant that CD40 activation of these cells at permissive, but not at restrictive, temperatures induced growth arrest and increased subG1 phase cells. These distinct outcomes of CD40 activation at restrictive rather than permissive conditions suggested a p53-dependent mechanism.
To confirm the role of p53 transactivation in the distinct responses to CD40 activation, we used p21-luciferase reporter assays. Results showed that culture of RPMI 8226 and HS Sultan cell lines at permissive temperatures increased p53 transactivation compared with cultures at restrictive conditions. Most important, CD40 triggering of these cells at permissive, but not at restrictive, temperatures demonstrated enhanced p53 transactivating activity. The use of p21-luciferase reporter constructs with mutated p53-binding sites completely abrogated the induction of luciferase activity on CD40 activation, confirming the specific requirement for p53 binding. Furthermore, RPA demonstrated that CD40L-triggered the induction of p53 and p21 mRNA expression, and immunoblotting confirmed increased p53 and p21 protein expression. These data provide strong evidence that CD40 regulates the cell cycle by a p53-dependent mechanism. Ongoing studies are defining the role of potential modulators of p53 action, such as MDM2,32 during CD40-mediated signal transduction.
XG-2, another human MM-derived cell line, was previously found to be hemizygous for the p53 gene37 and to have a missense mutation in codon 176 (Cys [TGC] to Tyr [TAC]; mtp53C176Y). XG-2 cells co-express wtp53 and mtp53 mRNA, but their p53 transactivation function is unknown. These cells strongly express cell surface CD40, and CD40 activation triggers growth arrest and apoptosis.13,14 Based on results of the current study of MM cells with similar responses to CD40 activation (ie, RPMI 8226 and HS Sultan cells cultured under permissive conditions), we postulate that mtp53C176Y in XG-2 cells confers a transactivating function. This was further supported by our observation that CD40 activation triggers apoptosis of ARH-77 MM cells, which have mtp53 that retains p53 transactivation function.38-41 These observations suggested that CD40 activation triggers growth and survival of MM cells in the absence of wtp53-like activity; however, in the presence of functional wt-like p53 activity, CD40 activation induced growth arrest and apoptosis of MM cells. Because CD40L is naturally expressed on activated T cells after antigen presentation, CD40 activation of MM cells in vivo could provide a mechanism for the selective and enhanced survival of those MM cells with mtp53.
In conclusion, we have demonstrated that CD40 activation of MM cell lines triggered a p53-dependent mechanism of cell cycle regulation. Our data may correlate with the tumor cell growth and resistance to apoptosis observed in late-stage MM, in which p53 abnormalities exist at high frequency.42 Our current study also emphasizes the potential importance of determining the p53 status of tumor cells before the initiation of novel treatment strategies targeting CD40 in patients with MM.
Supported by the National Institutes of Health grants CA50947 and CA78378; the Kraft Family Research Fund; the Health Manpower Development Plan Fellowship, Ministry of Health; the National Medical Research Council-Shaw Foundation Medical Research Fellowship; the National Medical Research Council-Singapore Totalisator Board Medical Research Fellowship; and the Singapore General Hospital Medical Research Fellowship, Singapore.
Reprints:Kenneth C. Anderson, Department of Adult Oncology, Dana-Farber Cancer Institute, 44 Binney Street, Boston, MA 02115; e-mail: kenneth_anderson@dfci.harvard.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal