BCL-6 mutations are accumulated during B-cell transit through the germinal center (GC) and provide a histogenetic marker for B-cell tumors. On the basis of a comprehensive analysis of 308 B-cell neoplasms, we (1) expand the spectrum of tumors associated withBCL-6 mutations; (2) corroborate the notion that mutations cluster with GC and post-GC B-cell neoplasms; and (3) identify heterogeneous mutation frequency among B-lineage diffuse large cell lymphoma (B-DLCL) subsets. Mutations are virtually absent in acute lymphoblastic leukemia (P < .001) and mantle cell lymphoma (P < .05), whereas they occur frequently in GC or post-GC neoplasms, including lymphoplasmacytoid lymphoma, follicular lymphoma, MALT lymphomas, B-DLCL and Burkitt lymphoma. Among B-DLCL, mutations occur frequently in systemic nodal B-DLCL, primary extranodal B-DLCL, CD5+ B-DLCL, CD30+ B-DLCL, and primary splenic B-DLCL, suggesting a similar histogenesis of these B-DLCL subsets. Conversely, mutations are rare in primary mediastinal B-DLCL with sclerosis (10.0%; P < .01), supporting a distinct histogenesis for this lymphoma. Longitudinal follow-up of B-DLCL transformed from follicular lymphoma shows that theyBCL-6 mutations may accumulate during histologic progression. Mutations also occur in some B-cell chronic lymphocytic leukemias, small lymphocytic lymphomas, and hairy cell leukemias, consistent with the hypothesis that a fraction of these lymphoproliferations are related to GC-like cells. Finally, the molecular pattern of 193 mutational events reinforces the hypothesis that mutations ofBCL-6 and immunoglobulin genes are caused by similar mechanisms.
The BCL-6 proto-oncogene has been originally identified because of its involvement in chromosomal translocations affecting band 3q27 in B-lineage diffuse large cell lymphoma (B-DLCL).1-3 The BCL-6 protein is a POZ/zinc finger trascriptional repressor which, in the B-cell lineage, is expressed selectively by germinal center (GC) B cells, but not by immature B-cell precursors or differentiated plasma cells.4 Experimental animal models have demonstrated that expression of BCL-6 is an absolute requirement for GC formation and function.5 6
The BCL-6 gene may be affected by 2 types of molecular alterations. The first type of BCL-6 alteration is represented by chromosomal translocations that lead to substitution of the gene promoter with heterologous sequences derived from the partner chromosome.1-3,7 Gross rearrangements of BCL-6 are virtually restricted to 30% B-DLCL.8-11 The second type of genetic alteration affecting BCL-6 is represented by point mutations of the 5′ noncoding region of the gene.12These mutations are somatic in nature, are often multiple in the same tumor, may be biallelic, and occur independent of cytogenetic alterations of band 3q27. The sequences affected by these mutations lie in the proximity of the BCL-6 promoter and overlap with the major cluster of chromosomal breakpoints. Mutations of BCL-6are regarded as a marker of B-cell transit through the GC because, in normal lymphoid tissues, they occur in approximately 30% to 50% of GC and memory B cells, whereas are absent in pre-GC, virgin B cells.13-15 On this basis, BCL-6 mutations have been proposed as a genetic marker for defining the histogenesis of B-cell lymphoproliferations.
The distribution of BCL-6 mutations in B-cell neoplasia has been investigated to a certain extent and, on the basis of available data, it has been suggested that these genetic lesions predominate in tumors displaying a GC or a post-GC phenotype.12,13,15However, a number of issues remain to be defined. For example, several histologic types of B-cell lymphomas recognized by the Revised European-American Lymphoma (REAL) classification have not been tested for BCL-6 mutations.12,13,15,16 Also, althoughBCL-6 mutations occur frequently in B-DLCL,12,13,15knowledge of their exact distribution among different subsets of the disease is lacking and may provide a clue to clarify the marked heterogeneity of this lymphoma.16
The aim of this study was a comprehensive analysis of BCL-6 mutations throughout the spectrum of B-cell neoplasia recognized by the REAL classification and, in particular, throughout distinct subsets of B-DLCL. Overall, our results (1) expand the spectrum of B-cell disorders associated with BCL-6 mutations; (2) corroborate the notion that mutations cluster with neoplasms of GC and post-GC B cells; and (3) identify heterogeneity of mutation frequency in different subsets of B-DLCL, suggesting heterogeneity in the histogenesis of this lymphoma. In addition, extensive analysis of the BCL-6 mutation pattern in B-cell neoplasia reinforces the hypothesis thatBCL-6 mutations may be caused by a mechanism similar to somatic hypermutation of immunoglobulin (Ig) genes.
Materials and methods
Tumor samples and DNA extraction
This study was based on 308 tumor samples representative of the spectrum of B-cell neoplasia recognized by the REAL classification.16 Tumor samples were derived from lymph nodes, bone marrow, or other involved organs obtained during routine diagnostic procedures. In all instances, with the exception of B-DLCL transformed from a follicular phase, the specimens were collected at diagnosis before specific therapy. Diagnosis was based on morphology and immunophenotypic analysis of cell surface markers and was complemented by immunogenotypic analysis of antigen receptor gene rearrangement.11 In most cases, the fraction of malignant cells was ≥ 70% and in all cases ≥ 30%.
On the basis of the REAL classification,16 samples were classified as precursor B-cell acute lymphoblastic leukemia (precursor B-cell ALL; n = 46), B-cell chronic lymphocytic leukemia (B-CLL; n = 30), small lymphocytic lymphoma (SLL; n = 9), lymphoplasmacytoid lymphoma (LPL; n = 5), mantle cell lymphoma (MCL; n = 20), follicular lymphoma (FL; n = 15), mucosa associated lymphoid tissue (MALT) lymphoma (n = 15), hairy cell leukemia (HCL; n = 8), B-DLCL (n = 125), and Burkitt lymphoma (BL; n = 35). Precursor B-cell ALL was representative of different molecular variants of the disease and included cases associated with hyperdiploidy (n = 5), rearrangement of BCR/ABL (n = 13), rearrangement of MLL (n = 9), rearrangement of TEL/AML-1 (n = 6), or no known genetic lesion (n = 13). MALT lymphomas originated in the gastrointestinal tract (n = 13) or in the thyroid (n = 2). B-DLCL samples were further subdivided into distinct subsets of the disease, which were either representative of specific B-DLCL categories formally recognized by the REAL classification16 or were arbitrarily identified on the basis of the phenotypic, clinical, or biologic peculiarities of the lymphoma. On these grounds, B-DLCL were subdivided into systemic B-DLCL arising de novo without clinical evidence of previous lymphoma (systemic de novo B-DLCL; n = 66), systemic B-DLCL transformed from a previous follicular lymphoma (transformed B-DLCL; n = 5), primary mediastinal B-DLCL with sclerosis (n = 10), CD5+ B-DLCL (n = 9), primary splenic B-DLCL (n = 15), primary extranodal B-DLCL (n = 12), CD30+ anaplastic B-DLCL (n = 5), and primary central nervous system B-DLCL (n = 3). Primary extranodal B-DLCL originated in the gastrointestinal tract (n = 8), thyroid (n = 2), testis (n = 1), or lung (n = 1) and included only cases without evidence of accompanying nodal involvement at diagnosis. A fraction of B-DLCL (n = 72), predominantly represented by systemic de novo B-DLCL, primary splenic B-DLCL, and primary extranodal B-DLCL, had been classified also according to the Working Formulation.17 Genomic DNA was purified by cell lysis followed by digestion with proteinase K, “salting out” extraction, and precipitation by ethanol.11
Oligonucleotides
All the oligonucleotides used in this study were synthesized by the solid phase triester method. The sequence of oligonucleotides used as primers for the mutational analysis of BCL-6 was as follows: E1.21B, 5′-CTCTTGCCAAATGCTTTG-3′ and E1.24, 5′-TAATTCCCCTCCTTCCTC-3′ (for fragment 1.10); E1.23, 5′-AGGAAGGAGGGGAATTAG-3′ and IP1.6, 5′-AAGCAGTTTGCAAGCGAG-3′ (for fragment 1.11); IP1.7, 5′-TTCTCGCTTGCAAACTGC-3′ and E1.26, 5′-CACGATACTTCATCTCATC-3′ (for fragment 1.12).12 Overall, these oligonucleotides amplify 3 partially overlapping PCR products spanning a 739 base pair (bp) region located downstream the first noncoding exon of BCL-6. The oligonucleotides used as primers for the mutational analysis ofp53 exons 5 through 8 have been previously reported.11 Primers used for analysis of Ig heavy chain variable (IgVH) genes included sense VHfamily-specific and 3′ antisense JH primers and have been reported previously.18 19
Polymerase chain reaction-single strand conformation polymorphism (PCR-SSCP)
PCR-SSCP was performed as previously reported.11 12Briefly, 100 ng of genomic DNA, 10 pmol of each primer, 2.5 μmol dNTPs, 1 μCi α-32P]dCTP (Amersham Life Sciences, Amersham, UK; specific activity, 3.000 Ci/mmol; 1 Ci = 37 Gbq), 10 mmol/L Tris-HCl (pH 8.8), 50 mmol/L KCl, 1 mmol/L MgCl2, 0.01% gelatin, and 0.5 U AmpliTaq polymerase (Taq gold, Perkin-Elmer, Norwalk, CT) were mixed in a final volume of 10 μL. Thirty-five cycles of denaturation (94°C), annealing (annealing temperatures were optimized for each pair of primers), and extension (72°C) were performed in a temperature controller (DNA Thermal Cycler Cetus; Perkin-Elmer, Norwalk, CT). Samples were heated at 95°C for 5 minutes, chilled on ice, and immediately loaded (3 μL) onto a 6% acrylamide/Tris-borate-EDTA (TBE) gel containing 10% glycerol. Gels were run at 8 W for 16 to 18 hours at room temperature, air dried, and analyzed by autoradiography using an intensifying screen (Quanta III; Dupont-NEN, Boston, MA) for 6 to 48 hours. By using the conditions described previously, reconstruction experiments have shown that the sensitivity of the PCR-SSCP method allows the detection of mutations present in 10% of the cell populations tested (sensitivity limit = 10%).
Sequencing procedures of BCL-6 gene
For DNA sequencing of BCL-6 5′ noncoding regions, a unique PCR product encompassing fragments E1.10, E1.11, and E1.12 (nucleotides +404 to +1142) was amplified by primers E1.21B and E1.26. For all samples subjected to BCL-6 DNA sequencing, the amplified PCR fragment was directly sequenced with appropriate primers using a commercially available kit (ThermoSequenase, Amersham Life Sciences). α-33P]-labeled terminator dideoxynucleotides (Amersham Life Science) were included in the sequencing mixture. For each DNA fragment analyzed, sequencing of both strands was performed on independent PCR reactions. In selected cases, PCR products showing mutations by direct sequencing were also sequenced after subcloning into the TA plasmid vector (Invitrogen, Leek, The Netherlands). DNA sequencing analysis of p53 exons 5-8 was performed as previously reported.11
Southern blot analysis and DNA probes
For Southern Blot analysis,11 6 to 10 μg of genomic DNA were digested with the appropriate restriction endonuclease, electrophoresed in a 0.8% agarose gel, denaturated, neutralized, transferred to Hybond C+ filters (Amersham Life Science), and hybridized to probes that had been α-32P]-labeled by the random priming extension method using a commercially available kit (Amersham Life Sciences). Filters were washed in 0.2 × NaCl-Na Citrate (SSC)/0.5% sodium dodecyl sulfate (SDS) for 1 hour at 60°C and then autoradiographed using intensifying screens. Ig gene rearrangement analysis was performed using a JH probe onBamHI, EcoRI, and HindIII digests and a Jκ probe on BamHI digests.11 The organization of theBCL-6, c-MYC, and BCL-2 loci was investigated as previously reported.11
Statistical analysis of the distribution and pattern of BCL-6 mutations
Mutation data were handled in a spreadsheet format using Excel (Microsoft Corp, Redmond, WA). The SPSS software (version 6.0 for Windows) was used for statistical elaboration. Statistical analysis was based on nonparametric methods; significant differences were considered when P-values < .05. The Fisher Exact test was used to compare the number of cases harboring BCL-6 mutations among the different categories of B-cell neoplasia. The relation betweenBCL-6 rearrangements and BCL-6 mutations was evaluated with χ2 test. Differences among all groups were evaluated with the Kruskal Wallis test (nonparametric 1-way ANOVA); differences between pairs of groups were evaluated with the Mann-Whitney test with Bonferroni adjustment for multiple comparison (significative levelP < .05). Mutation frequencies were normalized on the basis of the base composition of the sequence analyzed. The normalized mutation frequencies of each individual nucleotide were calculated by multiplying the mutation frequency of each single nucleotide by 0.25/f, where f is the frequency of occurrence of the specific nucleotide in the region sequenced.
To estimate the occurrence of mutations within specific nucleotide triplets, the number of mutations in each triplet was normalized for the frequency of the triplet in the sequence analyzed and compared with the expected mutation frequency by the goodness-of-fit χ2test. The expected mutation frequency for any triplet was obtained by multiplying the overall mononucleotide mutation frequency by 3. The expected frequency of mutations falling within RGYW (A/G G C/T A/T) motifs or the inverse sequence WRCY was obtained by multiplying the overall mononucleotide mutation frequency by 4, assuming no target preferences.
Sequencing analysis of IgVH genes
IgVH gene rearrangements were amplified with a set of 6 VH gene family-specific primers and a JH primer mix in separate reactions for each VH primer, as described.18 The VH primers used in this study hybridize to sequences in the framework region I of the respective VH families. PCR was performed for 38 cycles as described previously. The PCR buffer was represented by the Expand HF buffer (Boehringer-Mannheim, Monza, Italy). Annealing temperatures were 61°C for VH1, VH2, VH5, and VH6 reactions and 65°C for VH3 and VH4 reactions. PCR products were directly sequenced according to the sequencing procedures described previously. Because of the lack of an amplification product in some cases, the VHframework region I primers were replaced by VH leader specific primers, as previously reported.19 Sequences were compared with the V BASE sequence directory (MRC Centre for Protein Engineering, Cambridge, UK) using MacVector 6.0.1 software (Oxford Molecular Group PLC, Oxford, UK) for comparison of the rearranged IgV genes to the most homologous germline sequences.
Analysis of overall survival in B-DLCL with and without BCL-6 mutations
Analysis of overall survival (OS) in B-DLCL with and withoutBCL-6 mutations was performed in 72 patients with previously untreated de novo B-DLCL (either systemic de novo B-DLCL or primary extranodal B-DLCL), for whom complete clinical data were available. Patients had been diagnosed and treated at 3 Italian institutions from 1990 to 1995 and monitored through March 1999 or until death. The median follow-up duration from initiation of treatment for censored patients was 68 months. All patients were treated with an antracycline-containing regimen. Ten patients with localized stage of disease without adverse prognostic features were treated with 3 courses of either ACOPB (adriamycin, cyclophosphamide, vincristine, prednisone, bleomycin) or CHOP (cyclophosphamide, adriamycin, vincristine, prednisone), followed by locoregional radiotherapy. Twenty-one patients with localized stage and adverse prognostic features or advanced stage disease were treated with CHOP or a third-generation chemotherapy scheme such as MACOPB (methotrexate, adriamycin, cyclophosphamide, vincristine, prednisone, bleomycin) or VACOPB (etoposide, adriamycin, cyclophosphamide, vincristine, prednisone, bleomycin). Twenty-six elderly patients, over 65 years of age, received PVEBEC (prednisone, vinblastine, epirubicin, bleomycin, etoposide, cyclophosphamide) or VMP (etoposide, mitoxantrone, prednimustine) chemotherapy. Fifteen patients with advanced stage and adverse prognostic features were treated with MACOPB per 8 weeks plus MAD (mitoxantrone, high dose ara-C, dexamethasone) plus autologous stem cell transplantation with myeloablative chemotherapy BEAM (carmustine, etoposide, ara-C, melphalan) as preparative regimen. Assessment of response was performed as reported previously.20 All the patients who began treatment were considered assessable. OS includes all patients and was measured from the beginning of treatment to the date of death or last follow-up visit. OS curves were plotted according to the method of Kaplan-Meier and differences between curves were evaluated by the log-rank test.21 All calculations were made by applying the BMDP program (1985) developed at the Health Science Computing facility, UCLA (NIH) Special Research Resources.
Results
Characterization of the tumor panel
All cases of B-cell neoplasia displayed a major monoclonal B-cell population based on immunophenotypic and/or immunogenotypic analysis.
Distribution of BCL-6 mutations throughout the clinico-pathologic spectrum of B-cell neoplasia
All 308 samples of B-cell neoplasia were subjected to PCR-SSCP analysis of 3 partially overlapping fragments encompassing a 739 bp region within BCL-6 intron 1 (fragments E1.10, E1.11, E1.12). The selection of these PCR fragments was based on the observation that these sequences represent mutational hotspots of the gene, harboring > 90% mutations reported in B-cell neoplasia and that these sequences are consistently mutated in all cases carrying BCL-6mutations.12 Cases of B-cell neoplasia were scored positive for mutation when 1 or more PCR-SSCP fragments displayed a variant pattern that could not be attributed to a population polymorphism. Results are summarized in Table 1 and Table2 and representatively shown in Figure1 and Figure 2.
Distribution of BCL-6 mutations throughout the spectrum of B-cell neoplasia
| Histology . | BCL-6 Mutations (positive/tested)* . |
|---|---|
| Precursor B-cell neoplasms | |
| Acute lymphoblastic leukemia† | 0/46 (0%) (P < .001) |
| Peripheral B-cell neoplasms | |
| B-cell chronic lymphocytic leukemia | 9/30 (30.0%) |
| Small lymphocytic lymphoma | 2/9 (22.2%) |
| Lymphoplasmacytoid lymphoma | 2/5 (40.0%) |
| Mantle cell lymphoma | 2/20 (10.0%) (P < .05) |
| Follicular lymphoma | 9/15 (60.0%) |
| MALT lymphoma | 5/15 (33.3%) |
| Hairy cell leukemia | 2/8 (25.0%) |
| B-lineage diffuse large cell lymphoma‡ | 61/125 (48.8%) |
| Burkitt lymphoma | 13/35 (37.1%) |
| Histology . | BCL-6 Mutations (positive/tested)* . |
|---|---|
| Precursor B-cell neoplasms | |
| Acute lymphoblastic leukemia† | 0/46 (0%) (P < .001) |
| Peripheral B-cell neoplasms | |
| B-cell chronic lymphocytic leukemia | 9/30 (30.0%) |
| Small lymphocytic lymphoma | 2/9 (22.2%) |
| Lymphoplasmacytoid lymphoma | 2/5 (40.0%) |
| Mantle cell lymphoma | 2/20 (10.0%) (P < .05) |
| Follicular lymphoma | 9/15 (60.0%) |
| MALT lymphoma | 5/15 (33.3%) |
| Hairy cell leukemia | 2/8 (25.0%) |
| B-lineage diffuse large cell lymphoma‡ | 61/125 (48.8%) |
| Burkitt lymphoma | 13/35 (37.1%) |
Statistically significant differences in BCL-6 mutation frequency are indicated by the corresponding P-value.
Cases of precursor B-cell acute lymphoblastic leukemia were representative of different molecular variants of the disease and included cases associated with hyperdiploidy (n = 5), rearrangement of BCR/ABL (n = 13), rearrangement of MLL (n = 9), rearrangement of TEL/AML-1 (n = 6), or no known genetic lesion (n = 13).
The representation of the different subsets of B-lineage diffuse large cell lymphoma included in this study is reported in Table 2.
Distribution of BCL-6 mutations in different subsets of B-lineage diffuse large cell lymphoma (B-DLCL)
| B-DLCL Subset . | BCL-6 Mutations (positive/tested)* . |
|---|---|
| Systemic de novo B-DLCL | 33/66 (50.0%) |
| Transformed B-DLCL† | 3/5 (60.0%) |
| Primary mediastinal B-DLCL with sclerosis | 1/10 (10.0%) (P < .01) |
| CD5+ B-DLCL | 4/9 (44.4%) |
| Primary splenic B-DLCL | 9/15 (60.0%) |
| Primary extranodal B-DLCL‡ | 7/12 (58.3%) |
| CD30+ anaplastic B-DLCL | 2/5 (40.0%) |
| Primary central nervous system B-DLCL | 2/3 (66.6%) |
| B-DLCL Subset . | BCL-6 Mutations (positive/tested)* . |
|---|---|
| Systemic de novo B-DLCL | 33/66 (50.0%) |
| Transformed B-DLCL† | 3/5 (60.0%) |
| Primary mediastinal B-DLCL with sclerosis | 1/10 (10.0%) (P < .01) |
| CD5+ B-DLCL | 4/9 (44.4%) |
| Primary splenic B-DLCL | 9/15 (60.0%) |
| Primary extranodal B-DLCL‡ | 7/12 (58.3%) |
| CD30+ anaplastic B-DLCL | 2/5 (40.0%) |
| Primary central nervous system B-DLCL | 2/3 (66.6%) |
The criteria adopted for subdividing B-DLCL into distinct subsets are specified in “Materials and Methods.”
Statistically significant differences in BCL-6 mutation frequency are indicated by the corresponding P-value.
B-DLCL derived from histologic transformation of a previous follicular lymphoma.
B-DLCL presenting in extranodal sites without evidence of nodal involvement at diagnosis.
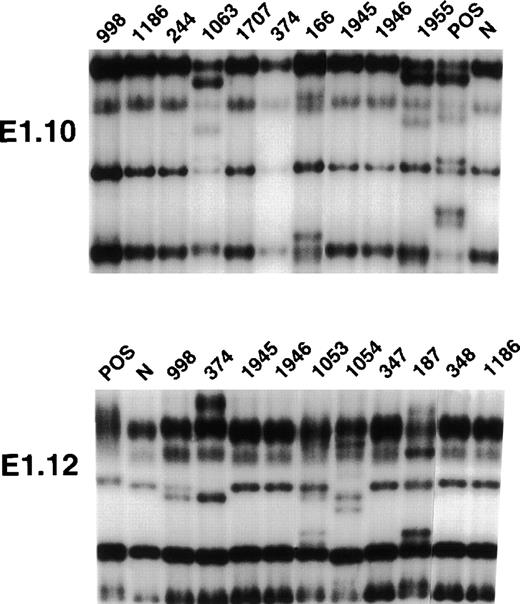
PCR-SSCP analysis of BCL-6 mutations in representative B-cell neoplasms.
Representative results obtained for PCR-SSCP fragments E1.10 and E1.12 are shown. Samples of B-cell neoplasms are indicated at the top of each lane by a numbered code. A positive control (POS), represented by a tumor sample known to harbor BCL-6 mutations, as well as a normal (N) sample, represented by a lymphoblastoid cell line, are also included for each PCR-SSCP fragment shown. Samples were scored positive when their migration pattern differed from the normal control (N) and the migration abnormalities could not be ascribed to population polymorphisms. Among the samples shown in the figure, cases scored as positive included cases 1063 (primary splenic B-DLCL), 166 (primary extranodal B-DLCL), 1955 (primary central nervous system B-DLCL) for PCR product E1.10, and cases 998 (systemic de novo B-DLCL), 374 (systemic de novo B-DLCL), 1053 (primary splenic B-DLCL), 1054 (primary splenic B-DLCL), and 187 (primary extranodal B-DLCL) for PCR product E1.12.
PCR-SSCP analysis of BCL-6 mutations in representative B-cell neoplasms.
Representative results obtained for PCR-SSCP fragments E1.10 and E1.12 are shown. Samples of B-cell neoplasms are indicated at the top of each lane by a numbered code. A positive control (POS), represented by a tumor sample known to harbor BCL-6 mutations, as well as a normal (N) sample, represented by a lymphoblastoid cell line, are also included for each PCR-SSCP fragment shown. Samples were scored positive when their migration pattern differed from the normal control (N) and the migration abnormalities could not be ascribed to population polymorphisms. Among the samples shown in the figure, cases scored as positive included cases 1063 (primary splenic B-DLCL), 166 (primary extranodal B-DLCL), 1955 (primary central nervous system B-DLCL) for PCR product E1.10, and cases 998 (systemic de novo B-DLCL), 374 (systemic de novo B-DLCL), 1053 (primary splenic B-DLCL), 1054 (primary splenic B-DLCL), and 187 (primary extranodal B-DLCL) for PCR product E1.12.
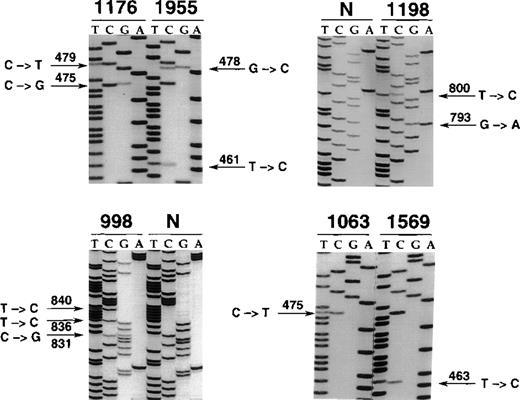
Nucleotide sequencing analyses of BCL-6 mutations in representative B-cell neoplasms.
The sequence of each case shown in the figure is matched to the sequence of a normal control (N) displaying germline BCL-6 alleles or to the sequence of a tumor sample harboring mutations at a different site. The position of mutations is indicated by the nucleotide number of the corresponding BCL-6 germline sequence (the first nucleotide of the BCL-6 cDNA was arbitrarily chosen as position +1). Cases included in the figure include systemic de novo B-DLCL (1176, 1198, 998), primary central nervous system B-DLCL (1955), and primary splenic B-DLCL (1063, 1569).
Nucleotide sequencing analyses of BCL-6 mutations in representative B-cell neoplasms.
The sequence of each case shown in the figure is matched to the sequence of a normal control (N) displaying germline BCL-6 alleles or to the sequence of a tumor sample harboring mutations at a different site. The position of mutations is indicated by the nucleotide number of the corresponding BCL-6 germline sequence (the first nucleotide of the BCL-6 cDNA was arbitrarily chosen as position +1). Cases included in the figure include systemic de novo B-DLCL (1176, 1198, 998), primary central nervous system B-DLCL (1955), and primary splenic B-DLCL (1063, 1569).
On these bases, mutations of 5′ noncoding regions ofBCL-6 were consistently absent among precursor B-cell neoplasms (n = 46; P < .001), independent of the molecular variant of the disease (Table 1). Among mature B-cell neoplasia, mutations were detected in 105/262 (40.0%) cases. In particular, mutations occurred in LPL (2/5; 40.0%), FL (9/15; 60.0%), B-DLCL (61/125; 48.8%), BL (13/35; 37.1%), MALT lymphomas (5/15; 33.3%) as well as in a subset of B-CLL (9/30; 30.0%), SLL (2/9; 22.2%), and HCL (2/8; 25.0%) (Table 1). Conversely, mutations in MCL (2/20; 10.0%) were significantly less frequent than in other mature B-cell neoplasms (P < .05) (Table 1).
In the case of B-DLCL, BCL-6 mutations occurred at different frequencies in different subsets of the disease (Table 2). In particular, BCL-6 mutations were virtually absent in primary mediastinal B-DLCL with sclerosis (1/10; 10%; P < .01), although occurred at a substantially similar frequency in all other B-DLCL variants, including systemic de novo B-DLCL (33/66; 50.0%), transformed B-DLCL (3/5; 60.0%), CD5+ B-DLCL (4/9; 44.4%), primary splenic B-DLCL (9/15; 60.0%), primary extranodal B-DLCL (7/12; 58.3%), CD30+ anaplastic B-DLCL (2/5; 40.0%), and primary central nervous system B-DLCL (2/3; 66.6%). In B-DLCL cases that had been also classified according to the Working Formulation (n = 72; predominantly represented by systemic de novo B-DLCL, primary splenic B-DLCL, and primary extranodal B-DLCL), mutations of BCL-6 distributed equally between Working Formulation category G (19/38; 50.0%), and category H (17/34; 50.0%).
Longitudinal follow-up of BCL-6 mutations
In an attempt to clarify the timing of BCL-6 mutation acquisition in transformed B-DLCL, we studied 5 B-DLCL transformed from a previous follicular phase before and after histologic progression. Mutations occurred in 1/5 follicular phases and in 3/5 transformed samples. In particular, 2 patients (cases 1931 and 1935 in Figure3) displayed BCL-6 mutations in the transformed, but not in the follicular phase, suggesting that mutations had been accumulated at the time of histologic progression. One patient displayed identical BCL-6 mutations in both phases of the disease.
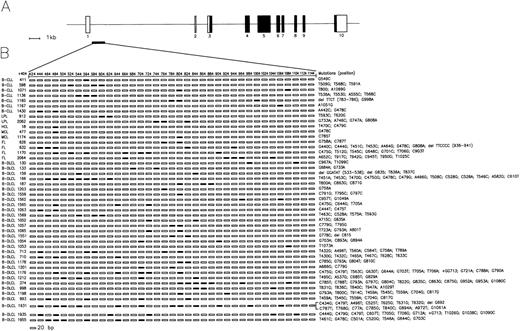
Characterization of BCL-6 mutations in 46 representative B-cell neoplasms.
(A) Schematic representation of the BCL-6 gene. Coding and non-coding exons are indicated by filled and empty boxes, respectively. The PCR fragment amplified for mutational analysis is approximately positioned below the BCL-6 gene map and expanded in panel B to show the distribution of mutations. (B) Each line of rectangles represent the BCL-6 sequence of a different B-cell neoplasm. Each case is indicated by a numbered code, together with the corresponding diagnosis (on the left; B-CLL, B-cell chronic lymphocytic leukemia; LPL, lymphoplasmacytoid lymphoma; HCL, hairy cell leukemia; MCL, mantle cell lymphoma; FL, follicular lymphoma; B-DLCL, B-lineage diffuse large cell lymphoma). Each rectangle represents a 20 bp interval of the BCL-6 sequence; nucleotide positions are indicated in the top line. The first nucleotide of the BCL-6 cDNA was arbitrarily chosen as +1. Filled rectangles indicate the presence of mutation(s) in the corresponding 20 bp of the BCL-6 sequence. The characteristics of individual mutations are detailed on the right (eg, G549C, G → C at nucleotide position 549).
Characterization of BCL-6 mutations in 46 representative B-cell neoplasms.
(A) Schematic representation of the BCL-6 gene. Coding and non-coding exons are indicated by filled and empty boxes, respectively. The PCR fragment amplified for mutational analysis is approximately positioned below the BCL-6 gene map and expanded in panel B to show the distribution of mutations. (B) Each line of rectangles represent the BCL-6 sequence of a different B-cell neoplasm. Each case is indicated by a numbered code, together with the corresponding diagnosis (on the left; B-CLL, B-cell chronic lymphocytic leukemia; LPL, lymphoplasmacytoid lymphoma; HCL, hairy cell leukemia; MCL, mantle cell lymphoma; FL, follicular lymphoma; B-DLCL, B-lineage diffuse large cell lymphoma). Each rectangle represents a 20 bp interval of the BCL-6 sequence; nucleotide positions are indicated in the top line. The first nucleotide of the BCL-6 cDNA was arbitrarily chosen as +1. Filled rectangles indicate the presence of mutation(s) in the corresponding 20 bp of the BCL-6 sequence. The characteristics of individual mutations are detailed on the right (eg, G549C, G → C at nucleotide position 549).
Characterization of BCL-6 mutations
The detailed characterization of the BCL-6 mutations in B-cell neoplasia was investigated by studying 193 mutational events observed in 46 samples representative of the clinico-pathologic spectrum of these disorders. Representative sequences are shown in Figure 2, whereas the characteristics of BCL-6 mutations are graphically represented in Figure 3. The overwhelming majority of mutations included single base-pair substitutions (n = 185), whereas single point deletions (n = 3), single point insertions (n = 1), and deletions/insertions of a short DNA stretch (n = 4) were observed only rarely. The frequency of mutation ranged from 0.0675 to 1.9 × 10-2/bp. Among cases harboring mutations, the mean frequency of mutation was overall similar in patients belonging to different categories of B-cell neoplasia.
Of a total of 185 single base-pair substitutions observed, 83 were transitions and 102 were transversions. The observed transition/transversion ratio was 0.81 (expected 0.5;P < .005). The frequency of each type of nucleotide substitution is shown in Table 3. The distribution of base substitutions was calculated using the coding strand and data were normalized for the nucleotide representation in the BCL-6 sequence analyzed. Analysis of the nature of the nucleotide substitutions indicated that T was mutated most frequently (29.4% of all mutations), followed by C (28.7%) and G (27.5%) (Table3). Conversely, A was mutated less frequently than expected by chance alone (14.4%; P < .01) (Table 3). Pyrimidine transitions were more frequent than expected (23.5% of all mutations;P < .02), whereas purine transversions were less frequent than expected (20.8%; P < .001). Preference for G → A transitions (P < .001), T → C transitions (P < .05), and C → G transversions (P < .01) was also present (Table 3). Conversely, nucleotide transversions C → A (P < .05), A → T (P < .05),A→C (P < .05), and G→T (P < .005) occurred less frequently than expected (Table 3).
Normalized frequency of BCL-6 mutations according to the type of single nucleotide substitution
| Type of Nucleotide Substitution . | Number of Mutations Observed . | % of Total Mutations Observed . | P3-150,3-151 . |
|---|---|---|---|
| Transitions and transversions | |||
| T → N | 68 | 29.4 | n.s. |
| C → N | 49 | 28.7 | n.s. |
| G → N | 51 | 27.5 | n.s. |
| A → N | 17 | 14.4 | <.01 |
| Transitions | |||
| G → A | 28 | 15.1 | <.001 |
| A → G | 7 | 5.90 | n.s. |
| T → C | 30 | 13.0 | <.05 |
| C → T | 18 | 10.5 | n.s. |
| Transversions | |||
| C → G | 24 | 14.0 | <.01 |
| G → C | 20 | 10.8 | n.s. |
| T → G | 20 | 8.70 | n.s. |
| T → A | 18 | 7.80 | n.s. |
| C → A | 7 | 4.10 | <.05 |
| A → T | 5 | 4.20 | <.05 |
| A → C | 5 | 4.20 | <.05 |
| G → T | 3 | 1.60 | <.005 |
| Type of Nucleotide Substitution . | Number of Mutations Observed . | % of Total Mutations Observed . | P3-150,3-151 . |
|---|---|---|---|
| Transitions and transversions | |||
| T → N | 68 | 29.4 | n.s. |
| C → N | 49 | 28.7 | n.s. |
| G → N | 51 | 27.5 | n.s. |
| A → N | 17 | 14.4 | <.01 |
| Transitions | |||
| G → A | 28 | 15.1 | <.001 |
| A → G | 7 | 5.90 | n.s. |
| T → C | 30 | 13.0 | <.05 |
| C → T | 18 | 10.5 | n.s. |
| Transversions | |||
| C → G | 24 | 14.0 | <.01 |
| G → C | 20 | 10.8 | n.s. |
| T → G | 20 | 8.70 | n.s. |
| T → A | 18 | 7.80 | n.s. |
| C → A | 7 | 4.10 | <.05 |
| A → T | 5 | 4.20 | <.05 |
| A → C | 5 | 4.20 | <.05 |
| G → T | 3 | 1.60 | <.005 |
Calculated with χ2 test; n.s., not significant.
The following nucleotide changes occurred more frequently than expected: G → A, T → C, and C → G. The following nucleotide changes occurred less frequently than expected: C → A, A → T, A → C, and G → T.
Clustering of BCL-6 mutations within specific DNA motifs
To determine whether the pattern of BCL-6 mutations exhibited sequence-specific preferences, we determined the mutation frequency in each of the 64 combinations of nucleotide triplets (Table4). The observed frequency of mutations in specific triplets was found to differ significantly from that expected based on chance alone (χ2 goodness-of-fit test). In particular, triplets AGC, GCT, TAC, CTA, GTT, CTG, TTA, and TGC were mutated at a frequency higher than expected (Table 4), whereas triplets CCC, CTC, GGG, TCC, CCT, GGA, GCC, AAA, ATG, and AAG were mutated at a frequency lower than expected (Table 4).
Frequency of BCL-6 mutations in specific nucleotide triplets and RGYW/WRCY motifs
| Nucleotide Sequence . | Occurrence4-150 . | Number of Mutations4-151 . | Frequency of Mutation . | P‡ . |
|---|---|---|---|---|
| Triplets mutated at higher than expected frequency | ||||
| AGC | 368 | 14 | 0.038 | <.0001 |
| GCT | 2208 | 44 | 0.020 | <.0001 |
| TAC | 92 | 4 | 0.043 | <.001 |
| CTA | 368 | 10 | 0.027 | <.001 |
| GTT | 1104 | 21 | 0.019 | <.001 |
| CTG | 1472 | 26 | 0.018 | <.001 |
| TTA | 1012 | 15 | 0.015 | <.025 |
| TGC | 1472 | 21 | 0.014 | <.025 |
| Triplets mutated at lower than expected frequency | ||||
| CCC | 1564 | 3 | 0.0019 | <.01 |
| CTC | 2116 | 8 | 0.0038 | <.025 |
| GGG | 1748 | 7 | 0.0040 | <.05 |
| TCC | 1840 | 7 | 0.0038 | <.05 |
| CCT | 1656 | 6 | 0.0036 | <.05 |
| GGA | 1656 | 6 | 0.0036 | <.05 |
| GCC | 1288 | 4 | 0.0031 | <.05 |
| AAA | 1288 | 4 | 0.0031 | <.05 |
| ATG | 736 | 1 | 0.0014 | <.05 |
| AAG | 736 | 1 | 0.0014 | <.05 |
| RGYW/WRCY mutated at higher than expected frequency | ||||
| AGCT | 92 | 10 | 0.109 | <.0001 |
| AGCA | 92 | 3 | 0.033 | <.05 |
| TGCT | 552 | 20 | 0.036 | <.0001 |
| Nucleotide Sequence . | Occurrence4-150 . | Number of Mutations4-151 . | Frequency of Mutation . | P‡ . |
|---|---|---|---|---|
| Triplets mutated at higher than expected frequency | ||||
| AGC | 368 | 14 | 0.038 | <.0001 |
| GCT | 2208 | 44 | 0.020 | <.0001 |
| TAC | 92 | 4 | 0.043 | <.001 |
| CTA | 368 | 10 | 0.027 | <.001 |
| GTT | 1104 | 21 | 0.019 | <.001 |
| CTG | 1472 | 26 | 0.018 | <.001 |
| TTA | 1012 | 15 | 0.015 | <.025 |
| TGC | 1472 | 21 | 0.014 | <.025 |
| Triplets mutated at lower than expected frequency | ||||
| CCC | 1564 | 3 | 0.0019 | <.01 |
| CTC | 2116 | 8 | 0.0038 | <.025 |
| GGG | 1748 | 7 | 0.0040 | <.05 |
| TCC | 1840 | 7 | 0.0038 | <.05 |
| CCT | 1656 | 6 | 0.0036 | <.05 |
| GGA | 1656 | 6 | 0.0036 | <.05 |
| GCC | 1288 | 4 | 0.0031 | <.05 |
| AAA | 1288 | 4 | 0.0031 | <.05 |
| ATG | 736 | 1 | 0.0014 | <.05 |
| AAG | 736 | 1 | 0.0014 | <.05 |
| RGYW/WRCY mutated at higher than expected frequency | ||||
| AGCT | 92 | 10 | 0.109 | <.0001 |
| AGCA | 92 | 3 | 0.033 | <.05 |
| TGCT | 552 | 20 | 0.036 | <.0001 |
The table includes only those nucleotide triplets and RGYW/WRCY motifs for which a statistically significant P-value was observed.
Occurrence refers to the total number of times that each nucleotide triplet or RGYW/WRCY motif appeared in the total BCL-6 sequence analyzed, ie, the number of nucleotides of the BCL-6 sequence analyzed in each single case multiplied by the number of samples investigated.
Number of mutations in each individual nucleotide triplet or RGYW/WRCY motif occurring in the total BCL-6 sequence analyzed.
To determine P values, the number of mutations observed in each nucleotide triplet or RGYW/WRCY motif was compared with the probability of mutation based on random chance using the goodness-of-fit χ2 test.
A variety of studies have suggested that the quadruplet motif RGYW (A/G G C/T T/A) and the inverse repeat WRCY are a target for increased mutational activity in Ig genes.22-24 To test whether such motifs are also preferentially targeted by the BCL-6 mutation mechanism, we analyzed the presence of mutations occurring in the RGYW quadruplet and in the inverse repeat WRCY. Together, RGYW/WRCY motifs represented the 17.7% of the total sequence. Mutations of RGYW/WRCY accounted for 31.0% of all nucleotide substitutions and occurred at a frequency higher than expected (P < .05). In particular,BCL-6 mutations preferentially targeted specific RGYW/WRCY motifs, including the AGCT, AGCA and TGCT nucleotide quadruplets (Table4).
Comparative analysis of mutations of BCL-6 and IgVH genes
The occurrence of mutations of BCL-6 and IgVHgenes was compared in a selected panel of samples, including FL, MALT lymphomas, and HCL. Results are summarized in Table5. Mutations of IgVH genes were detected in 15/15 (100%) FL, 15/15 (100%) MALT lymphomas, and 6/8 (75.0%) HCL. Comparison of BCL-6 and IgVHmutations in these cases revealed that all BCL-6 mutated samples harbored mutations of IgVH genes (Table 5). Conversely, only a fraction of IgVH mutated cases harbored mutations of BCL-6, including 9/15 FL (60.0%), 5/15 (33.3%) MALT lymphomas, and 2/8 (25.0%) HCL (Table 5). In all samples investigated, the frequency of IgVH mutations was approximately 10-fold higher than the frequency of BCL-6 mutations (Table 5).
Comparative analysis of BCL-6 and IgvH mutations in selected B-cell malignancies
| Histology . | BCL-6M/IgVM 5-150 . | BCL-6G/IgVM 5-150 . | BCL-6G/IgVG 5-150 . | Mean Frequency of Mutation (range) . | |
|---|---|---|---|---|---|
| BCL-6 . | IgVH . | ||||
| Follicular lymphoma (n = 15) | 9/15 (60.0%) | 6/15 (40.0%) | 0/15 | 0.83 × 10−2 bp | 9.8 × 10−2bp |
| (0.27-1.1) | (5.4-18.2) | ||||
| MALT lymphoma (n = 15) | 5/15 (33.3%) | 10/15 (66.7%) | 0/15 | 0.75 × 10−2 bp | 8.1 × 10−2bp |
| (0.27-1.9) | (3.2-15.5) | ||||
| Hairy cell leukemia (n = 8) | 2/8 (25.0%) | 4/8 (50.0%) | 2/8 (25.0%) | 0.27 × 10−2 bp | 6.9 × 10−2 bp |
| (0.13-0.40) | (3.3-10.8) | ||||
| Histology . | BCL-6M/IgVM 5-150 . | BCL-6G/IgVM 5-150 . | BCL-6G/IgVG 5-150 . | Mean Frequency of Mutation (range) . | |
|---|---|---|---|---|---|
| BCL-6 . | IgVH . | ||||
| Follicular lymphoma (n = 15) | 9/15 (60.0%) | 6/15 (40.0%) | 0/15 | 0.83 × 10−2 bp | 9.8 × 10−2bp |
| (0.27-1.1) | (5.4-18.2) | ||||
| MALT lymphoma (n = 15) | 5/15 (33.3%) | 10/15 (66.7%) | 0/15 | 0.75 × 10−2 bp | 8.1 × 10−2bp |
| (0.27-1.9) | (3.2-15.5) | ||||
| Hairy cell leukemia (n = 8) | 2/8 (25.0%) | 4/8 (50.0%) | 2/8 (25.0%) | 0.27 × 10−2 bp | 6.9 × 10−2 bp |
| (0.13-0.40) | (3.3-10.8) | ||||
BCL-6M/IgVM, cases harboring mutated BCL-6 and IgVH genes;BCL-6G/IgVM, cases harboring germlineBCL-6 genes and mutated IgVH genes;BCL-6G/IgVG, cases harboring germlineBCL-6 and IgVH genes. For each category, the number of positive/tested cases is indicated.
Relationship between BCL-6 mutations, BCL-6 rearrangements and other genetic lesions occurring in B-DLCL
Because B-DLCL is molecularly heterogeneous,10,11 16 we compared the distribution of BCL-6 mutations with that of several other genetic lesions of this lymphoma, including rearrangements of BCL-6, BCL-2, and c-MYC, as well as mutations of p53. Rearragements of BCL-6 were detected in 29/115 (25.2%) B-DLCL, including 20/66 (30.3%) systemic de novo B-DLCL, 2/15 (13.3%) primary splenic B-DLCL, 6/12 (50.0%) primary extranodal B-DLCL, and 1/4 (25.0%) CD30+anaplastic B-DLCL (Table 6). Notably, rearrangements of BCL-6 were absent in primary mediastinal B-DLCL with sclerosis (n = 8), transformed B-DLCL (n = 5), and primary central nervous system B-DLCL (n = 3) (Table 6). Comparison of the distribution of BCL-6 rearrangements and mutations confirmed that mutations can occur independent of the concomitant presence of rearrangements in all tested categories of B-DLCL (Table6). Comparison of the distribution of BCL-6 mutations with that of alterations of c-MYC, BCL-2, and p53 showed that BCL-6 mutations occurred in B-DLCL cases both positive and negative for these genetic alterations (Table 6).
Distribution of genetic lesions in different subsets of B-lineage diffuse large cell lymphoma (B-DLCL) with and without BCL-6 mutations
| B-DLCL Subset . | BCL-6 R6-150,6-151 . | c-MYC R6-150,6-151 . | BCL-2 R6-150,6-151 . | p53 M6-150,6-151 . | EBV6-150 . |
|---|---|---|---|---|---|
| Systemic de novo B-DLCL | |||||
| with BCL-6 mutations | 11/33 | 0/32 | 3/33 | 2/32 | 0/31 |
| withoutBCL-6 mutations | 9/33 | 1/32 | 9/33 | 4/31 | 0/31 |
| Transformed B-DLCL | |||||
| with BCL-6 mutations | 0/3 | 0/3 | 3/3 | 3/3 | 0/3 |
| without BCL-6 mutations | 0/2 | 0/2 | 2/2 | 2/2 | 0/2 |
| Primary mediastinal B-DLCL with sclerosis | |||||
| with BCL-6 mutations | n.d.§ | n.d. | 0/1 | 1/1 | 1/1 |
| without BCL-6 mutations | 0/8 | n.d. | 0/9 | 2/9 | 0/9 |
| CD5+ B-DLCL | |||||
| withBCL-6 mutations | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| without BCL-6 mutations | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Primary splenic B-DLCL | |||||
| with BCL-6 mutations | 1/9 | 0/9 | 0/9 | 1/9 | 0/9 |
| withoutBCL-6 mutations | 1/6 | 1/6 | 1/6 | 1/6 | 0/6 |
| Primary extranodal B-DLCL6-155 | |||||
| with BCL-6 mutations | 5/7 | 0/7 | 0/7 | 1/7 | 0/7 |
| withoutBCL-6 mutations | 1/5 | 0/5 | 0/5 | 1/5 | 0/5 |
| CD30+ anaplastic B-DLCL | |||||
| with BCL-6 mutations | 1/2 | 1/2 | 0/2 | 0/2 | 0/2 |
| withoutBCL-6 mutations | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 |
| Primary central nervous system B-DLCL | |||||
| with BCL-6 mutations | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 |
| withoutBCL-6 mutations | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| B-DLCL Subset . | BCL-6 R6-150,6-151 . | c-MYC R6-150,6-151 . | BCL-2 R6-150,6-151 . | p53 M6-150,6-151 . | EBV6-150 . |
|---|---|---|---|---|---|
| Systemic de novo B-DLCL | |||||
| with BCL-6 mutations | 11/33 | 0/32 | 3/33 | 2/32 | 0/31 |
| withoutBCL-6 mutations | 9/33 | 1/32 | 9/33 | 4/31 | 0/31 |
| Transformed B-DLCL | |||||
| with BCL-6 mutations | 0/3 | 0/3 | 3/3 | 3/3 | 0/3 |
| without BCL-6 mutations | 0/2 | 0/2 | 2/2 | 2/2 | 0/2 |
| Primary mediastinal B-DLCL with sclerosis | |||||
| with BCL-6 mutations | n.d.§ | n.d. | 0/1 | 1/1 | 1/1 |
| without BCL-6 mutations | 0/8 | n.d. | 0/9 | 2/9 | 0/9 |
| CD5+ B-DLCL | |||||
| withBCL-6 mutations | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| without BCL-6 mutations | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Primary splenic B-DLCL | |||||
| with BCL-6 mutations | 1/9 | 0/9 | 0/9 | 1/9 | 0/9 |
| withoutBCL-6 mutations | 1/6 | 1/6 | 1/6 | 1/6 | 0/6 |
| Primary extranodal B-DLCL6-155 | |||||
| with BCL-6 mutations | 5/7 | 0/7 | 0/7 | 1/7 | 0/7 |
| withoutBCL-6 mutations | 1/5 | 0/5 | 0/5 | 1/5 | 0/5 |
| CD30+ anaplastic B-DLCL | |||||
| with BCL-6 mutations | 1/2 | 1/2 | 0/2 | 0/2 | 0/2 |
| withoutBCL-6 mutations | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 |
| Primary central nervous system B-DLCL | |||||
| with BCL-6 mutations | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 |
| withoutBCL-6 mutations | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
Positive/tested;
R, rearrangement; M, mutation;
B-DLCL derived from histologic transformation of a previous follicular lymphoma;
n.d., not done;
B-DLCL presenting in extranodal sites without evidence of nodal involvement at diagnosis.
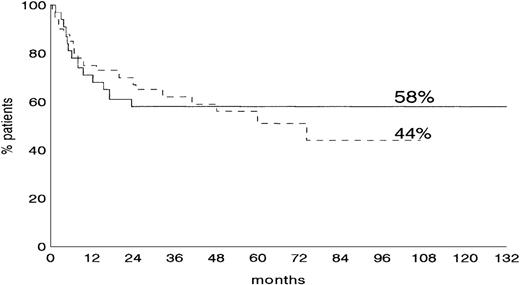
Overall survival in de novo B-DLCL with and without BCL-6 mutations
The OS according to BCL-6 mutations was assessed in 72 de novo B-DLCL for whom complete clinical data were available. At a median follow-up of 68 months, OS rate was 58% in cases harboringBCL-6 mutations and 44% in cases devoid of BCL-6 mutations (Figure 4), with no significant difference between the 2 groups.
Overall survival (OS) according to the presence or absence of BCL-6 mutation.
OS rates at 68 months 58% (BCL-6 mutated, solid line) versus 44% (BCL-6 nonmutated, dotted line).
Overall survival (OS) according to the presence or absence of BCL-6 mutation.
OS rates at 68 months 58% (BCL-6 mutated, solid line) versus 44% (BCL-6 nonmutated, dotted line).
Discussion
The aim of this study was a comprehensive investigation of the distribution and pattern of BCL-6 mutations throughout the spectrum of B-cell neoplasia. On the basis of the analysis of 300 B-cell tumors, we report that BCL-6 mutations cluster with B-cell neoplasms thought to originate from GC and post-GC B cells. In the case of B-DLCL, BCL-6 mutations associate with most, though not all, subsets of the disease, suggesting heterogeneity in the histogenesis of this lymphoma. Mutational analysis performed on almost 200 events indicates that BCL-6 mutations may be due to a molecular mechanism similar to somatic hypermutation of Ig genes.
Analysis of BCL-6 mutations throughout the spectrum of B-cell neoplasia confirms that mutations are virtually absent in B-cell tumors deriving from precursor and virgin B-cells, namely, precursor B-cell ALL and MCL. This observation is consistent with the fact thatBCL-6 mutations arise at the time of B-cell transit through the GC.13-15 Conversely, mutations occur at sustained frequency in LPL, FL, MALT lymphomas, B-DLCL, and BL for which a derivation from GC-related B cells has been either proven or postulated.18,25-30 Also, mutations are found in a fraction of HCL, which, in most cases, are thought to derive from post-GC B cells.31,32 The occurrence of BCL-6 mutations in a fraction of B-CLL and SLL is noteworthy, because these disorders have been traditionally viewed as proliferations of virgin B cells.16 In addition to BCL-6 mutations, however, other features of GC-related B cells have been recently reported in a subset of B-CLL,33 34 suggesting that the histogenesis of the disease may be more heterogeneous than previously thought.
According to the REAL classification,16 the termB-DLCL is thought to include more than 1 disease entity. The heterogeneity of B-DLCL reflects heterogeneity in the phenotype, clinical history, and primary site of the disease.16 With the exception of primary mediastinal B-DLCL with sclerosis,BCL-6 mutations distribute uniformly throughout most B-DLCL variants, including systemic de novo B-DLCL, systemic B-DLCL transformed from FL, CD5+ B-DLCL, primary splenic B-DLCL, primary extranodal B-DLCL, and primary central nervous system B-DLCL. Because BCL-6 mutations are a histogenetic marker of B-cell transit through the GC,13-15 it is conceivable that all B-DLCL subsets harboring BCL-6 mutations share a common origin from GC-related B cells. This notion is substantiated by data of somatic Ig gene mutation analysis.18,30,35 Remarkably, the association of CD5+ B-DLCL with both BCL-6 and somatic Ig gene mutations (this report and ref. 35) indicates that the histogenesis of this lymphoma differs from that of other malignancies of the CD5+ B-cell lineage, namely, MCL, B-CLL, and SLL which, in the majority of cases, display features of virgin B cells.16 In contrast to other B-DLCL subsets, the absence of BCL-6 mutations in primary mediastinal B-DLCL with sclerosis points to a different histogenetic pathway for this lymphoma, possibly related to a noncirculating B cell normally residing in the thymic medulla.16,36,37 The molecular and histogenetic peculiarities of primary mediastinal B-DLCL with sclerosis are also reinforced by the virtual absence of BCL-6 rearrangements (ref.38 and this report) and by the observation that these lymphomas fail to express the BCL-6 protein (A.Carbone et al, unpublished observation, 1999), a marker of GC-like phenotype that is commonly expressed by other B-DLCL variants.4
The finding of BCL-6 mutations in transformed B-DLCL, but not in the FL biopsy, which precedes histologic progression, is noteworthy. Despite the limited number of cases investigated, these results suggest that accumulation of BCL-6 mutations might be involved in the molecular mechanisms of FL transformation and mandate functional studies aimed at defining the precise biologic relevance ofBCL-6 mutations in FL histologic progression.
Our analysis of 193 mutational events corroborates and expands the characteristics of the mutational pattern of BCL-6. First, single nucleotide substitutions predominate, whereas insertions/deletions are rare. Second, nucleotide transitions occur more frequently than expected. Third, mutations cluster at high frequency in specific nucleotide triplets and RGYW/WRCY motifs. Finally, the pyrimidine T is more frequently mutated than the purine A, suggesting a mutational mechanism exhibiting strand polarity. Collectively, these features reinforce the hypothesis thatBCL-6 mutations may result from a mechanism similar to that causing somatic hypermutation of Ig genes.13-15,39-41Infact, both BCL-6 mutations and somatic Ig gene hypermutation display a net preference for single nucleotide substitutions, an excess of nucleotide transitions over transversions and a high degree of clustering in similar hotspots, including specific RGYW/WRCY motifs and selected nucleotide triplets (13-15,39-41 and this report). The hypothesis that the BCL-6 locus may be recognized by the somatic Ig gene hypermutation mechanism is consistent with results obtained in animal models showing that non-Ig sequences may be targeted by somatic Ig gene hypermutation.42 43
Although previous studies yielded controversial results regarding the strand polarity of BCL-6 mutations,13,14 the asimmetry of A and T mutations observed in the present report suggests that the BCL-6 mutation mechanism preferentially targets 1 DNA strand. At variance with strand polarity associated with somatic Ig hypermutation, which preferentially targets A over T,39,44the mutation mechanism of BCL-6 appears to preferentially target T over A. This discrepancy in strand polarity between Ig andBCL-6 mutations may be due to specific features of the gene sequences, as also suggested by experiments performed on other genes that have been artificially exposed to the somatic Ig hypermutation mechanism.42
Comparative analysis of BCL-6 and IgVH mutations in several B-cell malignancies deriving from GC or post-GC B cells has revealed that the proportion of cases harboring IgVHmutations is higher than the proportion of cases harboringBCL-6 mutations (this study and ref. 13). Also, the frequency of BCL-6 mutations in a given tumor sample is generally lower than that of IgVH mutations (this study and ref. 13). Notably, mutations of BCL-6 never occurred in the absence of IgVH mutations in B-cell malignancies analyzed to date for both mutations, including B-DLCL),13 FL, MALT lymphoma, HCL, and B-CLL. Absolute coincidence of BCL-6 and IgVH mutations should not be expected in view of the lower frequency of BCL-6 mutations, compared with IgVHmutations, in normal GC and post-GC B cells.13-15 In fact,BCL-6 mutations are found in approximately 30% to 50% normal GC and post-GC B cells, whereas IgVH mutations occur in virtually all such cells. Also, the mutation frequency of BCL-6 in normal GC and post-GC B cells is approximately 10-fold lower than that of IgVH genes. The precise reasons for the different incidence and frequency of BCL-6 versus IgVHmutations are not currently known. It is possible that the mutational mechanism targets BCL-6 with a lower efficiency compared with IgVH genes or that BCL-6 mutations occur relatively later than IgVH mutations in the progression of the cell transfer through the germinal center.
Currently, detection of BCL-6 mutations provides information on the B-cell compartment from which a given B-cell tumor originates and therefore bears implications for the disease histogenesis. Although a pathogenetic role for BCL-6 mutations has not been formally established, indirect observations suggest that mutations may carry functional consequences. In particular, it is remarkable thatBCL-6 mutations cluster in highly conserved genomic regions, suggesting that some mutations may affect regulatory domains ofBCL-6 and thus deregulate the physiologic expression of the gene.12 To define the precise pathogenicity of these genetic alterations, however, further studies are needed to test the ability of tumor-derived BCL-6 alleles to deregulate the expression of BCL-6.
Supported by ISS, II Programma nazionale di ricerca sull'AIDS 1998-Progetto Patologia clinica e terapia dell'AIDS, Rome, Italy; “Fondazione Piera, Pietro e Giovanni Ferrero,” Alba, Italy; by “Fondazione CRT,” Torino, Italy; and by Associazione Italiana per la Ricerca sul Cancro (A.I.R.C.), Milan, Italy. D.C. is being supported by a fellowship from A.I.R.C., Milan, Italy.
Reprints:Gianluca Gaidano, Division of Internal Medicine, Department of Medical Sciences, “Amedeo Avogadro” University of Eastern Piedmont, Via Solaroli 17, 28100 Novara, Italy.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal