Abstract
The major histocompatibility complex (MHC) class I genes are induced synergistically by interferons (IFN) and tumor necrosis factor (TNF) , a response thought to involve the cooperative action of Rel/NF-kB and interferon regulatory factor (IRF) transcription factors. The IFN-γ–inducible class II transcriptional activator (CIITA) has recently been shown to transactivate MHC class I as well as class II genes, and this investigation shows that CIITA synergizes strongly with RelA to stimulate HLA class I expression. The functional interaction of CIITA and RelA requires both promoter elements and the upstream Rel binding site and is not seen with a class II reporter. The promoter elements necessary for CIITA action are also required for induction by IFN-. HLA-A and HLA-B loci respond differentially to IFNs, and we identify locus-specific differences in critical promoter elements in addition to known polymorphisms in the Rel and IRF binding sites. The HLA-A promoter is transactivated relatively poorly by CIITA and does not interact detectably with CREB proteins implicated in CIITA recruitment, but the synergism with RelA can compensate for this weakness. The present findings illustrate that multiple transcription factors cooperate to regulate class I expression and that their relative importance differs according to the locus and cell type examined.
The major histocompatibility complex (MHC) cell surface glycoproteins are essential for the development and function of the immune system by virtue of their presentation of antigenic peptides to T cells. They are encoded by 2 main families of genes, class I and II, each of which consists of several polymorphic loci. Class I loci are expressed to varying degrees by most cell types, whereas constitutive class II expression is restricted to cells with an immune response function, but can be induced in other cell types by interferon-γ (IFN-γ).1,2 Class I expression is up-regulated by IFN-γ as well as by type I IFNs (α, β)1 but, paradoxically, type I IFNs can antagonize the induction of class II expression by IFN-γ.3
The MHC class I and II gene families are believed to have originated from a common precursor (Kaufman et al4) and the proteins retain very similar 3-dimensional structures,5 but their distinct patterns of constitutive and inducible expression suggest that their upstream control regions are distinct or at least highly diverged. However, a striking functional relationship of class I and II promoters has been demonstrated by recent reports that class II transcriptional activator (CIITA), a transcription factor necessary for constitutive and IFN-γ–inducible class II expression,6-8can also transactivate class I genes.9,10 CIITA does not bind directly to DNA but appears to interact with proteins bound to the X box region of the class II promoter.11,12 The X box consists of several discrete motifs: X1 binds the factor RFX13; X2 matches the consensus for a cyclic adenosine monophosphate (cAMP) response element (CRE) and binds members of the CRE binding protein (CREB)/activating transcription factor (ATF) family13; and X3 binds a helix-loop-helix (HLH) factor.14 Transactivation of the class I promoter by CIITA requires a site within the ENH-B region referred to as α which, like the class II X2 box, has the sequence of a CRE and has been shown to bind CREB/ATF family members.15,16 A class I X1 box homologue is also necessary for expression,17 and a putative X3 box has been noted in HLA-B promoters.18 Both class I and II promoters also have inverted CAAT boxes at identical distances from the X2/α motifs, which contribute to their expression.13 19
The class I genes are distinguished by additional upstream control elements, in particular the ENH-A region that is bound by Rel/NF-κB factors, and the adjacent interferon response element (IRE), which interacts with members of the IRF family.20 ENH-A was identified originally through its contribution to constitutive expression, but it is also needed for full induction via the IRE, which has only weak enhancer activity on its own.1 TNF and IFNs synergistically induce class I expression, a cooperativity thought to result from their respective activation of NF-κB and up-regulation of IRF1.21,22 IRF1 has also been suggested to cooperate with CIITA in class I induction by IFN-γ,10 although this conclusion was drawn from upstream deletions that removed both the ENH-A and IRE. We demonstrate in this report that CIITA synergizes strongly with RelA, and to a lesser extent with IRF1, and that this cooperativity can compensate for the relatively weak HLA-A promoter, which differs from HLA-B loci in the critical ENH-B α region.
Materials and methods
Cell lines, transfections, and chloramphenical acetyl transferase (CAT) assays
MOLT4, YHHH,23 and HeLa cells were maintained in RPMI 1640 Medium (Sigma, Poole, UK) supplemented with 4 mmol/L glutamine and 5% fetal calf serum (Gibco BRL, Paisley, UK) in a humidified incubator with 5% CO2. Cells in midlog phase were transfected by electroporation as described24 with 20 μg of reporter vector along with expression vectors or empty controls at the levels indicated in figures. Due to the effects of IFN and overexpression of transcription factors on the activity of standard reference plasmids, such internal standards were not used. Instead, multiple repeats of the transfections represented for each figure were performed in parallel and CAT expression for a given condition was designated as 100, as indicated in the figure legends. Recombinant IFN-α was a kind gift of M. Brunda (Hoffman-La Roche, Nutley, NJ) and was added at 1000 U/mL immediately after transfection. Cultures were grown 20 hours before preparation for CAT assays as described.24 Quantitation was performed with a PhosphorImager (Molecular Dynamics, Little Chalfont, UK).
Plasmids
Class I CAT reporters were based on the metallothionein promoter construct pMCAT3.25 The ENH-A/IRE-metallothionein construct has been described,24 and the class II DRA CAT reporter was kindly provided by Dr G. Andersson.26 The HLA-A and HLA-B upstream regions were generated by polymerase chain reaction (PCR) amplification from genomic clones for HLA-A127 and HLA-B5728 and cloned into pMCAT3 Hind III site in place of the metallothionein promoter. The primers for the full-length HLA-B reporter were: GGCATCAAGCTTTCCGTGATCAGT; GCATCTAAGCTTCTGAGGAGATTC. The upstream primer for the truncated HLA-B reporter was: CCGAAAGCTTTGTCTGCATTGGGG. The primers for the HLA-A reporter were: CCGAAAGCTTTGTATGGATTGGGGAG; CAT- CCTAAGCTTCTGGGGAGAATCTGAG. Base changes were introduced by dut-ung29 or PCR-mediated mutagenesis. The IRF-1 expression vector was generated by transfer of the human complementary (cDNA) from a baculovirus expression vector30into pCDNA3 (Invitrogen, Groningen, The Netherlands). Empty pCDNA3 was used as the control vector for transfections. Expression vectors for CIITA (in pCDNA3) and RelA (under a cytomegalovirus promoter) were kindly provided by Drs K. Gustaffson and A. Baldwin, respectively.
Electrophoretic mobility shift assay (EMSA)
Preparation of nuclear extracts and EMSA protocols have been described.31 The sense strand sequences for oligonucleotide probes are: HLA-B R1/α CCAGGATACTCGTGACGCGTCCCC; HLA-A R1/α CCTGGATACTCACGACGCGGACCC; H2-D R1/αGGACACAGATGACGCGCTGGCAGG. Super-shift assays were performed with affinity-purified antisera (Santa Cruz) for ATF-1/CREB-1 (sc-270) and CREB-1 (sc-240).
Results
Sequence comparisons of HLA class I and II promoters
Alignment of HLA-A, HLA-B, and DRA upstream regions (Figure1A) illustrates the strong homologies that have been noted between the class I and class II promoters.32,35 When read from the noncoding strand, the α site of most HLA-B alleles shows a 6 out of 7 bp match with the X2 box of the DRA gene, whereas some have a full match.36 Overlapping the HLA-B α site is the motif CTCGTG, which has been referred to as the R1 site because its sequence is complementary to the R motif identified in the IRE region of HLA-B57.18 The R site binds the HLH/leucine zipper factor USF, suggesting that R1 may be equivalent to the class II X3 box, which binds the HLH protein E47.14 The R1/X3 box in turn overlaps the X1 box, which is recognized by RFX and required along with the X2/α sequence for transactivation of class I and II genes by CIITA.13,17 Although HLA class I loci exhibit high homology over much of the X1 box, HLA-A promoters differ from the HLH and CRE consensus sequences of the X3 and α boxes, particularly in having a T to C change at a position critical for both elements. The HLA-A loci have an IRE that differs from other class I genes and interacts poorly with IRF proteins,30,37 whereas its ENH-A is similar to H2 class I loci in that it has a second Rel binding site.30
Homologies between class I and II promoters.
(A) The upstream regions of DRA,2 HLA-B57,28and HLA-A127 are aligned to illustrate similarities in sequence and structure. The DRA S, X1, X2, X3, and Y boxes2,13,14 and the class I ENH-A and ENH-B regions and previously identified Rel, R, IRE, R1, X1, α, and inverted CAAT (IC) box control elements are indicated.15,18,24 32-34 Asterisks indicate the residues of HLA-A loci that disrupt the consensus sequences of the IRE, R1, and α sites. (B) Lower-case letters indicate base changes introduced into the HLA-B57 sequence for mutant EMSA probes and HLA-B CAT reporters.
Homologies between class I and II promoters.
(A) The upstream regions of DRA,2 HLA-B57,28and HLA-A127 are aligned to illustrate similarities in sequence and structure. The DRA S, X1, X2, X3, and Y boxes2,13,14 and the class I ENH-A and ENH-B regions and previously identified Rel, R, IRE, R1, X1, α, and inverted CAAT (IC) box control elements are indicated.15,18,24 32-34 Asterisks indicate the residues of HLA-A loci that disrupt the consensus sequences of the IRE, R1, and α sites. (B) Lower-case letters indicate base changes introduced into the HLA-B57 sequence for mutant EMSA probes and HLA-B CAT reporters.
Control elements important for induction by CIITA and IFN-
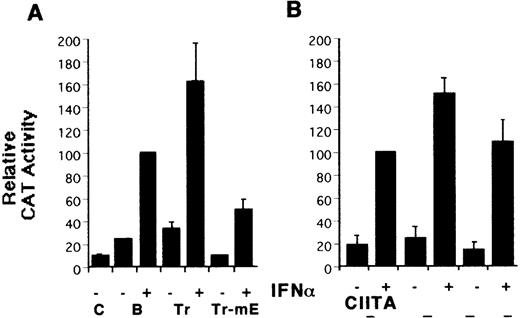
To identify the elements mediating class I induction by CIITA and to compare their effects on the response to IFN-α, a series of HLA-B mutant reporters was constructed (Figure 1B) and tested with the YHHH cell line, a variant of the MOLT4 human thymoma selected for its strong class I response to type I IFN.23 Deletion of sequences upstream of ENH-A led to increased induction by both IFN-α and CIITA (Figure 2A, B), similar to the de-repression reported for the H-2Ld promoter.9Mutation of the conserved Rel site in the truncated reporter led to a 25% decrease in CIITA transactivation and a much larger drop in the IFN-α response (Figure 2A, B). Mutation of the IRE eliminates the response to IFN-α as expected (Figure 2C), but only inhibits CIITA transactivation by about 15% (Figure 2D). Therefore, both the Rel site and IRE contribute to class I CIITA transactivation by CIITA, although not as strongly as for induction by IFN-α.
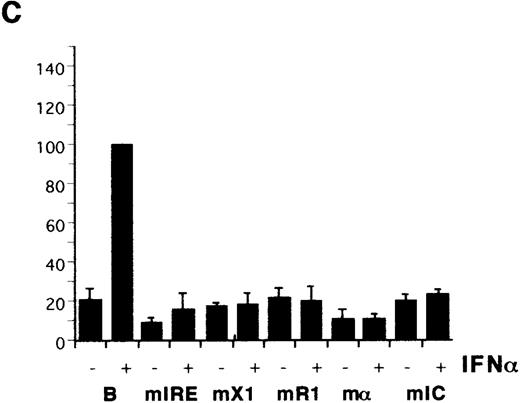
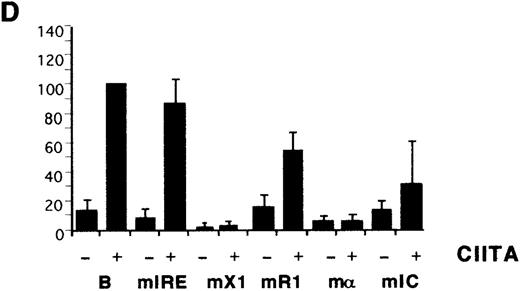
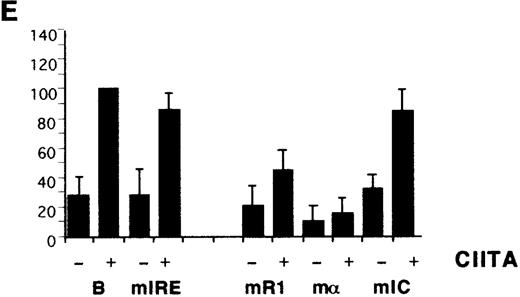
Identification of HLA-B upstream elements required for induction by IFN- and by CIITA.
(A) YHHH cells were transfected with CAT reporter vectors and cultured with (+) or without (−) IFN-α as described in “Materials and methods.” Transfections were performed in parallel and the CAT activity of the HLA-B vector in the presence of IFN-α was defined as 100 U. For this and subsequent figures, error bars represent SD on the mean from at least 3 independent transfections. “C” is the promoterless CAT vector control and “B” is the HLA-B reporter, with 500 bp of sequence upstream of the start codon of HLA-B57.28 Truncated (Tr) versions with 200 bp of HLA-B upstream sequence include the conserved Rel site in ENH-A, which was left intact, or mutated (Tr-mE). (B) Cotransfections with the HLA-B reporters were performed with 1 μg of a control (−) or CIITA (+) vector, and induced activity of the wild-type reporter was defined as 100 U. (C) The IFN-α inducibility of the full length HLA-BCAT vector, “B,” was compared with versions that had been mutated in the IRE, X1, R1, α, or IC elements (Figure 1B). (D) The HLA-B mutant reporter series was tested with the CIITA vector as described in panel B. (E) HeLa cells were transfected with the HLA-B reporter series −/+ CIITA as described for YHHH cells.
Identification of HLA-B upstream elements required for induction by IFN- and by CIITA.
(A) YHHH cells were transfected with CAT reporter vectors and cultured with (+) or without (−) IFN-α as described in “Materials and methods.” Transfections were performed in parallel and the CAT activity of the HLA-B vector in the presence of IFN-α was defined as 100 U. For this and subsequent figures, error bars represent SD on the mean from at least 3 independent transfections. “C” is the promoterless CAT vector control and “B” is the HLA-B reporter, with 500 bp of sequence upstream of the start codon of HLA-B57.28 Truncated (Tr) versions with 200 bp of HLA-B upstream sequence include the conserved Rel site in ENH-A, which was left intact, or mutated (Tr-mE). (B) Cotransfections with the HLA-B reporters were performed with 1 μg of a control (−) or CIITA (+) vector, and induced activity of the wild-type reporter was defined as 100 U. (C) The IFN-α inducibility of the full length HLA-BCAT vector, “B,” was compared with versions that had been mutated in the IRE, X1, R1, α, or IC elements (Figure 1B). (D) The HLA-B mutant reporter series was tested with the CIITA vector as described in panel B. (E) HeLa cells were transfected with the HLA-B reporter series −/+ CIITA as described for YHHH cells.
Previous CIITA studies used class I promoter mutations or deletions that affected several potential control elements or their relative spacings,9,10,17 so the HLA-B X1, R1, α, and IC boxes were mutated individually (Figure 1B). The X1 and α sites were critical for induction by IFN-α (Figure 2C) as well as for transactivation by CIITA (Figure 2D). R1 and IC were also necessary for the response to IFN-α and their mutation reduced, but did not eliminate, transactivation by CIITA. There is some cell-type specificity in the contribution of control elements, with the IC box important for CIITA transactivation in YHHH cells (Figure 2D), while having little effect in HeLa cells (Figure 2E). Therefore, although the X1 and α sites are critical for activity, as reported,17the Rel, IRE, R1, and IC sites also affect transactivation by CIITA. Furthermore, these same elements are necessary for class I induction by IFN-α.
Synergistic action of RelA, CIITA, and IRF-1
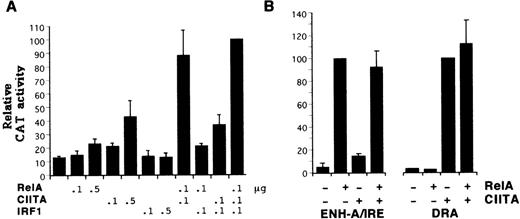
To investigate the influence of Rel and IRE sites on CIITA action, expression vectors for RelA and IRF1 were titrated with the CIITA vector (Figure 3A). At the levels used, CIITA transactivated the HLA-B reporter more strongly than RelA; IRF1 had no effect. CIITA demonstrated a striking synergy with RelA and to a lesser extent with IRF1. RelA and IRF1 have been shown previously to interact in class I induction,22 38 but under these conditions their combined action was relatively weak. Inclusion of all 3 factors leads to a slight increase over the level of expression achieved with RelA plus CIITA. Therefore, although CIITA can transactivate through the HLA-B promoter alone, under limiting conditions its activity is strongly enhanced by interaction with factors binding to the ENH-A or IRE or both.
Synergistic activity of CIITA with RelA and IRF1.
(A) MOLT4 cells were transfected with the HLA-B CAT reporter along with 0.1 or 0.5 μg of expression vectors for CIITA, RelA, or IRF1. The activity with all 3 expression vectors was defined as 100 U. (B) RelA (0.5 μg) and CIITA (0.1 μg) expression vectors were cotransfected with reporter plasmids consisting of the HLA-BENH-A/IRE region placed upstream of the basal metallothionein promoter24 (left panel) or the DRA promoter (right panel).
Synergistic activity of CIITA with RelA and IRF1.
(A) MOLT4 cells were transfected with the HLA-B CAT reporter along with 0.1 or 0.5 μg of expression vectors for CIITA, RelA, or IRF1. The activity with all 3 expression vectors was defined as 100 U. (B) RelA (0.5 μg) and CIITA (0.1 μg) expression vectors were cotransfected with reporter plasmids consisting of the HLA-BENH-A/IRE region placed upstream of the basal metallothionein promoter24 (left panel) or the DRA promoter (right panel).
To determine whether the interaction between CIITA and RelA requires class I promoter elements, the 2 factors were tested with a reporter that has the HLA-B ENH-A and IRE region upstream of a metallothionein promoter. This construct is sufficient to mediate strong induction by IFN-α,39 demonstrating that the class I promoter is not absolutely required for this response. As seen in Figure 3B, CIITA had no significant effect on the ability of RelA to transactivate the ENH-A/IRE-metallothionein construct, and RelA did not potentiate the action of limiting amounts of CIITA on the DRA reporter. Therefore, the class I ENH-A and promoter regions must both be present for the synergy of RelA and CIITA.
Differential regulation of HLA-A and HLA-B
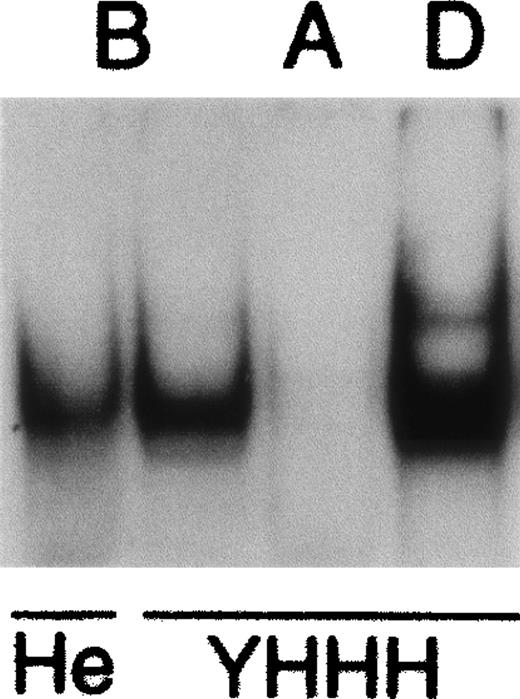
The HLA-A promoter has a poor match with the HLH (R1) and CRE (α) consensus sequences due to a conserved T to C change (Figure 1A), but has been shown to be transactivated strongly by CIITA.10The R1/α regions of HLA-B or H-2D generate complexes with nuclear extracts, but no significant binding was seen with the equivalent HLA-A promoter sequence (Figure 4). The inability of the HLA-A promoter to interact well with nuclear factors may be analogous to the situation with many class II promoters where cooperative binding of multiple proteins is required to overcome the weakness of individual control elements,40 but this remains to be investigated.
Interaction of nuclear factors with MHC promoter elements.
Nuclear extracts from HeLa (He) and YHHH cells formed complexes with DNA probes representing the R1/α region of HLA-B (B) and H-2D (D) genes, but not the equivalent region from HLA-A (A). Super-shift analysis indicated that the predominant nuclear factors were CREB1 and USF1 (data not shown).
Interaction of nuclear factors with MHC promoter elements.
Nuclear extracts from HeLa (He) and YHHH cells formed complexes with DNA probes representing the R1/α region of HLA-B (B) and H-2D (D) genes, but not the equivalent region from HLA-A (A). Super-shift analysis indicated that the predominant nuclear factors were CREB1 and USF1 (data not shown).
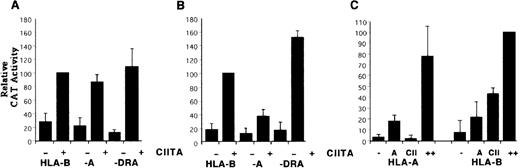
The HLA-A loci have an IRE sequence with relatively low affinity for IRF proteins,30 and the promoter region implicated in the recruitment of CIITA has now been shown to interact poorly with nuclear factors. However, we demonstrated previously that the ENH-A region of HLA-A loci mediates stronger transactivation by RelA by virtue of having 2 Rel binding sites, whereas HLA-B has only 1.30Given the strong interaction between RelA and CIITA demonstrated with the HLA-B reporter, we tested the possibility that the stronger ENH-A of HLA-A could help compensate for its weaker promoter. The hypothesis was first tested by comparing the relative induction by CIITA of reporters in YHHH and HeLa cells, which have different levels of basal class I expression mediated by ENH-A.24 The HeLa cell experiment of Gobin et al10 using HLA-A2 and HLA-B7 reporters was repeated using saturating levels of the CIITA expression vector with DRA, HLA-A1, and HLA-B57 reporters (Figure5A). The class I reporters both gave a level of transactivation roughly equal to that found with DRA. This is similar to the earlier report, although Gobin et al10 found that the HLA-B reporter activity was about a third lower than the other 2. Allelic differences between B7 and B57 might account for this discrepancy, but the strong induction of HLA-A is confirmed. The experiment was repeated in YHHH cells, which have a very low basal activity mediated by ENH-A and no detectable nuclear factors that bind specifically to the Rel site.39 In contrast to the similar activities seen in HeLa cells, the reporters showed substantially different levels of transactivation (Figure 5B). The HLA-A reporter was induced weakly in comparison with HLA-B, which in turn was lower than DRA. Therefore, there is a strong cell type dependence on the relative transactivation of class I promoters by CIITA.
Differential induction of HLA-A and -B reporters by CIITA.
HeLa cells (A) and YHHH cells (B) were transfected in parallel with HLA-A, HLA-B, and DRA CAT reporter vectors with (+) or without (−) 1 μg of CIITA expression vector. Induced activity of the HLA-B reporter for each cell type was defined as 100 U. (C) YHHH cells were transfected with HLA-A or HLA-B reporter vectors on their own (−) or with 0.5 μg of expression vectors for RelA (A), CIITA (CII), or both (++). Activity of the HLA-B reporter in the presence of both RelA and CIITA vectors was defined as 100 U.
Differential induction of HLA-A and -B reporters by CIITA.
HeLa cells (A) and YHHH cells (B) were transfected in parallel with HLA-A, HLA-B, and DRA CAT reporter vectors with (+) or without (−) 1 μg of CIITA expression vector. Induced activity of the HLA-B reporter for each cell type was defined as 100 U. (C) YHHH cells were transfected with HLA-A or HLA-B reporter vectors on their own (−) or with 0.5 μg of expression vectors for RelA (A), CIITA (CII), or both (++). Activity of the HLA-B reporter in the presence of both RelA and CIITA vectors was defined as 100 U.
To determine if Rel factor expression could overcome the cell type differences in HLA-A and HLA-B transactivation by CIITA, cotransfection experiments similar to those shown in Figure 3 were performed in YHHH cells. Again, CIITA and RelA act synergistically with the HLA-B reporter, and their interaction is even more striking with HLA-A (Figure 5C). At levels of CIITA that induce HLA-B expression significantly there is no effect on the HLA-A reporter, but RelA acts similarly on the 2. However, in the presence of both transcription factors, expression from the HLA-A reporter rises to about 80% of that seen with coinduction of HLA-B. This slightly lower activity is similar to that seen with these reporters in HeLa cells. Therefore, the differential induction of HLA-A and HLA-B by CIITA can be compensated for by interactions with Rel factors acting through ENH-A.
Discussion
The MHC class I loci are expressed constitutively by many cell types, but levels vary widely and the genes are subject to a number of developmental controls and cytokine signals that are mediated by multiple elements contained within a relatively short upstream region. Although more distal sequences have been implicated in developmental and viral controls on expression,41,42 transgenic studies indicate that a 150 bp region encompassing the ENH-A, IRE, and ENH-Β is sufficient to mediate relatively normal patterns of transcription and may constitute a locus control region (LCR) for HLA class I genes.43 The data presented here confirm the importance of individual control elements, and also demonstrate that interactions between the primary regions of the LCR are crucial for class I regulation by the IFN-γ–induced factor CIITA and by IFN-α.
Cooperativity between the ENH-A and IRE is known to be important for class I regulation by IFNs,1 and synergistic induction by TNF and IFNs involves functional and physical interactions between NF-κB and IRF proteins that bind to these sites.21,22,29It is now evident that CIITA is also an important contributor to IFN-γ–induced class I expression.9,10 Although promoter elements alone can mediate transactivation by overexpressed CIITA, upstream sequences are required for stimulation by IFN-γ.10 The IRE was suggested to be the upstream element involved in the IFN-γ response,10 but our demonstration of potent synergy between RelA and CIITA indicates that factors bound to the ENH-A contribute strongly to induction. The level of class I expression will depend therefore on the presence of factors bound to ENH-A, which will vary according to cell type or cytokine stimulation.
The ability of RelA to compensate for the relatively weak transactivation of the HLA-A promoter by CIITA illustrates the importance of functional interaction between these factors and also the locus-specific differences that exist between class I genes. HLA-A loci are induced poorly by IFNs in comparison with HLA-B, a deficit due at least in part to their atypical IRE, which has a lower affinity for IRF proteins.30,37 On the other hand, the ENH-A of HLA-A loci mediates stronger induction by Rel/NF-κB factors, which are typically activated by inflammatory cytokines such as TNF and interleukin-1.30 Therefore, the HLA loci have diverged such that they are regulated differentially by cytokine-responsive transcription factors. We have shown here that they also differ in the critical ENH-B region, with the HLA-A sequence unable to interact with nuclear factors, as detected by EMSA. CREB factors are important for CIITA activation of class II loci,44 and a dominant negative CREB145 inhibits CIITA induction of both HLA-A and HLA-B reporters (data not shown), suggesting that CREB1, or a highly related factor, is required for expression of both loci. The ability of HLA-A to be induced by CIITA despite a poor promoter is analogous to the situation described for some class II loci, where individual transcription factors do not interact with the promoter, but will bind cooperatively to the S-X-Y promoter region.40 Therefore, it seems likely that protein–protein interactions will also be important for assembly of appropriate factors on the HLA-A promoter that will allow CIITA to bind. In addition, the strong ENH-A of HLA-A loci serves to recruit NF-κB factors that can cooperate with CIITA, but it remains to be determined whether this involves direct physical interaction.
No synergy is seen between RelA and CIITA when they are coexpressed with a class II reporter, which lacks a Rel binding site, or when the class I Rel site is placed upstream of the metallothionein promoter, which lacks sequences equivalent to the ENH-B region. Therefore, if there is a physical interaction between RelA and CIITA it is of insufficient affinity to affect transcriptional activity, and it appears that they must be bound to their respective sites for functional cooperation to occur. RelA and CIITA have both been shown to interact with CBP/p300 proteins,46,47 so it is possible that they cooperatively recruit the transcriptional machinery through these coactivators. Transcription factors binding to the S-X-Y elements of class II genes and to the equivalent ENH-B class I region have been suggested to form an “enhanceosome,”17,40 but it is interesting to note that the 70-bp spacing between ENH-A and ENH-B places the Rel and CRE sites adjacent to each other if the class I LCR adopts a nucleosomal arrangement.48 With such a spatial organization, CIITA would be placed in close proximity to Rel dimers and conceivably able to bind the same coactivators.
It is now apparent that the regulated expression of class I genes involves the integrated action of multiple transcription factors. It will be important to determine which proteins can assemble into the ENH-B “enhanceosome,” and whether the composition differs according to cell type, cytokine signaling, and the HLA locus examined. The data presented here show that ENH-B elements are required for induction by IFN-α as well as for the action of CIITA, and it remains to be examined whether the same ENH-B components are involved in both responses. The relatively large number of transcriptional elements involved in basal and induced class I expression provides an array of control points for regulation according to cell type, differentiation stage, cell cycle, confluence, neural activity, and exposure to cytokines.1,18,49-51 However, the existence of several elements critical for expression also provides a number of targets through which class I loci can be silenced in virally infected or transformed cells, thereby promoting escape from immune surveillance.52-54 The regulation of class I genes involves a surprising degree of complexity, and further characterization of their transcriptional control will be important for understanding normal expression patterns and possible means of manipulation for clinical purposes.
Acknowledgments
I would like to thank Dean Gentle for technical support, and Drs G. Andersson, A. Baldwin, M. Brunda, K. Gustaffson, B. Mach, and C. Vinson for supplying reagents.
Supported by an MRC Senior Fellowship to J.G.
Reprints:John Girdlestone, Anatomy Dept, MRC Centre for Immune Regulation, The Medical School, University of Birmingham, Birmingham UK B15 2TT; e-mail: girdlesj@bham.ac.uk.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal