Abstract
Thirty-two natural killer (NK) and cytotoxic T-cell lymphomas and 14 noncytotoxic nodal T-cell lymphoma controls were immunostained with the use of monoclonal antibodies reactive against NK-cell receptor (NKR) molecules (CD94, NKG2A, p58.2, p58.1, p140, p70, p50.3). All NK-cell lymphomas (4 nasal/oral and 1 intestinal) expressed at least 1 NKR, the CD94/NKG2A complex. Two were positive for 1 or more killer immunoglobulin-like receptors. Of 15 extranodal cytotoxic T-cell lymphomas, 3 expressed CD94, including 2 intestinal and 1 hepatosplenic γδ T-cell lymphomas. In contrast, none of the nodal lymphomas were positive. Detection of NKRs may provide a useful tool to confirm the diagnosis of NK-cell lymphomas and to delineate a subgroup of cytotoxic T-cell lymphomas. Expression of NKRs only in extranodal cytotoxic T-cell lymphomas might reflect differences in the homing capabilities of cytotoxic T cells expressing NKRs in normal individuals and might be influenced in part by localized chronic immune reactions.
Interest in the natural killer (NK) cell has gathered momentum in the last decade, in particular, because many of the molecular mechanisms underlying its function have now been elucidated.1-3 Lymphomas arising from NK cells and the related extranodal T-cell lymphomas of cytotoxic phenotype (ETLCP) have also been recognized.4-9 These are a heterogeneous group of neoplasms, having in common the expression of 1 or more of the cytotoxic granule associated proteins, but the major types have been more clearly defined by hematopathologists in recent years.10,11 However, identification of NK-cell lymphomas and their distinction from ETLCP can still be problematic, depending on negative features, such as the lack of T-cell receptor (TCR) peptide expression, absence of CD3 on the cell membrane, and a germline configuration of the TCR genes. CD56 is a useful, but not specific, marker for NK cells, and its role in the function of the NK cell is as yet unknown.12
The discovery of major histocompatibility complex (MHC) class I–specific NK cell receptors (NKRs), of either the inhibitory or the activatory type, has provided much insight into the control of NK-cell function.13,14 Engagement of inhibitory NKRs by normal self-MHC class I alleles on potential target cells leads to inhibition of cytotoxicity, whereas loss of such normal alleles in virus-infected, transformed, or allogeneic cells leads to loss of inhibition and allows stimuli that activate NK cytotoxicity to come into play. Similar receptors have been detected on a small subset of cytotoxic T-lymphocytes, both in the peripheral blood and in lymphoid tissues,15 and there is evidence that these NKR+ T cells are oligoclonal populations of memory T cells generated as a consequence of chronic antigenic stimulation.16 Identification of NKRs on lymphomas of NK and T-cell lineage may therefore be used to confirm the cell type and has the potential to delineate a special biological group of tumors.
To our knowledge, there is no published study on the occurrence of NKRs on NK and T-cell lymphomas. The purpose of this brief report is to present the preliminary results of such a study.
Materials and methods
Case selection
A total of 46 lymphoid malignancies of T- and NK-cell origin were retrieved from the files of the Lymph Node Reference Center and the surgical pathology files of the Institute of Pathology at the University of Würzburg, Germany, and of the Institute of Clinical Pathology at the University of Vienna, Austria. Twenty of these were extranodal lymphomas (Table 1) and 12 were nodal lymphomas (8 peripheral T-cell lymphomas not otherwise specified and 4 anaplastic large-cell lymphomas). They were chosen to represent the major categories of cytotoxic lymphoid neoplasms, expressing at least 1 of the 2 cytotoxic granule–associated proteins, TIA1 or granzyme B. Five angioimmunoblastic T-cell lymphomas (AILs), 9 periperal T-cell lymphomas of noncytotoxic origin, and 13 specimens from various normal and reactive lymphoid tissues (5 lymph nodes, 5 spleens, and 3 tonsils) were included as controls.
Expression of different NK receptors in NK and T-cell lymphomas of cytotoxic phenotype
| Case no. . | Age/ sex . | Site . | Diagnosis . | Immunophenotype . | EA . | TCR γ gene . | Lineage . | CD94 (XA185) . | NKG2A (Z270) . | p58.2 (GL183) . | p58.1 (EB6) . | p140 (Q66) . | p70 (Z27) . | p50.3 (FST172) . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 10/M | Nasal | Nasal T/NK | TIA1+/GrB−/CD3−(p)/CD5−/CD56+/βF1−/TCRδ1− | GL | NK | + | + | − | − | − | − | − | |
| 2 | 30/M | Nasal | Nasal T/NK | TIA1−/GrB+/CD3−(p)/CD5−/CD56+/βF1−/TCRδ1− | GL | NK | + | + | − | − | + | − | − | |
| 3 | 57/M | Oropharynx | Nasal T/NK | TIA1+/GrB+/CD3−(f)/CD5−/CD56+/βF1−/TCRδ1− | GL | NK | + | + | − | − | − | − | − | |
| 4 | 64/M | Nasal | Nasal T/NK | TIA1+/GrB+/CD3+(p)/CD5−/CD56+/βF1−/TCRδ1− | GL | NK | + | + | − | − | − | − | − | |
| 5 | 17/F | Spleen | HS γ/δ TCL | TIA1+/GrB−/CD3+(f)/CD56+/βF1−/TCRδ1+ | mono | T (γδ) | + | + | + | + | − | − | − | |
| 6 | 59/M | Spleen | PTCL NOS | TIA1+/GrB+/CD3+(p)/CD56+/βF1+/TCRδ1− | mono | T (αβ) | − | − | − | − | − | − | − | |
| 7 | 50/F | Small intestine | INKL | TIA1+/GrB−/CD3+(p)CD3−(f)/CD5−/CD56+/βF1−/TCRδ1− | Y | GL | NK | + | + | + | + | − | − | − |
| 8 | 51/M | Small intestine | ITCL | TIA1+/GrB+/CD3+(p&f)/CD5−/CD56−/βF1−/TCRδ1− | Y | nd | T | − | − | − | − | − | − | − |
| 9 | 66/F | Small intestine | ITCL | TIA1+/GrB+/CD3+(p&f)/CD5−/CD56−/βF1−/TCRδ1− | N | nd | T | − | − | − | − | − | − | − |
| 10 | 61/F | Small intestine | ITCL | TIA1+/GrB−/CD3+(p&f)/CD5−/CD56+/βF1−/TCRδ1− | N | mono | T | − | − | − | − | − | − | − |
| 11 | 53/F | Small intestine | ITCL | TIA1+/GrB+/CD3+(p&f)/CD5+/CD56+/βF1+/TCRδ1− | Y | mono | T | − | − | − | − | − | − | − |
| 12 | 52/M | Small intestine | ITCL | TIA1+/GrB+/CD3+(p&f)/CD5−/CD56−/βF1+/TCRδ1− | N | nd | T | − | − | − | − | − | − | − |
| 13 | 66/M | Small intestine | ITCL | TIA1+/GrB−/CD3+(p&f)/CD5+/CD56−/βF1−/TCRδ1− | Y | nd | T | − | − | − | − | − | − | − |
| 14 | 60/M | Small intestine | ITCL | TIA1+/GrB+/CD3+(f)/CD5−/CD56−/βF1−/TCRδ1− | nd | nd | T | + | − | − | − | − | − | − |
| 15 | 63/M | Small intestine | ITCL | TIA1+/GrB−/CD3+(p&f)/CD5−/CD56−/βF1+/TCRδ1− | Y | nd | T | − | − | − | − | − | − | − |
| 16 | 70/F | Small intestine | ITCL | TIA1−/GrB+/CD3+(p&f)/CD5−/CD56−/βF1−/TCRδ1− | Y | nd | T | − | − | − | − | − | − | − |
| 17 | 39/M | Small intestine | ITCL | TIA1+/GrB+/CD3+(p&f)/CD5−/CD56−/βF1+/TCRδ1− | Y | nd | T | + | + | − | − | − | − | − |
| 18 | 35/M | Skin | ALCL | TIA1+/CD3+(p) | nd | T | − | − | − | − | − | − | − | |
| 19 | 57/F | Skin | CD30+ LPD | TIA1+/CD3+(p)/CD5− | nd | T | − | − | − | − | − | − | − | |
| 20 | 26/F | Skin | PTCL NOS | TIA1+/CD3−(p)/CD56+ | mono | T | − | − | − | − | − | − | − |
| Case no. . | Age/ sex . | Site . | Diagnosis . | Immunophenotype . | EA . | TCR γ gene . | Lineage . | CD94 (XA185) . | NKG2A (Z270) . | p58.2 (GL183) . | p58.1 (EB6) . | p140 (Q66) . | p70 (Z27) . | p50.3 (FST172) . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 10/M | Nasal | Nasal T/NK | TIA1+/GrB−/CD3−(p)/CD5−/CD56+/βF1−/TCRδ1− | GL | NK | + | + | − | − | − | − | − | |
| 2 | 30/M | Nasal | Nasal T/NK | TIA1−/GrB+/CD3−(p)/CD5−/CD56+/βF1−/TCRδ1− | GL | NK | + | + | − | − | + | − | − | |
| 3 | 57/M | Oropharynx | Nasal T/NK | TIA1+/GrB+/CD3−(f)/CD5−/CD56+/βF1−/TCRδ1− | GL | NK | + | + | − | − | − | − | − | |
| 4 | 64/M | Nasal | Nasal T/NK | TIA1+/GrB+/CD3+(p)/CD5−/CD56+/βF1−/TCRδ1− | GL | NK | + | + | − | − | − | − | − | |
| 5 | 17/F | Spleen | HS γ/δ TCL | TIA1+/GrB−/CD3+(f)/CD56+/βF1−/TCRδ1+ | mono | T (γδ) | + | + | + | + | − | − | − | |
| 6 | 59/M | Spleen | PTCL NOS | TIA1+/GrB+/CD3+(p)/CD56+/βF1+/TCRδ1− | mono | T (αβ) | − | − | − | − | − | − | − | |
| 7 | 50/F | Small intestine | INKL | TIA1+/GrB−/CD3+(p)CD3−(f)/CD5−/CD56+/βF1−/TCRδ1− | Y | GL | NK | + | + | + | + | − | − | − |
| 8 | 51/M | Small intestine | ITCL | TIA1+/GrB+/CD3+(p&f)/CD5−/CD56−/βF1−/TCRδ1− | Y | nd | T | − | − | − | − | − | − | − |
| 9 | 66/F | Small intestine | ITCL | TIA1+/GrB+/CD3+(p&f)/CD5−/CD56−/βF1−/TCRδ1− | N | nd | T | − | − | − | − | − | − | − |
| 10 | 61/F | Small intestine | ITCL | TIA1+/GrB−/CD3+(p&f)/CD5−/CD56+/βF1−/TCRδ1− | N | mono | T | − | − | − | − | − | − | − |
| 11 | 53/F | Small intestine | ITCL | TIA1+/GrB+/CD3+(p&f)/CD5+/CD56+/βF1+/TCRδ1− | Y | mono | T | − | − | − | − | − | − | − |
| 12 | 52/M | Small intestine | ITCL | TIA1+/GrB+/CD3+(p&f)/CD5−/CD56−/βF1+/TCRδ1− | N | nd | T | − | − | − | − | − | − | − |
| 13 | 66/M | Small intestine | ITCL | TIA1+/GrB−/CD3+(p&f)/CD5+/CD56−/βF1−/TCRδ1− | Y | nd | T | − | − | − | − | − | − | − |
| 14 | 60/M | Small intestine | ITCL | TIA1+/GrB+/CD3+(f)/CD5−/CD56−/βF1−/TCRδ1− | nd | nd | T | + | − | − | − | − | − | − |
| 15 | 63/M | Small intestine | ITCL | TIA1+/GrB−/CD3+(p&f)/CD5−/CD56−/βF1+/TCRδ1− | Y | nd | T | − | − | − | − | − | − | − |
| 16 | 70/F | Small intestine | ITCL | TIA1−/GrB+/CD3+(p&f)/CD5−/CD56−/βF1−/TCRδ1− | Y | nd | T | − | − | − | − | − | − | − |
| 17 | 39/M | Small intestine | ITCL | TIA1+/GrB+/CD3+(p&f)/CD5−/CD56−/βF1+/TCRδ1− | Y | nd | T | + | + | − | − | − | − | − |
| 18 | 35/M | Skin | ALCL | TIA1+/CD3+(p) | nd | T | − | − | − | − | − | − | − | |
| 19 | 57/F | Skin | CD30+ LPD | TIA1+/CD3+(p)/CD5− | nd | T | − | − | − | − | − | − | − | |
| 20 | 26/F | Skin | PTCL NOS | TIA1+/CD3−(p)/CD56+ | mono | T | − | − | − | − | − | − | − |
EA indicates enteropathy association (Y = yes, N = no); HS γδ TCL, hepatosplenic γδ T-cell lymphoma; PTCL NOS, peripheral T-cell lymphoma not otherwise specified; ITCL, intestinal T-cell lymphoma; INKL, intestinal NK cell lymphoma; ALCL, anaplastic large-cell lymphoma; CD30+ LPD, CD30+ lymphoproliferative disorder; p, paraffin sections; f, frozen sections; nd, not done; GL, germline; mono, clonal rearrangement.
Immunohistochemistry
Monoclonal antibodies against 7 NKRs (αCD94/XA185; αNKG2A/Z270; αEB6/p58.1; αp58.2/GL183; αp140/Q66; αp70/Z27; and αp50.3/FST172) were produced in the laboratory of L.M. Other primary antibodies were purchased from the following sources: CD3 (paraffin sections, DAKO, Copenhagen, Denmark; frozen sections, Becton Dickinson, Mountain View, CA); CD5 (DAKO); CD56 (1:20) (Novocastra, Newcastle, Great Britain); TIA1 (1:800) (Coulter Immunology, Hialeah, FL); granzyme B (1:20) (Monosan, Uden, The Netherlands), βF1 (1:10) and TCRγδ (1:10) (T-cell Diagnostics, Woburn, MA).
For NKR staining, acetone fixed cryostat sections were air dried and incubated with the primary antibody for 30 minutes (0.5% phosphate-buffered saline [PBS]) at room temperature, followed by horseradish peroxidase (HRP)–labeled rabbit antimouse antibody (1:50 in 30% human plasma with 70% PBS) for 30 minutes and subsequently with an HRP-labeled goat antirabbit antibody for 30 minutes at room temperature. Color was routinely developed with the use of diamino-benzidine (DAB). Paraffin sections were stained as described recently.17
Stains were scored as positive if more than 50% of the malignant (morphologically atypical) cells were positive.
Clonal TCRγ gene rearrangement was determined by means of a polymerase chain reaction assay amplifying the TCRγ chain gene as previously described.18
Results and discussion
Results of immunostaining for the NKRs in extranodal lymphomas.
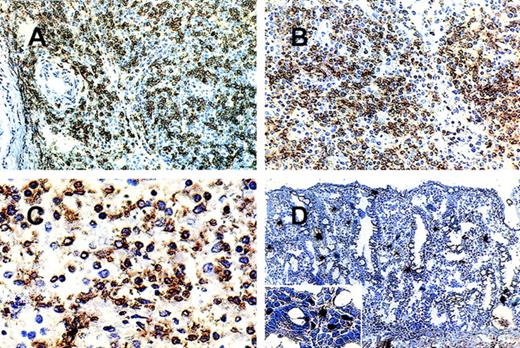
(A) (B) Nasal NK cell lymphoma (case 2): nasal biopsy. Most pleomorphic small-to- medium-sized cells are (A) CD94+ and (B) p140+. (C) Hepatosplenic γ/δ T-cell lymphoma (case 5): splenectomy specimen. Most of the medium-sized and large transformed cells are p58.2+. (D) Preserved intestinal mucosa adjacent to a lymphomatous infiltrate. Scattered lamina propria lymphocytes are p50.3+.
Results of immunostaining for the NKRs in extranodal lymphomas.
(A) (B) Nasal NK cell lymphoma (case 2): nasal biopsy. Most pleomorphic small-to- medium-sized cells are (A) CD94+ and (B) p140+. (C) Hepatosplenic γ/δ T-cell lymphoma (case 5): splenectomy specimen. Most of the medium-sized and large transformed cells are p58.2+. (D) Preserved intestinal mucosa adjacent to a lymphomatous infiltrate. Scattered lamina propria lymphocytes are p50.3+.
It is significant that all 5 NK lymphomas (Figure 1A and 1B), without exception, expressed CD94/NKG2A. These 2 molecules assemble to a heterodimer and belong to the family of lectin-type NKR. Its specific ligand has been shown to be HLA-E.18,19 NKRs of the lectin superfamily are expressed in the majority of NK cells.19This provides a clue as to how the indiscriminate killing of bystander cells by NK tumors may be kept in check.
In contrast, the total repertoire of NKRs of the immunoglobulin superfamily, so-called killer immunoglobulin-like receptors (KIRs), of both the inhibitory and the activatory type expressed by all the NK cells in an individual is variable, and each receptor is expressed only on a subset of the individual's NK cells.20 Most human NK-cell clones have been found to simultaneously express 2 or more NKRs of the KIR and CD94/NKG2 families combined.21 Expression of 1 or 2 KIRs was found in only 2 of the 5 NK lymphomas studied. The only activatory receptor tested (p50.3) was not detected in any of the lymphomas. The significance of this relatively low level of detection of NKRs of the immunoglobulin superfamily is not known at present
Of the nodal and extranodal cytotoxic T-cell lymphomas, only 3 expressed CD94. This is consistent with the fact that only a small subset of cytotoxic T cells are NKR-positive.15 All 3 positive cases were extranodal lymphomas. Of these, only the hepatosplenic γδT-cell lymphoma (HSTCL, Figure 1C) additionally expressed 2 KIRs (p58.2, p58.1). Two or more NKRs have been detected on a single T-cell.15,16 The other 2 positive cases were intestinal T-cell lymphomas, of which 1 was confirmed to be of αβT-cell lineage, on the basis of the expression of βF1 but not of TCRγδ. Both were CD56−. Three of the intestinal lymphoma samples contained preserved intestinal mucosa adjacent to the NKR-negative lymphomatous infiltrate. In these areas, scattered small lymphoid cells of the lamina propria stained for all NKRs tested, whereas no reactivity was observed on intraepithelially distributed cells. (Figure 1D). Studies are underway to investigate the NKRs' status on human mucosal lymphocytes in normal and inflamed intestine as these cells differ significantly from peripheral blood lymphocytes with respect to phenotypical and functional properties.22-24
Of the NKR-negative cases, 4 were CD56+ T-cell lymphomas (cases 6, 10, 11, and 20). It is evident that CD56 positivity need not be associated with NKR positivity. None of the nodal T-cell lymphomas of cytotoxic or noncytotoxic phenotype expressed any of the NKRs, although occasional NKR-positive cells were found scattered among the tumor cells. In the reactive lymphoid tissues, scattered NKR-positive cells were also found. In the normal spleen, CD94 positivity was found in up to 20% of all lymphoid cells, whereas KIR-positive cells were less numerous, accounting for less than 5% of all lymphoid cells. γδ T cells share many similarities with NK cells, including expression of KIRs at higher levels than αβ T cells.25It is therefore interesting to note that the HSTCL was the only T-cell lymphoma studied that expressed the KIRs. Moreover, in this case, more than 70% of all the tumor cells showed staining with 4 of the NKR antibodies, whereas none stained with the other 3 antibodies. The uniform expression of several KIRs on the vast majority of cells appears to be a definite hint for a clonal proliferation rather than a reactive process, although this is by no means as specific as TCR gene rearrangement studies for determining clonality, and reactive T-cell proliferations have also been shown to be monoclonal or oligoclonal in type.16 However, in the context of NK-cell lymphomas, the feasibility of using NKR staining patterns for identifying clonal populations of NK cells should be further explored as there is presently no tool available for assessing monoclonality in NK cell lymphomas.
Further studies are also required to determine whether the expression of NKRs delineates a biologically distinct group of T-cell lymphomas and to relate this to their pathogenesis, including possible association with sites of chronic antigenic stimulation.
Supported by the Sonderforschungsbereich 172 of the University of Würzburg; supported in part by Associazione Italiana per la Ricerca sul Cancro (AIRC) and Ministero dell' Università e della Ricerca Scientifica e Tecnologica (MURST).This study was done in part while F.C.S.H. was a guest professor at the University of Würzburg.
Reprints: Hans Konrad Müller-Hermelink, Pathologisches Institut, Universität Würzburg, Josef Schneider Str 2, D 97080 Würzburg, Germany; e-mail:path062@mail.uni-wuerzburg.de.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal