Abstract
Deletion of the multidrug resistance gene MRP1has been demonstrated in acute myeloid leukemia (AML) patients with inversion of chromosome 16 (inv[16]). These AML patients are known to have a relatively favorable prognosis, which suggests thatMRP1 might play an important role in determining clinical outcome. This study analyzed MRP1 deletion by fluorescent in situ hybridization (FISH), with a focus on inv(16) AML patients. Functional activity of multidrug resistance protein (MRP) was studied in a flow cytometric assay with the use of the MRP substrate carboxyfluorescein (CF) and the inhibitor MK-571. MRP1, MRP2, and MRP6 messenger RNA (mRNA) expression was determined with reverse transcriptase–polymerase chain reaction (RT-PCR). The results were compared with normal bone marrow cells. MRP1deletion was detected in 7 AML patients; 2 cases showed no MRP1FISH signals, and 5 cases had 1 MRP1 signal, whereas in 4 AML patients with inv(16) no MRP1 deletions were observed. A variability in MRP activity, expressed as CF efflux–blocking by MK-571, was observed (efflux-blocking factors varied between 1.2 and 3.6); this correlated with the number of MRP1 genes (r = 0.91, P < .01). MRP activity in the AML cases was not different from normal hematopoietic cells. MRP1 mRNA was detected in patients with 1 or 2 MRP1 FISH signals, but not in patients with no MRP1 signals. MRP2 and MRP6 mRNA were expressed predominantly in AML samples with 1 MRP1 signal, whereas in normal bone marrow cells no MRP2 and MRP6 mRNA was observed. In conclusion, this study shows that MRP activity varies among inv(16) AML cases and does not differ from that in normal hematopoietic cells; this might be in part due to the up-regulation of other MRP genes.
The occurrence of cross-resistance to structurally and functionally unrelated drugs, called multidrug resistance (MDR), is a main cause of failure in the chemotherapeutic treatment of malignant disorders. Several mechanisms of MDR have been identified1; one of these is the overexpression of adenosine triphosphate (ATP)–dependent membrane proteins that function as drug-efflux pumps. The multidrug resistance protein MRP12 is a member of the superfamily of ATP-binding cassette (ABC) transporters to which P-glycoprotein (P-gp), encoded by the MDR1 gene, also belongs. MRP1, a 190-kd protein, is encoded by the MRP1gene located on chromosome 16p13 and has been shown to transport a broad range of organic substrates, such as glutathione (GSH) conjugates3 and other anionic conjugates.4Overexpression of MRP1 results in resistance to different classes of chemotherapeutic agents.5 Recently 5 additional MRP family members, MRP2 through MRP6, have been identified.6-8 However, the role of these MRP1isoforms in MDR is not well defined yet. The most recently discovered member of the MRP family, MRP6, is located on chromosome 16p13, immediately next to the MRP1 gene. The 3′ end of theMRP6 gene was found to be identical with the anthracycline resistance–associated (ARA) gene.9Overexpression of the complete MRP6 gene or part of it was observed only in cell lines with MRP1 gene overexpression, suggesting a coamplification as a result of the location adjacent to MRP1.8
Studies describing MRP1 messenger RNA (mRNA) and protein expression in AML demonstrated variable results. In a study,10 MRP1 protein expression appeared to have no impact on treatment outcome in AML. Another study showed MRP function, but not MRP1 protein expression, to be a poor prognostic factor for achievement of complete remission in AML.11 Previously we reported that blasts of AML patients express MRP1 and MRP2 mRNA and that in these blasts MRP functional activity correlates with MRP1 protein expression.12 Recently it was demonstrated that knockout of the mrp1 gene (mrp1[-/-]) in murine embryonic stem cells and in mice leads to an increased in vitro sensitivity to chemotherapeutic agents that are commonly used in the treatment of AML.13 14
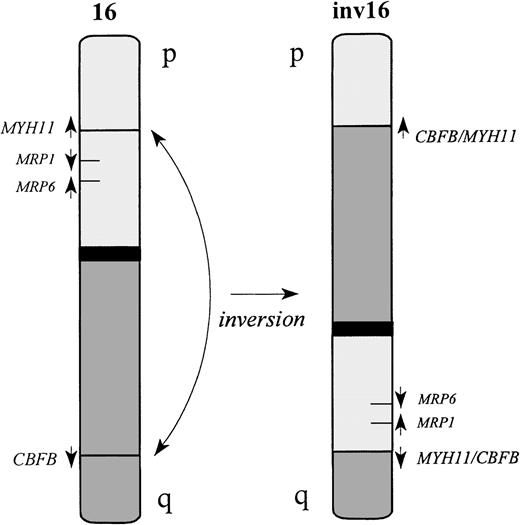
Inversion of chromosome 16 is a recurrent chromosomal rearrangement identified in a subgroup of AML patients, most commonly patients with the French-American-British (FAB) classification M4Eo.15These patients generally have a high response rate to chemotherapy and a relatively favorable prognosis.16,17 The fusion of the core binding factor B (CBFB) gene located at chromosome 16q22 and the myosin heavy chain (MYH11) gene located at 16p13 results in a chimeric translation product coding for N-terminal CBFB and C-terminal MYH11 sequences.18 It is considered likely that this fusion protein contributes to leukemogenesis and has a dominant effect since only 1 of the 2 chromosomes 16 is affected. Deletion of 1 MRP1 allele17 and also deletion of the ARA gene19 have been demonstrated in cells of patients with inv(16) and were associated with increased duration of disease-free survival, suggesting an important role for MRP1 in determining clinical outcome.17 This study did not evaluate MRP activity in these inv(16) AMLs, and the occurrence of MRP1deletion in additional subclasses of AML was not evaluated.
In the present study, we analyzed the functional activity of MRP in AML patients, with a focus on inv(16) patients versus normal bone marrow samples, using a flow cytometric assay with the MRP-specific substrate carboxyfluorescein (CF) in combination with the MRP inhibitor and leukotriene D4 receptor antagonist MK-571.20 In addition, the occurrence of MRP1deletions was studied with the fluorescent in situ hybridization (FISH) technique, and the expression of MRP1, MRP2, and MRP6 mRNA were detected by means of reverse transcriptase–polymerase chain reaction (RT-PCR).
Patients, materials, and methods
Patients
After informed consent, bone marrow aspirates were collected from 10 de novo AML patients, diagnosed as inv(16) in the participating hospitals between April 1986 and March 1998, and 1 bone marrow sample was taken from 1 of 15 de novo AML patient samples from a previous study12; the latter was the only sample that did not show any MRP1 protein and a very low MRP activity in that study. Patients were classified as AML according to the FAB classification.21 Leukemic blasts from bone marrow or peripheral blood were enriched by Ficoll-Isopaque (Nycomed, Oslo, Norway) density gradient centrifugation; cryopreserved in RPMI 1640 medium (BioWhittaker, Brussels, Belgium) supplemented with 10% fetal calf serum (FCS) (Hyclone, Logan, UT) and 10% dimethyl sulfoxide (DMSO) (Merck, Amsterdam, The Netherlands); and stored at −196°C. Upon analysis, AML cells were thawed, treated with DNase I (Boehringer Mannheim, Almere, The Netherlands), washed with RPMI 1640 medium, and incubated for 1 hour in RPMI 1640 medium, supplemented with 10% FCS at 37°C, 5% CO2. May-Grünwald Giemsa stainings were made or immunophenotyping was performed to determine the percentage of blast cells in the AML cells. For phenotyping, 0.5 × 106 were incubated with 5 μL of fluorescein isothiocyanate– or phycoerythrin-labeled mouse monoclonal antibodies, CD34, CD33, or immunoglobulin G isotype–matched control (Becton Dickinson, Mountain View, CA) for 20 minutes at 4°C, washed with RPMI 1640 medium, and analyzed with a FACStar flow cytometer (Becton Dickinson Medical Systems, Sharon, MA). Viability of the cells was determined by trypan blue exclusion.
Normal bone marrow samples
Normal bone marrow samples were obtained after informed consent from 5 patients who underwent cardiac surgery. Samples were cryopreserved and thawed as described above. T-lymphocyte depletion was performed by 2-aminoethylisothiouronium bromide-treated sheep red blood cell (SRBC) rosetting. T-cell–SRBC rosettes were removed by Ficoll Isopaque density gradient centrifugation. Viability of the cells was determined by trypan blue exclusion.
Cell lines
The human small cell lung cancer cell line GLC4 and its in vitro doxorubicin-selected, MRP1-overexpressing counterpart GLC4/ADR,22 were maintained in RPMI 1640 supplemented with 10% FCS. GLC4/ADR cells were cultured in the presence of 1.2 μmol/L doxorubicin (Pharmacia and Upjohn, Woerden, The Netherlands).
Cytogenetics
To study cytogenetics, bone marrow cells were cultured for 24 and 48 hours in RPMI 1640 supplemented with 15% FCS. The cultures were harvested and chromosome preparations were made according to standard cytogenetic techniques. The chromosomes were G-banded with the use of trypsin or pancreatin, and karyotypes were described according to the International System for Human Cytogenetic Nomenclature ISCN 1995.
Fluorescent in situ hybridization
Chromosome preparations were made according to standard cytogenetic techniques, and FISH was performed on the preparations according to standard laboratory protocols. An MRP1 complementary DNA fragment containing the basepairs (bp) 1036-5590, which was excised with BamHI from the expression vector pJ3Ω-MRP,23was used as a probe for hybridization. An H36 cosmid, which is located on chromosome 16q24, was used as a control of the hybridization procedure and as a detector of chromosome 16q. Detection was performed by fluorescence microscopy. The results of the FISH technique were described as follows: 0 signals when no MRP1 signal was detected on either of the chromosomes 16 in all observed metaphases; 1 signal when an MRP1 signal was observed on 1 of the 2 chromosomes 16 in all observed metaphases; and 2 signals when MRP1 was detected on both chromosomes 16 in at least 4 observed metaphases.
RNA extraction and RT-PCR
RNA extraction.
Total cellular RNA was isolated from 10 × 106 AML blasts or 5 × 106 cell line cells with the use of 1 mL Trizol reagent (Life Technologies, Breda, The Netherlands). RNA was extracted, precipitated, and washed according to the manufacturer's protocol.
RT-reaction.
RNA (1 μg) was reverse transcribed in 15 μL of RT buffer containing 1.8 mmol/L each of deoxyadenosine triphosphate, deoxyguanosine triphosphate, deoxycytidine triphosphate, and thymidine 5′-triphosphate (Promega Corp, Madison, WI); 9 mmol/L MgCl2; 68 mmol/L KCl; 45 mmol/L Tris/HCl (pH 8.3); 0.8 mg/mL bovine serum albumin; 28% glycerol; 10 U Moloney murine leukemia virus reverse transcriptase (M-MLV RT) (Phamacia, Woerden, The Netherlands); 4.8 U RNAguard (Pharmacia); 0.2 μg pd(N)6random primers (Pharmacia); and 3 mmol/L dithiothreitol (Life Technologies). The reaction conditions were 65°C for 10 minutes and 37°C for 60 minutes.
PCR analysis for CBFB/MYH11 mRNA fusion transcript.
CBFB (0.33 μL, 50 μmol/L) and MYH11(0.38 μL, 50 μmol/L) primers and 33.8 μL H2O were added, and the PCR reaction mix was incubated for 5 minutes at 96°C. Taq DNA polymerase (0.5 μL) (Pharmacia) was added, up to a final volume of 50 μL. The PCR reaction was subjected to 40 cycles of denaturation (94°C, 1 minute), annealing (57°C, 1 minute), and extension (72°C, 1 minute). Five μL of the reaction products were separated on a 1.5% agarose gel. The differentially spliced CBFB/MYH11 transcripts were visualized by ethidium bromide staining. The amplified product contains, depending on the nucleotide of splicing, 416 (type A), 1136 (type C), or 1343 (type D) bp. The sequences of the primers are as follows: CBFB 5′GCAGGCAAGGTATATTTGAAGG3′, MYH11 5′CTCTTCTCCTCATTCTGCTC3′.
PCR analysis for MRP1, MRP2, ARA/MRP6, and β-2 microglobin mRNA.
PCR analysis for MRP1 and MRP2 mRNA was performed as described previously.12 The primer pair chosen for MRP1 corresponds to bases 987 to 1007 (sense) and 1956 to 1976 (antisense); the amplified product contains 990 bp. The sequences of the MRP1 primers are as follows: sense, 5′AATGCGCCAAGACTAGGAAG3′; antisense, 5′ACCGGAGGATGTTGAACAAG3′. For MRP2, the amplified product consists of 1067 bp. Primer pairs corresponding to bases 961 to 981 (sense) and 2007 to 2027 (antisense) are chosen. MRP2 primer sequences are as follows: sense, 5′CTGGTTGATGAAGGCTCTGT3′, and antisense, 5′CTGCCATAATGTCCAGGTTC3′.
ARA/MRP6 PCR was performed as described previously24 with some modifications. MRP6 primer sequences are as follows: sense, 5′ACACCCATTGGTCACCTGCTA3′, antisense: 5′GGTCACCTGGAGGGCAGCAGAGAC3′. The PCR reaction was subjected to 40 cycles of denaturation (95°C, 1 minute), annealing (65°C, 1 minute), and extension (72°C, 1 minute). The amplified product contains 324 bp.
For β-2 microglobulin, the PCR reaction was subjected to 22 cycles of denaturation (95°C, 30 seconds), annealing (56°C, 30 seconds), and extension (72°C). Primer pairs chosen for β-2 microglobulin are as follows: sense, 5′CCAGCAGAGAATGGAAAGTC3′ and antisense, 5′GATGCTGCTTACATG-TCTCG3′; the amplified product consists of 268 bp.
Flow cytometric detection of functional drug efflux
Functional activity of the MRP and P-gp transporters was demonstrated as described previously.12 To determine MRP (MRP1 and homologues MRP2 and MRP6) activity, we used the compound carboxyfluorescein diacetate (CFDA) (Sigma Chemical, Bornem, Belgium), which permeates the plasma membrane and which is transformed into the fluorescent anion CF upon cleavage of the ester bonds. CFDA was used in combination with the leukotriene D4 receptor antagonist and the MRP inhibitor MK-571 (kindly provided by Dr Ford-Hutchinson, Merck Sharp, Kirkland, Quebec, Canada).25 For the detection of P-gp activity, Rh123 (Sigma) was used together with the P-gp–specific inhibitor PSC833 (provided by Sandoz, Basel, Switzerland).
Cells (0.5 × 106) were loaded for 20 minutes at 37°C, 5% CO2 with 0.1 μmol/L CFDA or 200 ng/mL Rh123 with or without inhibitor (20 μmol/L MK-571 or 2 μg/mL PSC833) in RPMI 1640 medium. Thereafter, cells were washed in ice-cold medium and incubated for 1 hour in drug-free medium with or without inhibitor at 37°C, 5% CO2. Efflux was stopped by pelleting the cells and adding ice-cold medium. Fluorescence of CF and Rh123 was analyzed with a FACStar flow cytometer, equipped with an argon laser. CF and Rh123 fluorescence of 10 000 events was logarithmically measured through a 530-nm band-pass filter at an excitation wavelength of 488 nm. The efflux-blocking factors of the inhibitors were expressed as the median relative fluorescence units in inhibitor blocked cells divided by the median relative fluorescence units in unblocked cells after 60 minutes of efflux. Each time the assay was performed, GLC4 cells were taken as control.
Statistical analysis
The paired Student t test was used to calculate significance, and correlations were calculated by means of the Pearson bivariate correlation test. A P of less than .05 was considered significant.
Results
Patient characteristics
Bone marrow samples were obtained from 11 de novo AML patients. Patient characteristics are described in Table 1. According to the FAB classification, 1 patient was M1, 1 was M2, and 9 were M4Eo. The patients were treated with intensive chemotherapy regimens according to the protocol of the Dutch-Belgian Hemato-Oncology Cooperative Group for AML (Hovon)26,27 or according to the protocol of the European Organization for the Research and Treatment of Cancer (EORTC).27 28
Cytogenetic analysis and RT-PCR
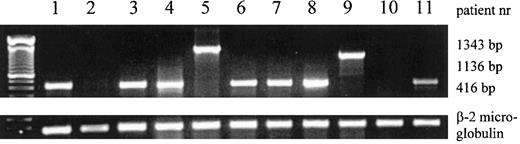
Cytogenetic analysis demonstrated that 10 patients had karyotypes showing clonal chromosomal abnormalities and that 1 patient had a normal karyotype. Relevant aberrations are shown in Table1. Inv(16)(Figure1) was demonstrated in all 9 patients with M4Eo and in 1 patient classified as M2. To confirm the cytogenetic results, RT-PCR was performed to detect the different fusion transcripts related to inv(16). In 9 patient samples, a fusion product was detected; 7 of these showed the 416-bp product of type A, 1 sample showed the 1136-bp product of type C, and 1 showed the 1343-bp product of type D (Table 1, Figure 2). In patient 10 with FAB classification M4Eo and cytogenetic karyotype inv(16), no fusion product was detected, even after 8 additional cycles of PCR.
Characteristics of inv(16) AML patients
| Patient no. . | Relevant FABcytogenetic classificationaberrations . | Percentage of blasts* . | Inv(16) PCR product (bp) . | Treatment† . | |
|---|---|---|---|---|---|
| 1 | M4Eo | inv(16) | 85 | 420 | Ara-C, DNR, VCR |
| 2‡ | M1 | N | 95 | — | Ara-C, Ida |
| 3 | M4Eo | inv(16) | 90 | 420 | Ara-C, Ida, Amsa |
| 4 | M4Eo | inv(16) | 80 | 420 | Ara-C, Ida, Amsa |
| 5 | M4Eo | inv(16) | 90 | 1400 | Ara-C, Ida, Amsa |
| 6 | M4Eo | inv(16) | 80 | 420 | Ara-C, DNR, Amsa |
| 7 | M4Eo | inv(16) | 90 | 420 | Ara-C, DNR, Amsa |
| 8 | M4Eo | inv(16) | 95 | 420 | Ara-C, Ida** |
| 9 | M2 | inv(16) | 85 | 1200 | Ara-C, Ida, Amsa |
| 10 | M4Eo | inv(16) | 95 | — | Ara-C, DNR |
| 11 | M4Eo | der(11)t(11;16), der(16)inv(16)t(11;16) | 95 | 420 | Ara-C, Ida, Amsa |
| Patient no. . | Relevant FABcytogenetic classificationaberrations . | Percentage of blasts* . | Inv(16) PCR product (bp) . | Treatment† . | |
|---|---|---|---|---|---|
| 1 | M4Eo | inv(16) | 85 | 420 | Ara-C, DNR, VCR |
| 2‡ | M1 | N | 95 | — | Ara-C, Ida |
| 3 | M4Eo | inv(16) | 90 | 420 | Ara-C, Ida, Amsa |
| 4 | M4Eo | inv(16) | 80 | 420 | Ara-C, Ida, Amsa |
| 5 | M4Eo | inv(16) | 90 | 1400 | Ara-C, Ida, Amsa |
| 6 | M4Eo | inv(16) | 80 | 420 | Ara-C, DNR, Amsa |
| 7 | M4Eo | inv(16) | 90 | 420 | Ara-C, DNR, Amsa |
| 8 | M4Eo | inv(16) | 95 | 420 | Ara-C, Ida** |
| 9 | M2 | inv(16) | 85 | 1200 | Ara-C, Ida, Amsa |
| 10 | M4Eo | inv(16) | 95 | — | Ara-C, DNR |
| 11 | M4Eo | der(11)t(11;16), der(16)inv(16)t(11;16) | 95 | 420 | Ara-C, Ida, Amsa |
AML indicates acute myeloid leukemia; FAB, French-American-British; PCR, polymerase chain reaction; N, normal; Ara-C, cytosine arabinoside; DNR, daunorubicin; VCR, vincristine; Ida, idarubicin; Amsa, amsacrine.
Percentage of blasts determined by May-Grünwald Giemsa staining, except for patient 4, where it was determined according to the percentage of CD33+ cells.
Patient 1 was treated according to the protocol of the European Organization for the Research and Treatment of Cancer; other patients were treated according to the protocol of the Dutch-Belgian Hemato-Oncology Cooperative Group (Hovon).
This patient sample showed no MRP1 protein expression and a low MRP activity in a previous study.12
Schematic representation of the inv(16).
Both CBFB and MYH11 genes are transcribed in the centromeric to telomeric direction. The inversion fuses the 5′ end of the CBFB gene, located on 16q22, upstream of the 3′ end of MYH11 on 16p13. The reciprocal fusion geneMYH11/CBFB is also generated in most of the cases. However, deletion of MYH11 sequences upstream of the 5′ breakpoint is reported in some cases.29
Schematic representation of the inv(16).
Both CBFB and MYH11 genes are transcribed in the centromeric to telomeric direction. The inversion fuses the 5′ end of the CBFB gene, located on 16q22, upstream of the 3′ end of MYH11 on 16p13. The reciprocal fusion geneMYH11/CBFB is also generated in most of the cases. However, deletion of MYH11 sequences upstream of the 5′ breakpoint is reported in some cases.29
Expression of inv(16) mRNA fusion products and β-2 microglobulin mRNA in AML patient samples, determined by RT-PCR.
The 416-bp, 1136-bp, and 1343-bp products of type A, C, and D are shown. The patient numbers correspond to the numbers in Table 1.
Expression of inv(16) mRNA fusion products and β-2 microglobulin mRNA in AML patient samples, determined by RT-PCR.
The 416-bp, 1136-bp, and 1343-bp products of type A, C, and D are shown. The patient numbers correspond to the numbers in Table 1.
Fluorescent in situ hybridization
The FISH technique showed no MRP1 signals in 2 of the 11 AML patient samples (Table 2). One of these 2 patients was classified as M4Eo and showed inv(16) in both the cytogenetic analysis and the RT-PCR. The other patient was classified as M1, having a normal karyotype and showing no fusion transcript. In patients 3 through 7, 1 MRP1 signal was demonstrated on 1 of the 2 chromosomes 16; the other chromosome 16 did not show an MRP1 signal. Two MRP1 signals were observed in patients 8 through 11. These patients showed inv(16); 1 MRP1signal was observed on the q arm of 1 of the chromosomes 16, while the other chromosome showed an MRP1 signal on the p arm. As a control, 3 normal bone marrow samples were studied. Two signals were observed in the normal hematopoietic cells.
AML patient results
| Patient no. . | FAB classification . | MRP1 FISH no. signals . | MRP activity* . | P-gp activity† . | Overall survival (m) . | Present status . |
|---|---|---|---|---|---|---|
| 1 | M4Eo | 0 | 1.2 | 1.5 | 11 | relapse |
| 2 | M1 | 0 | 1.6 | 6.5 | — | toxic death |
| 3 | M4Eo | 1 | 2.3 | 2.4 | 26 | CR |
| 4 | M4Eo | 1 | 2.7 | 1.8 | — | toxic death |
| 5 | M4Eo | 1 | 2.3 | 1.8 | — | death in CR |
| 6 | M4Eo | 1 | 1.9 | 1.6 | 74 | CR |
| 7 | M4Eo | 1 | 2.4 | 2.1 | 81 | CR |
| 8 | M4Eo | 2‡ | 3.0 | 1.6 | 95 | CR |
| 9 | M2 | 2‡ | 3.6 | 2.5 | 23 | CR |
| 10 | M4Eo | 2‡ | 2.7 | 1.6 | 14 | CR |
| 11 | M4Eo | 2‡ | 3.3 | 2.2 | — | toxic death |
| NB | 2 | 2.8 ± 0.5 | 1.8 ± 0.4 | |||
| (n = 3) | (n = 9) | (n = 9) |
| Patient no. . | FAB classification . | MRP1 FISH no. signals . | MRP activity* . | P-gp activity† . | Overall survival (m) . | Present status . |
|---|---|---|---|---|---|---|
| 1 | M4Eo | 0 | 1.2 | 1.5 | 11 | relapse |
| 2 | M1 | 0 | 1.6 | 6.5 | — | toxic death |
| 3 | M4Eo | 1 | 2.3 | 2.4 | 26 | CR |
| 4 | M4Eo | 1 | 2.7 | 1.8 | — | toxic death |
| 5 | M4Eo | 1 | 2.3 | 1.8 | — | death in CR |
| 6 | M4Eo | 1 | 1.9 | 1.6 | 74 | CR |
| 7 | M4Eo | 1 | 2.4 | 2.1 | 81 | CR |
| 8 | M4Eo | 2‡ | 3.0 | 1.6 | 95 | CR |
| 9 | M2 | 2‡ | 3.6 | 2.5 | 23 | CR |
| 10 | M4Eo | 2‡ | 2.7 | 1.6 | 14 | CR |
| 11 | M4Eo | 2‡ | 3.3 | 2.2 | — | toxic death |
| NB | 2 | 2.8 ± 0.5 | 1.8 ± 0.4 | |||
| (n = 3) | (n = 9) | (n = 9) |
AML indicates acute myeloid leukemia; FAB, French-American-British; FISH, fluorescent in situ hybridization; m, months; NB, normal bone marrow; CR, complete remission.
Expressed as CF efflux-blocking factor of MK-571.
Expressed as Rh123 efflux-blocking factor of PSC833.
In these cases, 1 MRP1 signal was observed on the q arm of chromosome 16.
MRP1, MRP2, and MRP6 mRNA expression
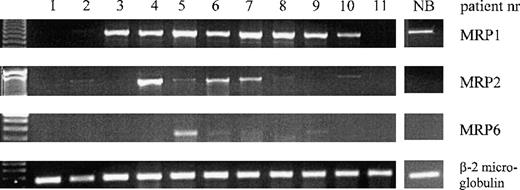
To study whether the MRP1 deletion in AML patient samples causes a diminished MRP1 mRNA expression, semiquantitative RT-PCR analysis was performed. MRP1 mRNA was observed in all patients with 1 or 2 MRP1 FISH signals except for patient 11. MRP1 mRNA expression was not observed in the patients with no MRP1 FISH signals (patients 1 and 2, Figure3). Since the MRP6 gene is located immediately next to the MRP1 gene at chromosome 16p13, RT-PCR was performed to study MRP6 mRNA expression in the AML samples. Patient 5 showed a distinct MRP6 mRNA band, while patients 6, 7, and 9 had a faint MRP6 mRNA expression (Figure 3). All of these patients, except for patient 9, had 1 MRP1 gene in the FISH study.
Expression of MRP1, MRP2, MRP6, and β-2 microglobulin mRNA in AML patient samples and in a representative normal bone marrow control (NB), determined by RT-PCR.
The patient numbers correspond to the numbers in Table 1.
Expression of MRP1, MRP2, MRP6, and β-2 microglobulin mRNA in AML patient samples and in a representative normal bone marrow control (NB), determined by RT-PCR.
The patient numbers correspond to the numbers in Table 1.
To analyze whether MRP2 mRNA might be up-regulated in the cases with an MRP1 deletion, MRP2 mRNA was assessed in the patient samples. The results for MRP2 mRNA are depicted in Figure 3. One of the patients with no MRP1 FISH signals (patient 2) expressed MRP2 mRNA at a low level. The patients with 1 MRP1FISH signal (patients 4 through 7) showed MRP2 mRNA expression at various levels, except for patient 3 where no detectable levels of MRP2 mRNA could be observed. In the patient samples with 2MRP1 FISH signals (patients 8 through 11), no or very low MRP2 mRNA expression was detected.
Normal hematopoietic bone marrow cells demonstrated a low MRP1 mRNA expression, while no detectable expression of MRP2 and MRP6 mRNA was found in 3 independent experiments. The results of a representative experiment are shown in Figure 3.
MRP and Pgp activity
To test whether the deletion of an MRP1 gene causes a diminished MRP functional activity in these AML samples, we performed a flow cytometric assay using CFDA in combination with the MRP inhibitor MK-571. A variability in CF efflux–blocking of MK-571 was observed in the 11 AML patients studied (Table 2, Figure4); efflux-blocking factors varied between 1.2 and 3.6. A significant correlation was observed between the number of MRP1 FISH signals and MRP activity (r = 0.91,P < .01) (Figure 4). To test whether the same variability could be observed with regard to the functionality of P-gp, a flow cytometric assay was performed with the use of Rh123 in combination with PSC833 to determine P-gp activity. The Rh123 efflux–blocking factors of PSC833 in these AML patient samples varied between 1.1 and 6.5. No correlation was observed between P-gp activity and MRP activity or between P-gp activity and MRP1 FISH signals. Finally, to correlate these findings with normal hematopoietic cells, MRP and P-gp activity were determined in normal bone marrow samples. The median CF efflux–blocking factor of MK-571 was 2.8 ± 0.5 (n = 9), and the P-gp activity, as determined by Rh123 efflux–blocking of PSC833, was 1.8 ± 0.4 (n = 9) (Table 2) in the normal hematopoietic bone marrow cells. These median values are not significantly different from the median values of the total group of AML patients. Furthermore, all CF efflux–blocking factors by MK-571 of the AML patient samples with 1 or 2 MRP1 FISH signals are within the range of the normal bone marrow values, whereas the 2 patient samples with noMRP1 FISH signals show lower MRP activity than the normal bone marrow samples (Figure 4).
MRP activity, expressed as CF efflux–blocking factors of MK-571.
The activity is expressed in relation to the number ofMRP1 FISH signals in AML patient samples and in normal bone marrow (NB) samples.
MRP activity, expressed as CF efflux–blocking factors of MK-571.
The activity is expressed in relation to the number ofMRP1 FISH signals in AML patient samples and in normal bone marrow (NB) samples.
Treatment outcome
At the time of evaluation, 6 patients were alive and in continuous complete remission (CR) (follow-up time, 14 to 95 months). The median overall survival of this patient group was 52.2 months ± 35.0. Two patients achieved CR but relapsed; 1 of these died. Three additional patients died during the remission-induction phase of chemotherapy (Table 2). No distinct correlations were observed between treatment outcome and MRP1 deletion or MRP activity in this small group of AML patients.
Discussion
The MRP1 gene is located at 16p13, centromeric to the MYH11 gene at a distance of approximately 150 kb.30 In the study of Kuss et al,17 a different treatment outcome in 5 inv(16) patients with MRP1 deletion and 7 inv(16) patients without MRP1 deletion was reported, suggesting that MRP1 has a critical role in determining clinical outcome in patients with inv(16). An additional study reported a deletion of between 150 kb and 350 kb, centromeric to the short-arm inversion breakpoint in the MYH11 gene, in a subgroup of inv(16) patients (6 of 38 patients studied), which did not affect the clinical outcome.29 In the present study, in a small group of AML patients we observed no correlations between treatment outcome and MRP1 deletion.
In 2 of the 11 patients studied, an MRP1 deletion on both chromosomes 16 was observed, 1 with inv(16). This patient sample showed, besides the deletion of MRP1 on the inverted chromosome, a deletion of MRP1 on the other chromosome 16. Even the 2 patients with 2 MRP1 deletions showed a significant but low MRP activity in the flow cytometric assay, which suggests that additional MRP1 homologues, such as MRP2, could function as transporters for certain anionic substrates in these AML samples. This might be the case in the present study with respect to the CF transport, since CF is not a specific substrate for MRP1, but may also be transported by MRP2 or MRP6. An additional explanation for the CF efflux in the 2 patient samples might be the presence of another, yet undescribed, pump. In these samples, MRP2 mRNA is expressed at a low level, and MRP6 mRNA is not expressed at a detectable level, while the normal bone marrow controls showed no MRP2 or MRP6 mRNA expression.
Two junctions are reported in CBFB31 and 4 inMYH11,32 resulting in 6 different in-frame fusion proteins (type A-F).31 In the present study, 7 of 10 inv(16) patients showed the most commonly occurring 416-bp product of type A, whereas in 1 patient sample the 1136-bp product of type C was detected; 1 patient had the 1343-bp product of type D, and 1 inv(16) patient showed no inv(16) RT-PCR product, in spite of the findings of the cytogenetic analysis and the MRP1 FISH technique. It has been reported that a breakpoint can occur at position 16q24, rather than at the usual q22 position.33 However, according to the cytogenetics of the AML patients, this seemed not to be the case in the present study. Another possibility is the occurrence of an additional CBFB/MYH11 fusion transcript,34 which cannot be detected with the primer sets used in this study.
A faint or no MRP2 and MRP6 mRNA expression was observed in AML samples with 2 MRP1 FISH signals and in normal bone marrow cells, whereas the samples with 1 MRP1 FISH signal showed distinct MRP2 or MRP6 mRNA expression. In the patient samples with no MRP1FISH signals, a faint or no MRP2 mRNA expression and no MRP6 mRNA expression were detected. However, 1 of these patients showed a high P-gp activity. These findings indicate that besides the deletion ofMRP1 in inv(16) patients, a variable expression of additional genes that are involved in MDR can be observed. Since a rapidly increasing number of ABC transporters that play a role in MDR have been identified (for the most recent update, seehttp://www.med.rug.nl/mdl/humanabc.htm), it seems likely that in the case of a deletion of 1 of these genes, the function is compensated for by other transporter proteins. Therefore, the relatively favorable prognosis of inv(16) AML patients seems to be caused by factors other than the deletion of the MRP1 gene. The clinical relevance of these findings has to be determined in a larger group of patients.
Reprints:E. Vellenga, Division of Hematology, Department of Internal Medicine, University Hospital Groningen, Hanzeplein 1, 9713 GZ Groningen, The Netherlands; e-mail: e.vellenga@int.azg.nl.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal