Abstract
The human t(14;18) chromosomal translocation is assumed to result from illegitimate rearrangement between BCL-2 and DH/JH gene segments during V(D)J recombination in early B cells. De novo nucleotides are found inserted in most breakpoints and have been thus far interpreted as nontemplated N region additions. In this report, we have analyzed both direct (BCL-2/JH) and reciprocal (DH/BCL-2) breakpoints derived from 40 patients with follicular lymphoma with t(14;18). Surprisingly, we found that more than 30% of the breakpoint junctions contain a novel type of templated nucleotide insertions, consisting of short copies of the surrounding BCL-2, DH, and JH sequences. The features of these templated nucleotides, including multiplicity of copies for 1 template and the occurrence of mismatches in the copies, suggest the presence of a short-patch DNA synthesis, templated and error-prone. In addition, our analysis clearly shows that t(14;18) occurs during a very restricted window of B-cell differentiation and involves 2 distinct mechanisms: V(D)J recombination, mediating the breaks on chromosome 14 during an attempted secondary DH to JH rearrangement, and an additional unidentified mechanism creating the initial breaks on chromosome 18. Altogether, these data suggest that the t(14;18) translocation is a more complex process than previously thought, involving the interaction and/or subversion of V(D)J recombination with multiple enzymatic machineries.
The t(14;18) (q32;q21) chromosomal translocation and ensuing overexpression of the proto-oncogene BCL-2 are assumed to be the initial steps of the malignant transformation to follicular lymphoma (FL).1,2 Analysis of the breakpoint regions have shown that the BCL-2 gene on chromosome 18 is fused to 1 of the JH gene segments from the immunoglobulin (Ig)H locus on chromosome 14,3-6 whereas the reciprocal junction consists in most cases of the fusion of a DH gene segment from the IgH locus on chromosome 14 with the remaining 3′ BCL-2untranslated region on chromosome 18.7-10 The involvement of the Ig DH and JH gene segments in the recombination process, together with the presence of N regions at the breakpoints, prompted early interpretations of the t(14;18) translocation as a mistake in the normal mechanism of V(D)J recombination.4-6,8,11 12
V(D)J recombination is a highly orchestrated lymphoid-specific mechanism, regulated throughout differentiation and cell cycle. Recombination is directed by recombination signal sequences (RSS), which are flanking each gene segment (for review see Lewis13). The initiation of the V(D)J recombination consists of a single strand cut at the precise coding end/RSS border, followed by transesterification on the opposite strand, generating 2 covalently sealed hairpin coding ends (for review see Gellert14). DNA hairpins are subsequently resolved into free ends. In case of nicks occurring on only 1 strand, the resulting protruding strand ends with palindromic nucleotides coming from the opposite strand. These nucleotide additions are termed P nucleotides and are a characteristic of V(D)J recombination because they are the direct result of DNA hairpin resolution. Alternatively, nicks could also happen on both strands, giving rise to deletion of part of the coding ends. Once coding ends are opened up, more modifications can take place. One of these modifications is the addition of nontemplated nucleotides (termed N nucleotides) to 3′OH free ends by the terminal deoxynucleotidyl transferase (TdT). The sum of those modifications of the coding ends before religation (P and N addition and nucleotide deletion) is referred to as “coding end processing” and constitutes the hallmark of the V(D)J recombination process. Consistent with the V(D)J recombination mechanism, DNA sequence analysis of the direct (BCL-2/JH) and reciprocal (DH/BCL-2) junctions revealed a normal processing of the DH and JH segments.4-10,15,16 It is therefore likely that at least the DH and JHcounterparts of the translocation are generated by V(D)J recombination. The involvement of the V(D)J recombination mechanism in theBCL-2 counterpart is yet more obscure. On chromosome 18, most breaks occur in the major breakpoint region (mbr), located in the 3′ untranslated region of the BCL-2 gene.6,7Within the mbr, Wyatt et al10 have subdefined 3 clusters of 15 to 20 base pairs (bp), in which 80%-90% of the mbr breaks occur. Despite the remarkable clustering of the breakpoints in BCL-2, compared with other similar types of translocations, no proper RSSs were found, arguing against a role of a RAG-1/2-mediated process. Nevertheless, the presence of many potential cryptic RSSs in the mbr leaves the possibility of very low levels of V(D)J recombination at those sites.17
As a consequence of the absence of RSS-mediated cuts and in contrast to the DH and JH coding ends, the mbr breaks do not occur at 1 precise location. Therefore, simultaneous analysis of both direct and reciprocal breakpoints from the same translocation event is necessary to infer the initial location of the mbr breakpoint and potential subsequent processing of its 5′ mbr and 3′ mbr ends. To date, only few t(14;18) translocations have been characterized at both direct and reciprocal breakpoints, giving a somewhat confusing picture of the possible mechanism responsible for the initial break at the mbr locus.7-11 Initially, Bakhshi et al7 noted a 3-bp duplication of the mbr sequence and proposed that this could be the result of a staggered double-strand break. However, in all cases reported so far, the duplications were short and could also be attributed to N additions. The issue of the presence of duplications as a general feature of the t(14;18) translocation is of importance, since both precise breaks and deletions are compatible with V(D)J recombination mechanism, but duplications are not. To get a better understanding of the molecular mechanism involved in the t(14;18) translocation, we report in this study a detailed analysis of the first comprehensive DNA sequence library of both direct and reciprocal breakpoint regions derived from 40 t(14;18) translocation-positive FL patients. Our results clearly show that 2 distinct mechanisms generate the breaks at the immunoglobulin and mbr loci and reveal the presence of an unexpected new type of error-prone templated nucleotide insertion at the breakpoints. The implications of these new features of the t(14;18) breakpoints shed new light on possible mechanisms involved in the translocation process and ensuing lymphomagenesis.
Materials and methods
Source of DNA samples
Samples in this study are derived from consecutive patients with follicular non-Hodgkin lymphoma from 2 independent sources: the University Hospital, Vienna, Austria, and the University Clinic, Goettingen, Germany. DNA was prepared from routine peripheral blood mononuclear cells, bone marrow aspirates, lymph node, or lymph node biopsies according to standard procedures.
Polymerase chain reaction amplification and sequencing
Direct (mbr/JH) breakpoints were amplified from 100 ng of genomic DNA with mbr-6A and Jex-B primers for 30 cycles with the following conditions: 30 seconds at 94°C, 30 seconds at 58°C, and 30 seconds at 72°C. The mbr-6A primer is located 90 bp 5′ of the mbr cluster 1, and Jex-B primer is a JH consensus primer located in and 3′ of the JH coding sequences. Double-nested secondary amplifications were performed with mbr-7A and JHcoB primers from 1 μL of the primary polymerase chain reaction (PCR), under the same conditions as above except for an annealing temperature of 61°C. The DH locus consists of 27 DH segments, spread over 60 kilobase (kb) of genomic DNA18 and grouped into 7 families with large intronic regions of complete homology. We therefore designed a restricted set of 7 DH primers mapping all members of each family (D1 to D7). Reciprocal (DH/mbr) breakpoints were amplified from 100 ng of genomic DNA with 1 of the D1 to D7 primers and the mbr5-B primer, with the same conditions as above (annealing temperature: 61°C). D1 to D7 primers are located 80 to 180 bp 5′ of the coding end, and mbr5-B primer is located 180 bp 3′ of mbr cluster 3. One μL of each of the 7 amplifications was used for the double-nested secondary PCR with the corresponding D1N1 to D7N1 primer and the mbrN1-B primer, with the same conditions as for the primary PCR (annealing temperature: 61°C). D1-7N1 primers are located 55 to 110 bp 5′ of the coding end, and mbrN1-B primer is located 120 bp 3′ of cluster 3. Of 45 samples that were positive for the direct junction, 5 remained negative for the reciprocal junction (not shown). In 1 sample, this is due to the presence of an unusual break 3′ of our mbr primers. For the 4 remaining samples, we subsequently tried several other primer combinations, including further 3′ mbr primers and a VH consensus primer,19 but none of the combinations resulted in the amplification of the reciprocal breakpoint.
Sequencing was performed directly from the PCR products, using the RPN2438 Thermo Sequenase kit (Amersham/Pharmacia/Biotech) and a LI-COR DNA Sequencer 4000 (MWG-Biotech) under conditions recommended by the manufacturer. Direct breakpoints were sequenced with the use of an IRD-800 labeled JHcoB primer, and reciprocal breakpoints were sequenced with the use of an IRD-800 labeled mbrN1-B primer.
Primers (5′ to 3′)
JH primers.
JHCo-B: ACCTGAGGAGACGGTGACC; JHex-B: GGACTCACCTGAGGAGAC.
mbr primers.
mbr6-A: CCAGCAGATTCAAATCTATGGT; mbr7-A: GAGTTGCTTTACGTGGCCTGTT; mbr5-B:GGAGGATCTTACCACGTGGAG; mbrN1-B: GGATAGCAGCACAGGATTGG.
DH primary.
D1: GGCCTCGGTCTCTGTGGGTG; D2: GTACAGCACTGGGCTCAGAG; D3: TGAGAGCGCTGGGCCCACAG; D4: CTGAGATCCCCAGGACGCAG; D5: TGGGAAGCTCCTCCTGACAG; D6: TTCCAGACACCAGACAGAGG; D7: ACATCAGCCCCCAGCCCCAC.
DH secondary.
D1N1: CACCCAGGAGGCCCCAGAG; D2N1: TGCACAGTCTCAGCAGGAG; D3N1: GACATCCCGGGTTTCCCCAG; D4N1: GACGCCTGGACCAGGGCCTG; D5N1: CCCGCCTCCAGTTCCAGGTG; D6N1: TGAGCCCAGCAAGGGAAGG; D7N1: AGGCCCCCTACCAGCCGCAG.
Statistics
T nucleotides are defined as short sequences in the breakpoint insertions, which present enough sequence identity with adjacent flanking sequences to exclude their concomitant presence by chance alone. The significance of each T-nucleotide observation in each sample was estimated with the use of a binomial test. If we consider as an approximation that each of the 4 bases has an equiprobability of representation, the “null probability” (ie, the probability to find a given sequence of length “h” by chance) is P0 = (1/4)h. T nucleotides are found by searching all possible sequences of length h in 1 given breakpoint de novo insertion (of length “n”) and attempting to match them to homologous sequences in adjacent flanking regions (of length “N”) in both direct and reverse-complement orientations. A T nucleotide observed in 1 of the breakpoint de novo insertion (n1) is either homologous to a sequence in the adjacent mbr, DH, or JH flanking sequences, or to a sequence in the other breakpoint de novo insertion of the same sample (n2). In the former case, N corresponds to the total length of the adjacent sequences looked at (∼ 200 nucleotides) and in the latter case N = n2. We only considered sequences of length h of at least 5 in breakpoint de novo insertions of length n of at least 5. The expected number of perfect matches occurring by chance is: e0 = P0 × (n + 1 − h) × 2 × N. In case of homologous but not identical sequences, the null probability to find a given sequence h with “m” mismatches (and h − m identities) is: Pm = [h!/{m! × (h − m)!}] × (1/4)(h − m) × (3/4)m. In this case, the expected number of matches occurring by chance is: em = Pm × (n + 1 − h) × 2 × N. The significance of the T-nucleotide observation is then calculated using a test statistic: Z = (observed − expected)/SDe, where “observed” is the number of copies of a given T nucleotide in a given sample, expected is e0 or em, and SDe is the standard deviation of the expected number calculated according to SDe = [e × (1 − Pe)]. In some samples, T nucleotides are observed in N, n1, and n2 with or without mismatches. In case of perfect matches, the only term changing in the test statistic formula is “observed” (observed = 2). In presence of mismatches, the 2 expected values em1 and em2, and their corresponding SDem are different. The test statistic is then calculated according toZ = (2 − Σem)/ΣSDem. Finally, the P value is calculated by comparing the Zvalue to a standard Normal (Gaussian) distribution (mean = 0, SD = 1). A Z value of at least 1.96 corresponds to aP value of no more than .05, and indicates that the T-nucleotide observation is significantly different from chance.
Results
Distinct mechanisms generate the breaks at the Ig and mbr loci
Sequence libraries obtained for the direct (mbr/JH) and reciprocal (DH/mbr) breakpoints of 40 t(14;18) FL samples are represented in Tables 1 and2, respectively. To investigate which mechanisms generate breaks at the Ig and mbr loci, we analyzed and compared the nucleotide processing of DH/JH coding ends and mbr 3′/5′ ends. As previously described, inspection of DH and JH coding ends confirmed a coding end processing typical of V(D)J recombination: The coding ends involved in the breakpoints are compatible with an initial break initiated at the precise RSS-coding end border, followed by subsequent coding end processing (Table 1, JH sequences, and Table 2, DH sequences). In agreement with normal human DJH junctions, numerous deletions and virtual absence of P regions were observed.19The only atypical observation is sample #38, in which the reciprocal junction consists of a fusion between the JH6 RSS spacer and the 3′ mbr (Table 2). Because the corresponding direct junction is prototypical and uses JH6 (Table 1), this observation suggests that illegitimate recombination might have occurred during an open-and-shut break.
Sequence library of the direct (mbr/JH) junctions
| Clone* . | mbr† . | S‡ . | BCL-2 mbr sequence1-153 . | De novo nucleotide additions (D regions)1-155 . | JH sequence . | JH . | S . |
|---|---|---|---|---|---|---|---|
| Germline | 1a | ACGTGGCCTGTTTCAACACAGACCCACCCAGAGC | ATTACTACTACTACTACGGTATGG | JH6b | |||
| Germline | 1b | CCTCCTGCCCTCCTTCCGCGGGG | ATTACTACTACTACTACTACATGG | JH6c | |||
| Germline | 2a | GCTTTCTCATGGCTGTCCTTCAGGGTCTTCCTGAAATG | ACAACTGGTTCGACCCCTGG | JH5 | |||
| Germline | 2b | CAGTGGTCGTTACGCTCC | ACTACTTTGACTACTGG | JH4 | |||
| Germline | 3a | ACCAAGAAAGCAGGAAACCTGTGGTATGAAGC | TGATGCTTTTGATATCTGG | JH3 | |||
| Germline | 3b | CAGACCTCCCCGGCGGGCCTCAGGGAACAGAATGATCAGAC | |||||
| #1*+ | 1a | −11 | ACGTGGCCTGTTTCAACACAGACCC | GGCTTCCTAGGGGTCCGG | ACTACTACTACTACGGTATGG | JH6 | −3 |
| #2*+ | 1b | −8 | CCTCCTGCC | TCGCGGGGACCAGGAGTTGAGTCCCGAAGG | TTGACTACTGGGGCCAAGG | JH4 | −6 |
| #3*+ | 1b | −7 | CCTCCTGCCC | CAAGTAGGGAGTCAGGG | TACTGGGGCCAAGG | JH4 | −11 |
| #4*+ | 1b | +34 | CCTCCTGCCCTCCT | AGGGCTGCCCAGACGAAA | ACTACTACTACTACGGTATGG | JH6 | −3 |
| #5* | 1b | −2 | CCTCCTGCCCTCCTTC | AGTGAGGATT{TCACGGTAG} | ACTACTACTACTACGGTATGG | D4-17/JH6 | −3/−3 |
| #6*+ | 1b | +1 | CCTCCTGCCCTCCTTCC | ACACCAACTC | ACTACTACTACGGTATGG | JH6 | −6 |
| #7 | 1b | +2 | CCTCCTGCCCTCCTTCC | T | CTACGGTATGG | JH6 | −13 |
| #8*+ | 1b | −2 | CCTCCTGCCCTCCTTCC | AAGAAGA | GG | JH6 | −22 |
| #9*+ | 1b | +4 | CCTCCTGCCCTCCTTCCGC | CCCACTTTCCGGATG | ACTACTACTACGGTATGG | JH6 | −6 |
| #10+ | 1b | 0 | CCTCCTGCCCTCCTTCCGC | CGATAAA | TACTACTACGGTATGG | JH6 | −8 |
| #11*+ | 1b | 0 | CCTCCTGCCCTCCTTCCGC | CGGACGTCTAGGA | ACTACTGG | JH4 | −9 |
| #12*+ | 1b | +1 | CCTCCTGCCCTCCTTCCGCGG | TTCGCACACATCCAGGGGAGG | ATTACTACTACTACTACGGTATGG | JH6 | 0 |
| #13*+ | 1b | +4 | CCTCCTGCCCTCCTTCCGCGG | AT | CTACGGTATGG | JH6 | −13 |
| #14* | 2a | −9 | GC | C | CTACTACGGTATGG | JH6 | −10 |
| #15 | 2a | +8 | TGTCCTTCAGGGTCTTCCT | TCAGAAGTAGTTTCCC | CTACTTTGACTACTGG | JH4 | −1 |
| #16*+ | 2b | −11 | CA | TCTCGCACCGGG | GG | JH? | |
| #17*+ | 2b | −5 | CAGTGGTCGT | CCCCTTT | tACTACTTTGACTACTACTGGGGC | JH4 | P + 1 |
| #18*+ | 2b | +17 | CAGTGGTCGTT | GAG | CTACTACTACTACTACATGG | JH6c | −4 |
| #19 | 2b | −14 | CAGTGGTCGTT | TTTAGGGCTCGTAGGCCTGAAAAAAC{GTATTACGATTTTTGGAGTGGTTATTAT} CC | ATTACTACTACTACTACTACATGG | D3-3/JH6c | 0/0 |
| #20 | 2b | P + 1 | CAGTGGTCGTTAt | CAGGTAGGGGG | CGGTATGG | JH6 | −16 |
| #21 | 2b | −7 | CAGTGGTCGTTA | AAGCCCGCACGGGCG | CTACGGTATGG | JH6 | −13 |
| #22*+ | 2b | −2 | CAGTGGTCGTTA | GGGTGTGGGGG | CTGG | JH? | |
| #23*+ | 2b | −4 | CAGTGGTCGTTA | TTGGCGTAGGTTCAACGGCCACCCCTCCGAAACCCG | CTACTACTACTACGGTATGG | JH6 | −4 |
| #24 | 2b | 0 | CAGTGGTCGTTA | GGGCTTAACTTCTACGGCATGGGC | ACGTCTGG | JH6 | −24 |
| #25*+ | 2b | −2 | CAGTGGTCGTTA | GAAAGG | AACTGGTTCGACCCCTGG | JH5 | −2 |
| #26*+ | 2b | 0 | CAGTGGTCGTTA | A | AACCCTGGTCACCGTCTC | JH4/5 | |
| #27*+ | 2b | +1 | CAGTGGTCGTTA | TTGTGTCCATAAAGCCCTATCTGA | TACTACTACTACGGTATGG | JH6 | −5 |
| #28*+ | 2b | 0 | CAGTGGTCGTTAC | CACCAAAGGAGGAA{GAATTACTATGGTTCAGGGAGTTATT}CGGT | TACTACTACGGTATGG | D3-10/JH6 | −5/−8 |
| #29 | 2b | −3 | CAGTGGTCGTTAC | TCCCCCTTCTCGGCAAGTGAA | TACTACTACTACGGTATGG | JH6 | −5 |
| #30*+ | 2b | −2 | CAGTGGTCGTTAC | AGACCGAGGGCCC | CTACATGG | JH6c | −16 |
| #31*+ | 3a | −10 | ACCAAGAAAG | AATCCGAATGG | ACTACTACTACTACGGTATGG | JH6 | −3 |
| #32*+ | 3a | −6 | ACCAAGAAAGCAG | CATCTCCGAAG | ACAACTGGTTCGACCCCTGG | JH5 | 0 |
| #33* | 3a | −2 | ACCAAGAAAGCAGG | C{GGTGTTATGACTAC} | ATGG | D3-22/JH6 | 0/−20 |
| #34+ | 3a | +18 | ACCAAGAAAGCAGGAA | TGAGGCGGTGCGGGGGGCAGGA | TGGTTCGACCCCTGG | JH5 | −5 |
| #35*+ | 3b | −12 | CA | CGGCCCTTTAGGATCCCCCATTGGTTC | CTACTACTACTACTACATGG | JH6c | −4 |
| #36 | 3b | −22 | CAGACCTCCC | TGG | ACGGTATGG | JH6 | −15 |
| #37 | 3b | 0 | CAGACCTCCCC | CTTGCTTGCCGAC | ATTACTACTACTACTACGGTATGG | JH6 | 0 |
| #38*+ | 3b | +4 | CAGACCTCCCCGGCGG | TTCCCCGGGACCCCTGAGATCAAGG | ACTACTACTACGGTATGG | JH6 | −6 |
| #39*+ | 3b | −13 | CAGACCTCCCCGGCGG | TAAAGGAAA | TGACTACTGG | JH4 | −7 |
| #40 | 3b | +56 | CAGACCTCCCCGGCGGGCCTCAGGGAACAGAAT | TAGGGACACCCACCAAATACTAGGGCATCAACCGATACCCCGGGAAGA | GTATGG | JH6 | −18 |
| Clone* . | mbr† . | S‡ . | BCL-2 mbr sequence1-153 . | De novo nucleotide additions (D regions)1-155 . | JH sequence . | JH . | S . |
|---|---|---|---|---|---|---|---|
| Germline | 1a | ACGTGGCCTGTTTCAACACAGACCCACCCAGAGC | ATTACTACTACTACTACGGTATGG | JH6b | |||
| Germline | 1b | CCTCCTGCCCTCCTTCCGCGGGG | ATTACTACTACTACTACTACATGG | JH6c | |||
| Germline | 2a | GCTTTCTCATGGCTGTCCTTCAGGGTCTTCCTGAAATG | ACAACTGGTTCGACCCCTGG | JH5 | |||
| Germline | 2b | CAGTGGTCGTTACGCTCC | ACTACTTTGACTACTGG | JH4 | |||
| Germline | 3a | ACCAAGAAAGCAGGAAACCTGTGGTATGAAGC | TGATGCTTTTGATATCTGG | JH3 | |||
| Germline | 3b | CAGACCTCCCCGGCGGGCCTCAGGGAACAGAATGATCAGAC | |||||
| #1*+ | 1a | −11 | ACGTGGCCTGTTTCAACACAGACCC | GGCTTCCTAGGGGTCCGG | ACTACTACTACTACGGTATGG | JH6 | −3 |
| #2*+ | 1b | −8 | CCTCCTGCC | TCGCGGGGACCAGGAGTTGAGTCCCGAAGG | TTGACTACTGGGGCCAAGG | JH4 | −6 |
| #3*+ | 1b | −7 | CCTCCTGCCC | CAAGTAGGGAGTCAGGG | TACTGGGGCCAAGG | JH4 | −11 |
| #4*+ | 1b | +34 | CCTCCTGCCCTCCT | AGGGCTGCCCAGACGAAA | ACTACTACTACTACGGTATGG | JH6 | −3 |
| #5* | 1b | −2 | CCTCCTGCCCTCCTTC | AGTGAGGATT{TCACGGTAG} | ACTACTACTACTACGGTATGG | D4-17/JH6 | −3/−3 |
| #6*+ | 1b | +1 | CCTCCTGCCCTCCTTCC | ACACCAACTC | ACTACTACTACGGTATGG | JH6 | −6 |
| #7 | 1b | +2 | CCTCCTGCCCTCCTTCC | T | CTACGGTATGG | JH6 | −13 |
| #8*+ | 1b | −2 | CCTCCTGCCCTCCTTCC | AAGAAGA | GG | JH6 | −22 |
| #9*+ | 1b | +4 | CCTCCTGCCCTCCTTCCGC | CCCACTTTCCGGATG | ACTACTACTACGGTATGG | JH6 | −6 |
| #10+ | 1b | 0 | CCTCCTGCCCTCCTTCCGC | CGATAAA | TACTACTACGGTATGG | JH6 | −8 |
| #11*+ | 1b | 0 | CCTCCTGCCCTCCTTCCGC | CGGACGTCTAGGA | ACTACTGG | JH4 | −9 |
| #12*+ | 1b | +1 | CCTCCTGCCCTCCTTCCGCGG | TTCGCACACATCCAGGGGAGG | ATTACTACTACTACTACGGTATGG | JH6 | 0 |
| #13*+ | 1b | +4 | CCTCCTGCCCTCCTTCCGCGG | AT | CTACGGTATGG | JH6 | −13 |
| #14* | 2a | −9 | GC | C | CTACTACGGTATGG | JH6 | −10 |
| #15 | 2a | +8 | TGTCCTTCAGGGTCTTCCT | TCAGAAGTAGTTTCCC | CTACTTTGACTACTGG | JH4 | −1 |
| #16*+ | 2b | −11 | CA | TCTCGCACCGGG | GG | JH? | |
| #17*+ | 2b | −5 | CAGTGGTCGT | CCCCTTT | tACTACTTTGACTACTACTGGGGC | JH4 | P + 1 |
| #18*+ | 2b | +17 | CAGTGGTCGTT | GAG | CTACTACTACTACTACATGG | JH6c | −4 |
| #19 | 2b | −14 | CAGTGGTCGTT | TTTAGGGCTCGTAGGCCTGAAAAAAC{GTATTACGATTTTTGGAGTGGTTATTAT} CC | ATTACTACTACTACTACTACATGG | D3-3/JH6c | 0/0 |
| #20 | 2b | P + 1 | CAGTGGTCGTTAt | CAGGTAGGGGG | CGGTATGG | JH6 | −16 |
| #21 | 2b | −7 | CAGTGGTCGTTA | AAGCCCGCACGGGCG | CTACGGTATGG | JH6 | −13 |
| #22*+ | 2b | −2 | CAGTGGTCGTTA | GGGTGTGGGGG | CTGG | JH? | |
| #23*+ | 2b | −4 | CAGTGGTCGTTA | TTGGCGTAGGTTCAACGGCCACCCCTCCGAAACCCG | CTACTACTACTACGGTATGG | JH6 | −4 |
| #24 | 2b | 0 | CAGTGGTCGTTA | GGGCTTAACTTCTACGGCATGGGC | ACGTCTGG | JH6 | −24 |
| #25*+ | 2b | −2 | CAGTGGTCGTTA | GAAAGG | AACTGGTTCGACCCCTGG | JH5 | −2 |
| #26*+ | 2b | 0 | CAGTGGTCGTTA | A | AACCCTGGTCACCGTCTC | JH4/5 | |
| #27*+ | 2b | +1 | CAGTGGTCGTTA | TTGTGTCCATAAAGCCCTATCTGA | TACTACTACTACGGTATGG | JH6 | −5 |
| #28*+ | 2b | 0 | CAGTGGTCGTTAC | CACCAAAGGAGGAA{GAATTACTATGGTTCAGGGAGTTATT}CGGT | TACTACTACGGTATGG | D3-10/JH6 | −5/−8 |
| #29 | 2b | −3 | CAGTGGTCGTTAC | TCCCCCTTCTCGGCAAGTGAA | TACTACTACTACGGTATGG | JH6 | −5 |
| #30*+ | 2b | −2 | CAGTGGTCGTTAC | AGACCGAGGGCCC | CTACATGG | JH6c | −16 |
| #31*+ | 3a | −10 | ACCAAGAAAG | AATCCGAATGG | ACTACTACTACTACGGTATGG | JH6 | −3 |
| #32*+ | 3a | −6 | ACCAAGAAAGCAG | CATCTCCGAAG | ACAACTGGTTCGACCCCTGG | JH5 | 0 |
| #33* | 3a | −2 | ACCAAGAAAGCAGG | C{GGTGTTATGACTAC} | ATGG | D3-22/JH6 | 0/−20 |
| #34+ | 3a | +18 | ACCAAGAAAGCAGGAA | TGAGGCGGTGCGGGGGGCAGGA | TGGTTCGACCCCTGG | JH5 | −5 |
| #35*+ | 3b | −12 | CA | CGGCCCTTTAGGATCCCCCATTGGTTC | CTACTACTACTACTACATGG | JH6c | −4 |
| #36 | 3b | −22 | CAGACCTCCC | TGG | ACGGTATGG | JH6 | −15 |
| #37 | 3b | 0 | CAGACCTCCCC | CTTGCTTGCCGAC | ATTACTACTACTACTACGGTATGG | JH6 | 0 |
| #38*+ | 3b | +4 | CAGACCTCCCCGGCGG | TTCCCCGGGACCCCTGAGATCAAGG | ACTACTACTACGGTATGG | JH6 | −6 |
| #39*+ | 3b | −13 | CAGACCTCCCCGGCGG | TAAAGGAAA | TGACTACTGG | JH4 | −7 |
| #40 | 3b | +56 | CAGACCTCCCCGGCGGGCCTCAGGGAACAGAAT | TAGGGACACCCACCAAATACTAGGGCATCAACCGATACCCCGGGAAGA | GTATGG | JH6 | −18 |
Samples marked by an asterisk (*) were PCR-amplified and sequenced twice. Samples marked by a plus (+) were amplified with a 4:1 proportion of Taq DNA polymerase and Vent high-fidelity DNA polymerase.
For practical purposes, the mbr sequence was broken into 6 immediately adjacent pieces (1a to 3b). Clusters 1, 2, and 3 are underlined.
S: status of coding end or mbr end processing (0 = precise, −n = deletion, +n = duplication, P + n = P nucleotide).
P nucleotides are shown in italics.
Sequence library of the reciprocal (DH/mbr) junctions
| Clone* . | DH . | S† . | DH sequence . | De novo nucleotide additions‡ . | BCL-2 mbr sequence2-153 . | mbr2-155 . | S . |
|---|---|---|---|---|---|---|---|
| Germline | D1-7 | GGTATAACTGGAACTAC | ACGTGGCCTGTTTCAACACAGACCCACCCAGAGC | 1a | |||
| Germline | D1-26 | GGTATAGTGGGAGCTACTAC | CCTCCTGCCCTCCTTCCGCGGGG | 1b | |||
| Germline | D2-2 | AGGATATTGTAGTAGTACCAGCTGCTATACC | GCTTTCTCATGGCTGTCCTTCAGGGTCTTCCTGAAATG | 2a | |||
| Germline | D2-8 | AGGATATTGTACTAATGGTGTATGCTATACC | CAGTGGTCGTTACGCTCC | 2b | |||
| Germline | D2-15 | AGGATATTGTAGTGGTGGTAGCTGCTACTCC | ACCAAGAAAGCAGGAAACCTGTGGTATGAAGC | 3a | |||
| Germline | D2-21 | AGCATATTGTGGTGGTGACTGCTATTCC | CAGACCTCCCCGGCGGGCCTCAGGGAACAGAATGATCAGAC | 3b | |||
| Germline | D3-3 | GTATTACGATTTTTGGAGTGGTTATTATACC | |||||
| Germline | D3-9 | GTATTACGATATTTTGACTGGTTATTATAAC | |||||
| Germline | D3-10 | GTATTACTATGGTTCGGGGAGTTATTATAAC | |||||
| Germline | D3-16 | GTATTATGATTACGTTTGGGGGAGTTATCGTTATACC | |||||
| Germline | D3-22 | GTATTACTATGATAGTAGTGGTTATTACTAC | |||||
| Germline | D4-4 | TGACTACAGTAACTAC | |||||
| Germline | D4-17 | TGACTACGGTGACTAC | |||||
| Germline | D5-5/18 | GTGGATACAGCTATGGTTAC | |||||
| Germline | D7-27 | CTAACTGGGGA | |||||
| #1 | D3-3 | −4 | GTATTACGATTTTTGGAGTGGTTATTA | ACT | TCCTGCCCTCCTTCCGCGGGG | 1b | −11 |
| #2*+ | D3-22 | 0 | TTACTATGATAGTAGTGGTTATTACTAC | TGTGGGTTGA | GCAGGG | 1b | −8 |
| #3*+ | D2-2 | −6 | AGGATATTGTAGTAGTACCAGCTGC | CCACCGT | GCGGGG | 1b | −7 |
| #4+ | D5-5/18 | −5 | GTGGATACAGCTATG | AGGTGAAAAACCACCCCCCAAG | AACACAGACCCACCCAGAGC | 1a | +34 |
| #5 | D2-2 | −9 | AGGATATTGTAGTAGTACCAGC | CCAGCCTTC | CGGGG | 1b | −2 |
| #6*+ | D3-3 | −8 | GTATTACGATTTTTGGAGTGGTT | TAACCAACTCT | CGCGGGG | 1b | +1 |
| #7* | D7-27 | 0 | CTAACTGGGGA | CACCCTTACCTATA | CCGCGGGG | 1b | +2 |
| #8+ | D3-9 | −6 | GTATTACGATATTTTGACTGGTTAT | AAGATCCAGG | GGGG | 1b | −2 |
| #9*+ | D2-2 | −5 | AGGATATTGTAGTAGTACCAGCTGCT | TCCGACCCCTGTGATCTAC | CCGCGGGG | 1b | +4 |
| #10*+ | D5-5/18 | −2 | GTGGATACAGCTATGGTT | CCTTTTCGGTGGCCAACCGACAG | GGGG | 1b | 0 |
| #11 | D3-10 | −12 | GTATTACTATGGTTCGGGG | GGGG | 1b | 0 | |
| #12 | D3-3 | −3 | GTATTACGATTTTTGGAGTGGTTATTAT | TGTTCGGCCAAATAC | GGG | 1b | +1 |
| #13+ | D3-9 | −7 | GTATTACGATATTTTGACTGGTTA | GCAATTCGGGGATTGGTAATGAGAAA | GCGGGG | 1b | +4 |
| #14 | D3-3 | −7 | GTATTACGATTTTTGGAGTGGTTA | CCTGGCGCCGCTATTCGGTAGGCGGACCCAAAAAGATAAGGGCCCCGACGAGTTTGCATGA | GCTGTCCTTCAGGGTCTTCCTGAAATG | 2a | −9 |
| #15 | D4-4 | −7 | TGACTACAG | GTCTTCCTGAAATG | 2a | +8 | |
| #16 | D5-5/18 | −1 | GTGGATACAGCTATGGTTA | TTGGCCT | GCTCC | 2b | −11 |
| #17* | D2-8 | −3 | AGGATATTGTACTAATGGTGTATGCTAT | GTAGGGG | TCC | 2b | −5 |
| #18 | D2-15 | −1 | ATTGTAGTGGTGGTAGCTGCTACTC | GTGTACTACTCTTGGGGGGGC | GAAATG | 2a | +17 |
| #19 | D2-2 | −12 | AGGATATTGTAGTAGTACC | GGGTCGGTAGGG | AAGCAGGAAACCTGTGGTATGAAGC | 3a | −14 |
| #20 | D4-17 | −1 | TGACTACGGTGACTA | GCCTCCCTGCCATACCAACGCCCTTC | CGCTCC | 2b | 0 |
| #21 | D2-15 | −19 | AGGATATTGTAG | CCCAAGCATAGGGTG | CCAAGAAAGCAGGAAACCTGTGGTAT | 3a | −7 |
| #22*+ | D2-21 | −10 | AGCATATTGTGGTGGTGA | TTTGAAGCAGGGGGGCTT | CTCC | 2b | −2 |
| #23+ | D3-3 | −4 | GTATTACGATTTTTGGAGTGGTTATTA | CGGGTCGGTCCGAACCGCAACAGGGGGTTCTTTGTC | CC | 2b | −4 |
| #24 | D2-2 | −11 | AGGATATTGTAGTAGTACCA | CCG | CGCTCC | 2b | 0 |
| #25+ | D1-26 | −5 | GGTATAGTGGGAGCT | CTACAAGGGGACTCTCCC | CTCC | 2b | −2 |
| #26+ | D4-4 | 0 | TGACTACAGTAACTAC | TTTC | CGCTCC | 2b | 0 |
| #27+ | D2-2 | −4 | AGGATATTGTAGTAGTACCAGCTGCTA | CCCAACCCCATG | ACGCTCC | 2b | +1 |
| #28 | D3-3 | −32 | cactgt | TTCGGAAGTTGTGCCAACGACCACA | GCTCC | 2b | 0 |
| #29 | D3-10 | −6 | GTATTACTATGGTTCGGGGAGTTAT | CGTGTGGGTTTTGGGAGACGCAGTCCC | CC | 2b | −3 |
| #30+ | D1-7 | 0 | GGTATAACTGGAACTAC | CTCACCTTCC | TCC | 2b | −2 |
| #31+ | D3-9 | −6 | GTATTACGATATTTTGACTGGTTAT | AACCTCTTAAG | GTGGTATGAAGC | 3a | −10 |
| #32 | D2-2 | −3 | AGGATATTGTAGTAGTACCAGCTGCTAT | TTGTCTTTGTTACCTCTTTCATTTT | TGTGGTATGAAGC | 3a | −6 |
| #33*+ | D2-2 | −8 | AGGATATTGTAGTAGTACCAGCT | TCGGTCC | ACCTGTGGTATGAAGC | 3a | −2 |
| #34 | D2-2 | −8 | AGGATATTGTAGTAGTACCAGCT | CC | 2b | +18 | |
| #35*+ | D3-3 | −6 | GTATTACGATTTTTGGAGTGGTTAT | CAGGGTAG | GGGCCTCAGGGAACAGAATGATCAGAC | 3b | −12 |
| #36 | D3-3 | −3 | GTATTACGATTTTTGGAGTGGTTATTAT | TTCGATCACGCCGG | TGATCAGAC | 3b | −22 |
| #37 | D3-16 | −3 | TTACGTTTGGGGGAGTTATCGTTAT | GTGGTCCCTTCCCGCTATA | GGCGGGCCTCAGGGAACAGAATGATCAG | 3b | 0 |
| #38*+ | JH6sp | ggtttttgtggggtgaggatggaca | CCCTACAACGCGCGCACAAGACGT | GCGGGCCTCAGGGAACAGAATGATCAG | 3b | +4 | |
| #39 | D3-9 | −7 | GTATTACGATATTTTGACTGGTTA | CGGGCTTCC | GAATGATCAGAC | 3b | −13 |
| #40 | D3-10 | −6 | GTATTACTATGGTTCGGGGAGTTAT | GTTTG | GCAGGAAACCTGTGGTATGAAGC | 3a | +56 |
| Clone* . | DH . | S† . | DH sequence . | De novo nucleotide additions‡ . | BCL-2 mbr sequence2-153 . | mbr2-155 . | S . |
|---|---|---|---|---|---|---|---|
| Germline | D1-7 | GGTATAACTGGAACTAC | ACGTGGCCTGTTTCAACACAGACCCACCCAGAGC | 1a | |||
| Germline | D1-26 | GGTATAGTGGGAGCTACTAC | CCTCCTGCCCTCCTTCCGCGGGG | 1b | |||
| Germline | D2-2 | AGGATATTGTAGTAGTACCAGCTGCTATACC | GCTTTCTCATGGCTGTCCTTCAGGGTCTTCCTGAAATG | 2a | |||
| Germline | D2-8 | AGGATATTGTACTAATGGTGTATGCTATACC | CAGTGGTCGTTACGCTCC | 2b | |||
| Germline | D2-15 | AGGATATTGTAGTGGTGGTAGCTGCTACTCC | ACCAAGAAAGCAGGAAACCTGTGGTATGAAGC | 3a | |||
| Germline | D2-21 | AGCATATTGTGGTGGTGACTGCTATTCC | CAGACCTCCCCGGCGGGCCTCAGGGAACAGAATGATCAGAC | 3b | |||
| Germline | D3-3 | GTATTACGATTTTTGGAGTGGTTATTATACC | |||||
| Germline | D3-9 | GTATTACGATATTTTGACTGGTTATTATAAC | |||||
| Germline | D3-10 | GTATTACTATGGTTCGGGGAGTTATTATAAC | |||||
| Germline | D3-16 | GTATTATGATTACGTTTGGGGGAGTTATCGTTATACC | |||||
| Germline | D3-22 | GTATTACTATGATAGTAGTGGTTATTACTAC | |||||
| Germline | D4-4 | TGACTACAGTAACTAC | |||||
| Germline | D4-17 | TGACTACGGTGACTAC | |||||
| Germline | D5-5/18 | GTGGATACAGCTATGGTTAC | |||||
| Germline | D7-27 | CTAACTGGGGA | |||||
| #1 | D3-3 | −4 | GTATTACGATTTTTGGAGTGGTTATTA | ACT | TCCTGCCCTCCTTCCGCGGGG | 1b | −11 |
| #2*+ | D3-22 | 0 | TTACTATGATAGTAGTGGTTATTACTAC | TGTGGGTTGA | GCAGGG | 1b | −8 |
| #3*+ | D2-2 | −6 | AGGATATTGTAGTAGTACCAGCTGC | CCACCGT | GCGGGG | 1b | −7 |
| #4+ | D5-5/18 | −5 | GTGGATACAGCTATG | AGGTGAAAAACCACCCCCCAAG | AACACAGACCCACCCAGAGC | 1a | +34 |
| #5 | D2-2 | −9 | AGGATATTGTAGTAGTACCAGC | CCAGCCTTC | CGGGG | 1b | −2 |
| #6*+ | D3-3 | −8 | GTATTACGATTTTTGGAGTGGTT | TAACCAACTCT | CGCGGGG | 1b | +1 |
| #7* | D7-27 | 0 | CTAACTGGGGA | CACCCTTACCTATA | CCGCGGGG | 1b | +2 |
| #8+ | D3-9 | −6 | GTATTACGATATTTTGACTGGTTAT | AAGATCCAGG | GGGG | 1b | −2 |
| #9*+ | D2-2 | −5 | AGGATATTGTAGTAGTACCAGCTGCT | TCCGACCCCTGTGATCTAC | CCGCGGGG | 1b | +4 |
| #10*+ | D5-5/18 | −2 | GTGGATACAGCTATGGTT | CCTTTTCGGTGGCCAACCGACAG | GGGG | 1b | 0 |
| #11 | D3-10 | −12 | GTATTACTATGGTTCGGGG | GGGG | 1b | 0 | |
| #12 | D3-3 | −3 | GTATTACGATTTTTGGAGTGGTTATTAT | TGTTCGGCCAAATAC | GGG | 1b | +1 |
| #13+ | D3-9 | −7 | GTATTACGATATTTTGACTGGTTA | GCAATTCGGGGATTGGTAATGAGAAA | GCGGGG | 1b | +4 |
| #14 | D3-3 | −7 | GTATTACGATTTTTGGAGTGGTTA | CCTGGCGCCGCTATTCGGTAGGCGGACCCAAAAAGATAAGGGCCCCGACGAGTTTGCATGA | GCTGTCCTTCAGGGTCTTCCTGAAATG | 2a | −9 |
| #15 | D4-4 | −7 | TGACTACAG | GTCTTCCTGAAATG | 2a | +8 | |
| #16 | D5-5/18 | −1 | GTGGATACAGCTATGGTTA | TTGGCCT | GCTCC | 2b | −11 |
| #17* | D2-8 | −3 | AGGATATTGTACTAATGGTGTATGCTAT | GTAGGGG | TCC | 2b | −5 |
| #18 | D2-15 | −1 | ATTGTAGTGGTGGTAGCTGCTACTC | GTGTACTACTCTTGGGGGGGC | GAAATG | 2a | +17 |
| #19 | D2-2 | −12 | AGGATATTGTAGTAGTACC | GGGTCGGTAGGG | AAGCAGGAAACCTGTGGTATGAAGC | 3a | −14 |
| #20 | D4-17 | −1 | TGACTACGGTGACTA | GCCTCCCTGCCATACCAACGCCCTTC | CGCTCC | 2b | 0 |
| #21 | D2-15 | −19 | AGGATATTGTAG | CCCAAGCATAGGGTG | CCAAGAAAGCAGGAAACCTGTGGTAT | 3a | −7 |
| #22*+ | D2-21 | −10 | AGCATATTGTGGTGGTGA | TTTGAAGCAGGGGGGCTT | CTCC | 2b | −2 |
| #23+ | D3-3 | −4 | GTATTACGATTTTTGGAGTGGTTATTA | CGGGTCGGTCCGAACCGCAACAGGGGGTTCTTTGTC | CC | 2b | −4 |
| #24 | D2-2 | −11 | AGGATATTGTAGTAGTACCA | CCG | CGCTCC | 2b | 0 |
| #25+ | D1-26 | −5 | GGTATAGTGGGAGCT | CTACAAGGGGACTCTCCC | CTCC | 2b | −2 |
| #26+ | D4-4 | 0 | TGACTACAGTAACTAC | TTTC | CGCTCC | 2b | 0 |
| #27+ | D2-2 | −4 | AGGATATTGTAGTAGTACCAGCTGCTA | CCCAACCCCATG | ACGCTCC | 2b | +1 |
| #28 | D3-3 | −32 | cactgt | TTCGGAAGTTGTGCCAACGACCACA | GCTCC | 2b | 0 |
| #29 | D3-10 | −6 | GTATTACTATGGTTCGGGGAGTTAT | CGTGTGGGTTTTGGGAGACGCAGTCCC | CC | 2b | −3 |
| #30+ | D1-7 | 0 | GGTATAACTGGAACTAC | CTCACCTTCC | TCC | 2b | −2 |
| #31+ | D3-9 | −6 | GTATTACGATATTTTGACTGGTTAT | AACCTCTTAAG | GTGGTATGAAGC | 3a | −10 |
| #32 | D2-2 | −3 | AGGATATTGTAGTAGTACCAGCTGCTAT | TTGTCTTTGTTACCTCTTTCATTTT | TGTGGTATGAAGC | 3a | −6 |
| #33*+ | D2-2 | −8 | AGGATATTGTAGTAGTACCAGCT | TCGGTCC | ACCTGTGGTATGAAGC | 3a | −2 |
| #34 | D2-2 | −8 | AGGATATTGTAGTAGTACCAGCT | CC | 2b | +18 | |
| #35*+ | D3-3 | −6 | GTATTACGATTTTTGGAGTGGTTAT | CAGGGTAG | GGGCCTCAGGGAACAGAATGATCAGAC | 3b | −12 |
| #36 | D3-3 | −3 | GTATTACGATTTTTGGAGTGGTTATTAT | TTCGATCACGCCGG | TGATCAGAC | 3b | −22 |
| #37 | D3-16 | −3 | TTACGTTTGGGGGAGTTATCGTTAT | GTGGTCCCTTCCCGCTATA | GGCGGGCCTCAGGGAACAGAATGATCAG | 3b | 0 |
| #38*+ | JH6sp | ggtttttgtggggtgaggatggaca | CCCTACAACGCGCGCACAAGACGT | GCGGGCCTCAGGGAACAGAATGATCAG | 3b | +4 | |
| #39 | D3-9 | −7 | GTATTACGATATTTTGACTGGTTA | CGGGCTTCC | GAATGATCAGAC | 3b | −13 |
| #40 | D3-10 | −6 | GTATTACTATGGTTCGGGGAGTTAT | GTTTG | GCAGGAAACCTGTGGTATGAAGC | 3a | +56 |
Samples marked with an asterisk (*) were PCR-amplified and sequenced twice. Samples marked by a plus (+) were amplified with a 4:1 proportion of Taq DNA polymerase and Vent high-fidelity DNA polymerase.
S: status of coding end or mbr end processing (0 = precise, −n = deletion, +n = duplication, P + n = P nucleotide).
De novo nucleotide additions are represented in bold character, DH segments found in the direct breakpoints are shown between brackets. Mutations in the DH sequences are underlined. These sequence data are available from Genbank under accession numbers AF147979 to AF148063.
P nucleotides are shown in italics.
For practical purposes, the mbr sequence was broken into 6 immediately adjacent pieces (1a to 3B). Clusters 1, 2, and 3 are underlined.
At the mbr locus, inspection of 5′ and 3′ mbr ends revealed 3 types of breaks (5′ mbr ends, Table 1; 3′ mbr ends Table2): (1) precise: The mbr sequence shows no gain or loss of nucleotide (eg, #10, CCGC↓GGGG). This type represents a small proportion of breakpoints (7 of 40, 17.5%); (2) deletion: A short fragment of the mbr is missing and present neither at the direct nor at the reciprocal breakpoint (eg, #3, GCCC↓TCCTTCC↓GCGG). Deletions range from 2 to 22 bp and represent the majority of the breakpoint types (21 of 40, 52.5%); (3) duplications: A fragment of the mbr is present at both the direct and the reciprocal junctions (eg, #9, CCTTCCGC↓CCGCGGGG). Duplications range from 1 to 56 bp and are also frequent (12 of 40, 30%). Although both deletion and precise ends are compatible with RAG-mediated coding end formation and processing, the generation of duplications is not compatible with the hairpin-intermediate step. RAG-1/2 initial nick takes place at the precise coding-end/RSS border and generates a full-length coding-end hairpin for each partner to be recombined. Duplications are supposedly generated by staggered breaks, consisting of a single-strand nick on opposite DNA strands some nucleotides apart. This indicates that, although very likely responsible for the break and subsequent processing at the Ig locus on chromosome 14, V(D)J recombination is not responsible for the initial breaks at the mbr locus on chromosome 18.
The “de novo nucleotide additions” in the t(14;18) breakpoints show templated insertions
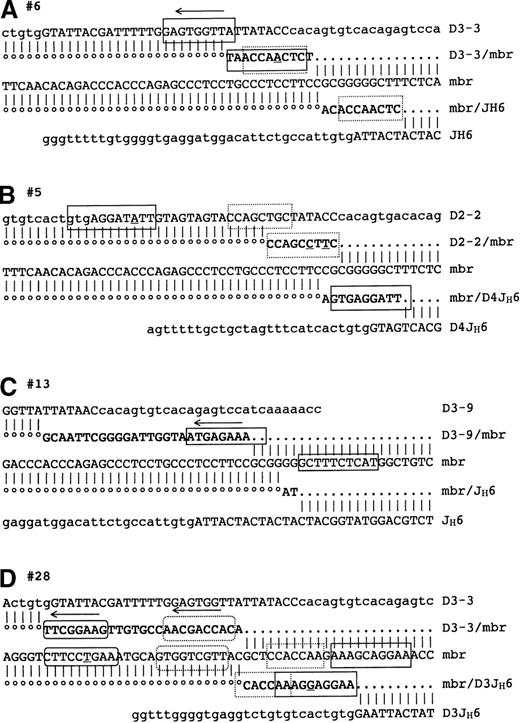
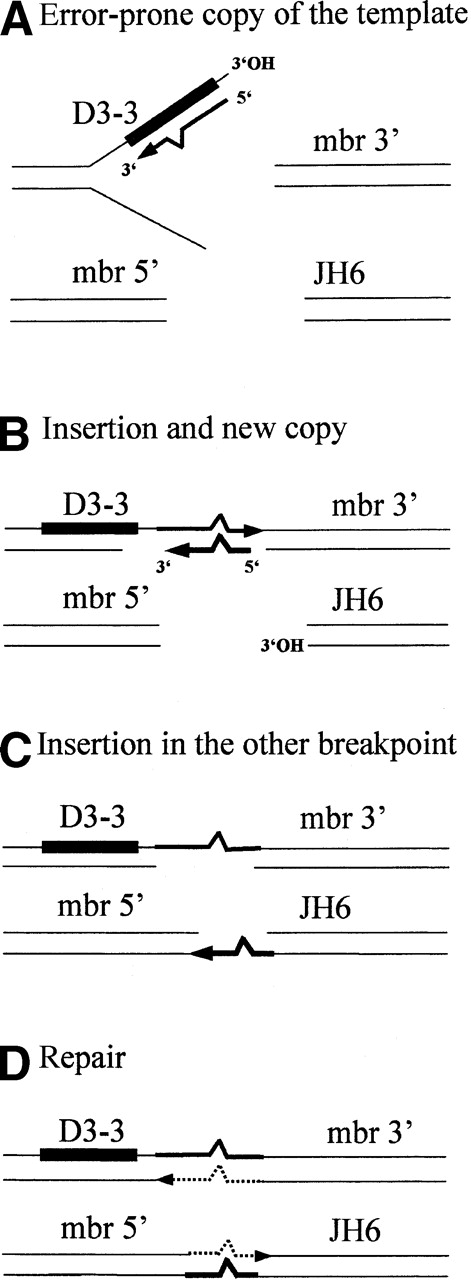
We carried out a detailed analysis of the de novo nucleotide additions present in the direct and reciprocal breakpoints (Tables 1and 2). Figure 1A shows the reciprocal (D3-3/mbr) and direct (mbr/JH6) breakpoint junctions of sample #6 and their homology to the original D3-3, mbr and JH6 genomic sequences. De novo nucleotide additions are shown between the regions of homology. Surprisingly, we found that the insertions at both breakpoints contain an identical sequence: 5′ ACCAACTC 3′ (broken-line boxed sequences). Moreover, the sequence of the D3-3/mbr junction, 5′ TAACCAACTC 3′, is also present as reverse-complement in the adjacent D3-3 genomic sequence, with 1 nucleotide mismatch (A7) (solid-line boxed sequences): 5′ GAG-TGGTTA 3′ on the plus strand, or 5′ TAACCA-CTC 3′ on the minus strand. To exclude the possibility of Taq-polymerase introduced mistakes, those sequences were confirmed by a second PCR amplification and sequencing. Such long stretches of identity in such short sequences are very unlikely to be coincidental (see statistics below). The presence of a similar sequence motif in the D3-3 segment and in both breakpoints implies therefore the occurrence of a templated DNA synthesis. Insertions at the t(14;18) breakpoints were so far interpreted as N-nucleotide additions. However, N regions are nontemplated nucleotides, added to free 3′OH ends (preferentially protruding ends) by the TdT.20-23 Although N-nucleotide synthesis is not completely random, displaying a marked preference for Gs, TdT does not use any template for polymerase extension. It is therefore clear that the nucleotide insertions observed in these breakpoints are not generated by the TdT. Palindromic (P) nucleotides are also frequently found in normal V(D)J junctions. However, P nucleotides and variations thereof24 25 all result from resolution of the hairpin intermediate and, by definition, cannot be present more than once. Furthermore, P nucleotides do not require de novo synthesis and are consequently also devoid of mismatches. It is therefore clear that the templated nucleotides observed here are not derived from the hairpin-opening mechanism generating P nucleotides. Which mechanism could account for the generation of these templated insertions? The multiplicity of “copies” for 1 template together with the occurrence of mismatches in the template/copy pair strongly suggest the presence of a short-patch DNA synthesis consisting of an error-prone copy of a template, followed by its insertion at a breakpoint. This possibility is illustrated in Figure 2: The D3-3 sequence provides a template for an error-prone synthesis (Figure2A), followed by insertion of the copy at the reciprocal breakpoint (Figure 2B). Since the presence of multiple copies containing identical mismatches is more likely issued from vertical than horizontal lineage, this new sequence could in turn provide the template for another copy (Figure 2B) subsequently inserted in the other breakpoint (Figure 2C).
Nucleotide sequence comparison of the germline chromosomes 14 (DH, JH) and 18 (mbr), and direct (mbr/JH) and reciprocal (DH/mbr) breakpoints resulting from the translocation of the indicated sample.
Nucleotide identity is shown by vertical lines and dots. DHand JH segment RSSs are represented in small cases. De novo nucleotide insertions in the breakpoints are shown in bold characters between dots. Regions of homology are boxed. Mismatches are underlined. Arrows indicate a reverse complement orientation relative to the corresponding other boxed sequence(s).
Nucleotide sequence comparison of the germline chromosomes 14 (DH, JH) and 18 (mbr), and direct (mbr/JH) and reciprocal (DH/mbr) breakpoints resulting from the translocation of the indicated sample.
Nucleotide identity is shown by vertical lines and dots. DHand JH segment RSSs are represented in small cases. De novo nucleotide insertions in the breakpoints are shown in bold characters between dots. Regions of homology are boxed. Mismatches are underlined. Arrows indicate a reverse complement orientation relative to the corresponding other boxed sequence(s).
Model of T-nucleotide formation involving a putative error-prone DNA synthesis.
Several ends with 3′OH hydroxyl groups could possibly provide primers for polymerase extension: (1) One of the broken ends, either by a “snap-back” mechanism (eg, step A, intra-strand priming from D3.3 top strand) or by “strand invasion” (eg, step B, priming from JH6, bottom strand); (2) template-directed capture of filler RNA/DNA, annealing in front of the copy.37
Model of T-nucleotide formation involving a putative error-prone DNA synthesis.
Several ends with 3′OH hydroxyl groups could possibly provide primers for polymerase extension: (1) One of the broken ends, either by a “snap-back” mechanism (eg, step A, intra-strand priming from D3.3 top strand) or by “strand invasion” (eg, step B, priming from JH6, bottom strand); (2) template-directed capture of filler RNA/DNA, annealing in front of the copy.37
Templated nucleotides are a general feature of the t(14;18) breakpoint insertions
To see if this observation was an isolated case or a general feature of the t(14;18) breakpoint insertions, we searched our sequence library for similar observations. Strikingly, we found numerous examples in which the de novo additions present in the breakpoints are templated. In sample #5 for example (Figure 1B), the D2-2 11-bp sequence 5′ GTGAGGATATT 3′ is found in the direct breakpoint with 1 nucleotide deletion (solid-line boxes), and the D2-2 neighboring 8-bp sequence 5′ CCAGC-TGC 3′ is found in the reciprocal breakpoint with 2 mismatches (broken-line boxes). In this example, the templated nucleotides constitute the quasi totality of the breakpoint insertions. In sample #13 (Figure 1C), the 10-bp sequence 5′ GCTTTCTCAT 3′ is found in the mbr and in the reciprocal breakpoint in reverse-complement orientation. The origin of 2 of the 10 nucleotides in the reciprocal breakpoint sequence is ambiguous because these nucleotides could either belong to the mbr 3′ end or to the templated insert or both. The presence of a short stretch of homology at both the insert and the mbr ends could in fact provide an anchoring site for the insertion of the copy, a mechanism extensively used during nonhomologous recombination and V(D)J recombination.26 27 Another typical example is sample #28 (Figure 1D), in which 4 immediately adjacent sequences in the mbr are also found in both direct and reciprocal breakpoint inserts. Two of those contiguous mbr sequences (solid- and dash-line rectangular boxes) are found overlapping in the direct breakpoint. One possibility is that those mbr sequences would have constituted a unique template for a long error-prone copy of the sequence 5′ CCACCAAGAAAGCAGGAA 3′, subsequently inserted in the direct breakpoint. Alternatively, those 2 templates could have generated 2 copies (1 from the plus strand and 1 from the minus strand of the mbr) with the AA/TT overlapping doublet providing an anchoring site for a tandem insertion. This last type of “patchwork” insertion displaying the assembly of fragmented pieces is observed in the reciprocal breakpoint of the same sample. Here, the 2 adjacent mbr sequences 5′ CTTCCTGAA 3′ and 5′ GTGGTCGTT 3′ (solid- and dash-line ovoid boxes) are found inserted in reverse-complement orientation, but in inverse order (dash-line ovoid followed by solid-line ovoid, 5′ to 3′, minus strand), excluding the possibility of a single copy.
Most templated nucleotide insertions constitute highly significant observations
We have found many more examples of templated insertions of various length. However, it is clear that the shorter the identity between the sequences, the less obvious the identification and the higher the risk of fortuitous comparisons. To avoid such fortuitous comparisons, the significance of each sequence homology in each sample was estimated using a binomial test as described in the “Materials and methods” section. This test was designed with a conservative approach and is therefore very stringent (ie, more likely to accept than to reject the null hypothesis that the observation is due to chance only). As a reference point, the average length of the breakpoint insertions in this survey is n = 15 nucleotides. Although the “null” probability to find a given sequence of length h = 7 nucleotides by chance is ∼61 × 10−6—in other words, an event happening by chance only once every 16 kb—the calculatedP value is P = .1. The observation is in this case considered not significantly different from chance. Here, only a perfect match of at least 8 nucleotides would be considered as highly significant (P < .0001). For example, the sequence CCAGC-TGC in sample #5 and the sequence CTTCCTGAA in sample #28 have associated P values of .30 and .37, respectively, and are therefore considered not significantly different from chance under this test.
Nevertheless, the occurrence of highly significant templated nucleotides is remarkably high in the breakpoints. Of 67 breakpoint de novo nucleotide insertions (≥ 5 nucleotides), 23 (34%) sequences (≥ 5 nucleotides) presented highly significant identity with adjacent flanking sequences (P ≤ .05, Table3). Overall, this corresponds to 42% of the samples. This figure is probably still an underestimation of the real frequency because of the stringent binomial test applied.
Highly significant templated nucleotides found in the direct and reciprocal breakpoints
| Sample . | Sequence3-150 . | Template . | →3-151 . | Copy 13-152 . | → . | Copy 2 . | P value . |
|---|---|---|---|---|---|---|---|
| #2 | GTGGgTTgA | DH | → | Reciprocal | |||
| GGaGTTGAG | → | Direct | .03 | ||||
| #3 | AAGtAGGGagtcAGGGcTaCTGGG | mbr | → | Direct | <.0001 | ||
| #4 | CCCAGACG | JH | → | Direct | .0006 | ||
| #5 | GTGAGGAT | DH | → | Direct | .001 | ||
| #6 | GAGTtGGTTA | DH | → | Reciprocal | |||
| GAGTTGGT | → | Direct | <.0001 | ||||
| #10 | TTTaTCGG | Direct | ↔ | Reciprocal | <.0001 | ||
| #13 | GCTTTCTCAT | mbr | → | Reciprocal | <.0001 | ||
| #15 | ACTACTTcTGA | JH | → | Direct | <.0001 | ||
| #17 | CCCCTT | JH | → | Direct | |||
| CCCCT | → | Reciprocal | .036 | ||||
| #19 | GGGcTCgGTAGG | Direct | ↔ | Reciprocal | <.0001 | ||
| #21 | AGCCCaaGCAcaGGGcGC | Direct | ↔ | Reciprocal | <.0001 | ||
| #22 | GGGGGCTT | mbr | → | Reciprocal | |||
| GGGGGCT | → | Direct | .01 | ||||
| #23 | CaGGGTCggTCCtGAAccGCA | mbr | → | Reciprocal | |||
| TCCGAAaCCcGC | → | Direct | <.0001 | ||||
| #24 | ACTtCTACGGcATGG | JH | → | Direct | <.0001 | ||
| #28 | GTGGTCGTT | mbr | → | Reciprocal | <.0001 | ||
| CCACCaagAAAGgAGGAA | mbr | → | Direct | <.0001 | |||
| #29 | AAAACCCA | DH | → | Reciprocal | .01 | ||
| #35 | TCAGGGT | mbr | → | Reciprocal | .001 |
| Sample . | Sequence3-150 . | Template . | →3-151 . | Copy 13-152 . | → . | Copy 2 . | P value . |
|---|---|---|---|---|---|---|---|
| #2 | GTGGgTTgA | DH | → | Reciprocal | |||
| GGaGTTGAG | → | Direct | .03 | ||||
| #3 | AAGtAGGGagtcAGGGcTaCTGGG | mbr | → | Direct | <.0001 | ||
| #4 | CCCAGACG | JH | → | Direct | .0006 | ||
| #5 | GTGAGGAT | DH | → | Direct | .001 | ||
| #6 | GAGTtGGTTA | DH | → | Reciprocal | |||
| GAGTTGGT | → | Direct | <.0001 | ||||
| #10 | TTTaTCGG | Direct | ↔ | Reciprocal | <.0001 | ||
| #13 | GCTTTCTCAT | mbr | → | Reciprocal | <.0001 | ||
| #15 | ACTACTTcTGA | JH | → | Direct | <.0001 | ||
| #17 | CCCCTT | JH | → | Direct | |||
| CCCCT | → | Reciprocal | .036 | ||||
| #19 | GGGcTCgGTAGG | Direct | ↔ | Reciprocal | <.0001 | ||
| #21 | AGCCCaaGCAcaGGGcGC | Direct | ↔ | Reciprocal | <.0001 | ||
| #22 | GGGGGCTT | mbr | → | Reciprocal | |||
| GGGGGCT | → | Direct | .01 | ||||
| #23 | CaGGGTCggTCCtGAAccGCA | mbr | → | Reciprocal | |||
| TCCGAAaCCcGC | → | Direct | <.0001 | ||||
| #24 | ACTtCTACGGcATGG | JH | → | Direct | <.0001 | ||
| #28 | GTGGTCGTT | mbr | → | Reciprocal | <.0001 | ||
| CCACCaagAAAGgAGGAA | mbr | → | Direct | <.0001 | |||
| #29 | AAAACCCA | DH | → | Reciprocal | .01 | ||
| #35 | TCAGGGT | mbr | → | Reciprocal | .001 |
Mismatches are represented by small cases. Point mutations are underlined, nucleotide insertions are specified above the line, and deletions are specified below the line. Sequence shown is the one of the copy.
Single arrow indicates the probable template/copy relationship. Double arrow indicates that either sequence could be the template or the copy.
Reciprocal = nucleotide insertions in the reciprocal junction; Direct = nucleotide insertions in the direct junction.
Features of templated nucleotides
Combining the features of all highly significant templated nucleotides in Table 3, we could extract the following as general rules. In most cases, the “template/insert” pair contains mismatches (represented in small cases), including point mutations (eg, #3, underlined nucleotides), insertions (eg, #2, nucleotides above the line), and deletions (eg, #28, nucleotides under the line). Templates can be found in the adjacent Ig and mbr loci and in similar proportions. Moreover, 1 of the breakpoints can also constitute a template for copy and insertion in the other breakpoint (eg, #10). Genealogical relationship between 1 template and 2 copies can tentatively be reconstituted in some cases (eg, #2). Importantly, copies found in 1 of the breakpoints can be issued from templates located in a sequence segment involved in the other breakpoint (eg, #5, DH provides the template for the copy in the direct breakpoint). Finally, the whole breakpoint insertion region can be constituted of a patchwork of templated nucleotides, generated from different templates, and inserted in both direct or reverse-complement orientations (eg, #28, Figure 1D). Insertion of the copies in the breakpoints might be facilitated by anchoring to 1 of the broken ends through regions of microhomology (eg, #13, Figure 1C).
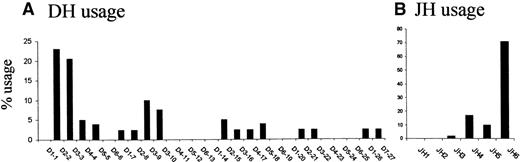
Biased usage of 5′ DH and 3′ JHgene segments associated with t(14;18) breakpoints
To investigate if particular DH and JH gene segments are preferentially associated with the translocation process, we analyzed the frequency of DH and JH gene usage in DH/mbr and mbr/JH breakpoints (Figure3). As shown in Figure 3A, the overall use of DH segments is nonrandom, D2-2 (23%) and D3-3 (20.5%) contributing to the majority of segments found in the reciprocal breakpoints. Similarly, JH segment usage is strikingly biased toward JH6 (71%) (Figure 3B). Biased usage of gene segments could be due to difference in the RSS sequences. However, 3′ RSS sequences are very conserved between members of the D2 or D3 family.18 In addition, the marked predominance for D2-2, D3-3, and JH6 usage contrasts with the distribution observed in normal V(D)J junctions at all stages of differentiation.18,19 28 Therefore, preferential usage of the most 5′ DH segments together with preferential usage of the most 3′ JH segments strongly suggests that the t(14;18) translocation process occurs during an attempted secondary DH to JH rearrangement.
Comparison of DH segment usage and of JH segment usage.
(A) Comparison of DH segment usage in the direct breakpoints; (B) comparison of JH segment usage in the reciprocal breakpoints. DH and JH segments are represented 5′ to 3′ (left to right) as ordered in their respective loci (not to scale).
Comparison of DH segment usage and of JH segment usage.
(A) Comparison of DH segment usage in the direct breakpoints; (B) comparison of JH segment usage in the reciprocal breakpoints. DH and JH segments are represented 5′ to 3′ (left to right) as ordered in their respective loci (not to scale).
Somatic mutations are observed on the DH segment of rare mbr to DJH direct breakpoints
In the majority of samples, we found a prototypic mbr/JH fusion in the direct breakpoint and DH/mbr fusion in the reciprocal breakpoint (Tables 1 and2). However, we observed 4 samples containing a mbr/DH/JH fusion (#5, #19, #28, #33, Table 1). Compatible with an attempted secondary D to DJHrearrangement on the same chromosome, all samples used a more 5′ DH in the reciprocal breakpoint than the 1 used in the DJH junction (eg, D3-3 to D3-10/JH6, sample #28). Unexpectedly, the DH segments used in the mbr/DH/JH direct breakpoint of 3 of those 4 samples (#5, #28, and #33) contained somatic mutations (underlined in Table 1). To exclude the possibility that those mutations are due to Taq-polymerase introduced mistakes, the sequence of those samples were confirmed by a second PCR-amplification and sequencing. Remarkably, those DH segments constitute the only sequences in which somatic mutations are found, since none of the sequences of the flanking mbr in the direct breakpoint (Table 1) or DHand mbr regions in the reciprocal breakpoints (Table 2) presented a single-point mutation, including the ones corresponding to those 4 cases. If the somatic hypermutation process had happened posttranslocation, it would not have been limited to 3 of these 4 already infrequent cases or to the sequence of the DH segments. On the contrary, mutations would be expected in the adjacent mbr and in at least some of the other direct breakpoints. Furthermore, in the recombined der 14 chromosome, the closest promoter 5′ of the mbr/JH breakpoint is the BCL-2 promoter. However, the BCL-2 promoter has not been described to target somatic hypermutation and is not located at a proper distance from the mbr breakpoint. It is therefore very unlikely that the somatic hypermutation happened posttranslocation. Thus, since bystander DJH rearrangements on the nonfunctional allele have been shown to undergo low levels of somatic hypermutation along with the VHDJH rearrangement on the functional allele,29 30 this suggests that both alleles underwent at least 1 round of somatic hypermutation before the translocation took place.
Discussion
Fundamental questions concerning the mechanism of t(14;18) translocation remain to be answered. Is the V(D)J recombination mechanism involved in the generation of both Ig and mbr breaks? If not, which mechanisms are responsible for the initial breaks at the mbr locus and for subsequent illegitimate joining? The goal of this study was to extend current understanding of the molecular mechanism involved in the t(14;18) translocation. We report here a detailed analysis of the first comprehensive DNA sequence library of both direct and reciprocal breakpoint regions derived from 40 t(14;18) translocation-positive FL patients. Our survey confirms that the JH and DH coding ends engaged in the direct and reciprocal breakpoints of t(14;18) translocation show features of normal V(D)J recombination. This implies the presence and involvement of a functional and active V(D)J recombination machinery at the time of the translocation. However, our analysis also clearly shows that the formation of duplications is a general feature of the mechanism creating breaks at the mbr locus. Although both deletion and precise ends are still compatible with RAG-mediated coding end formation and processing, the presence of duplications is not compatible with RAG-mediated hairpin formation. Furthermore, precise breaks and deletions are not a particular feature of V(D)J recombination and are also found together with duplications during nonhomologous end-joining and somatic hypermutation mechanisms.31-34 Altogether, the absence of proper RSS signals in the mbr, the presence of a distinct mechanism from V(D)J recombination for a substantial fraction of the breaks, and the presence of a break signature compatible with other types of mechanisms strongly suggest that V(D)J recombination is not responsible for the initial breaks at the mbr locus. Thus, 2 distinct mechanisms are creating the initial breaks at the Ig and mbr loci, as previously proposed.7
One potential candidate for the initiation of breaks at the mbr locus is the recently described 2-ended transposition mechanism.35 As Agrawal et al36 and Hiom et al35 have shown in a cell-free system, RAG-1 and RAG-2 proteins can drive the coupled insertion of the signal ends into new DNA sites in a transpositional reaction. In the 2-ended transposition, each of the 2 signal ends makes a strand exchange on the opposite side of the target sequence—for example, the mbr locus in the case of t(14;18)—3 to 5 nucleotides apart. In the translocation model, the nick left on each side is then converted into a hairpin by the same mechanism used during V(D)J recombination to generate coding ends. The prediction of such a model is therefore the presence in the signal junctions of a 3- to 5-bp piece of the mbr and a corresponding deletion of the mbr in the breakpoints. In this study, we found precise breaks, deletions, and duplications of more or less than 3-5 bp. Thus, although double-ended transposition might in vivo result in various types of breaks—as for example additional processing of the potential mbr hairpin intermediates—our results do not support this model's predictions stricto sensu.
To gain information on other potential mechanisms involved in the translocation process, we did a detailed analysis of the nucleotide insertions present in most direct mbr/JH and reciprocal DH/mbr breakpoints junction (de novo nucleotide additions, Tables 1 and 2). Surprisingly, our analysis revealed that the de novo insertions, previously thought to be N nucleotides, contain recurrent templated nucleotides consisting of short error-prone copies of the surrounding mbr, DH, and JH sequences (Figure 1 and Table 3). These templated nucleotides (called here T nucleotides) are clearly distinct from N and P nucleotides and represent a novel type of t(14;18) breakpoint insertions. The occurrence of T nucleotides in the breakpoints is remarkably high. In our survey, over a third of the breakpoints contained clearly identifiable and highly significant T nucleotides. The general features of the T nucleotides, including the multiplicity of copies for 1 template and the occurrence of mismatches in the copies, strongly suggest the presence of a short-patch DNA synthesis, templated and error-prone (illustrated in Figure 2).
Short nucleotide insertions, termed “filler DNA,” have been observed with various frequencies at immune, nonimmune, and oncogenic rearrangements in mammalian cells.37 However, the features of T nucleotides described here have not been observed in any of those junctions and are not easily explained by the mechanisms involved. Nonhomologous end joining, an error-prone repair pathway activated in response to DNA double-strand breakage, could be used in the translocation process to facilitate end joining through the presence of microhomologies.26 However, T-nucleotide formation per se cannot be accounted for by the DNA slip-mispair synthesis model generating direct repeats in nonhomologous end joining.31 The features of T nucleotides also clearly differ from the damage-repair type of mechanism recently described for t(4;11) translocations in acute lymphoblastic leukemia, in which the deleted pieces from the breakpoints are used as fill-in insertions.38 39 Although definitive answers must await the characterization of other translocation breakpoints, T-nucleotide formation might be unique to the t(14;18) junctions.
The origin of T nucleotide insertions and the ground for their presence in t(14;18) breakpoints are yet to be elucidated. Are T nucleotides involved in the translocation mechanism or only a by-product of unusual combinations of enzymatic activities, accidentally present at the moment of the translocation process? In this respect, the putative presence of an error-prone synthesis is puzzling. Generation of uncorrected DNA misincorporations is very risky for the genetic material of a cell, and very few mechanisms in mammalian cells use this process purposefully. The prominent example in which error-prone DNA synthesis has been proposed to be involved is the somatic hypermutation mechanism, an additional mechanism of diversification of Ig genes occurring in the germinal centers (for review see Storb40and references therein).41 During this mechanism, point mutations are introduced in the rearranged V(D)J genes (and surprisingly also in the non-Ig gene BCL-6). Interestingly, recent observations have revealed that somatic hypermutation mechanism is not restricted to the introduction of point mutations and that nucleotide deletions, duplications, and insertions corresponding to nucleotides of unknown origin are also frequently seen.32-34 Some of the duplications contain mutations and are separated from their template by stretches of nucleotides, suggesting the occurrence of DNA strand breaks that could provide a focus for error-prone DNA synthesis. It is interesting to note that these features are similar to the ones observed in this report for the mbr breaks and could represent a common pathway of DNA breakage and end processing. The involvement of the somatic hypermutation mechanism in chromosomal translocation has already been proposed for the c-myc/IgH t(8;14) associated with Burkitt lymphoma.33,42 The recent observations of continued RAG expression in late stages of B-cell development43-48 raised the interesting possibility that t(14;18) translocation could take place in the germinal centers and could involve both V(D)J recombination and somatic hypermutation mechanisms. Several additional observations in our survey support this possibility: First, there is a striking biased usage toward the most 5′ DH and the most 3′ JH gene segments in the breakpoints, suggesting that t(14;18) translocation occurs during an attempted secondary DH to JH rearrangement; second, cases of somatic hypermutation are found exclusively on the DHregions of rare cases of mbr/DH/JH fusion, suggesting that rounds of somatic hypermutation occurred before translocation. Thus, although to date no more than correlative evidences are available for its involvement in the t(14;18) translocation process, the somatic hypermutation mechanism—or part of its components—is an interesting potential candidate.
Therefore, although the mechanisms creating the initial breaks at the mbr locus and generating the T-nucleotide insertions remain to be elucidated, the features of the breakpoints described here suggest that t(14;18) translocation is a complex process, which might involve the interaction and/or subversion of the V(D)J recombinase with multiple enzymatic machineries.
Acknowledgments
We are grateful to Jim Koziol for advice on the statistical analysis and to Ann J. Feeney, David Schatz, Rolf Marschalek, and Rodrig Marculescu for helpful comments and suggestions on the manuscript.
Supported by a grant for the Interdisciplinary Cooperation Project (ICP) “Molecular Medicine” from the University of Vienna.
Reprints:Bertrand Nadel, Department of Internal Medicine I, Division of Hematology, University of Vienna, Waehringer Guertel 18-20, A-1090 Vienna, Austria; e-mail: bertrand.nadel@akh-wien.ac.at.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal