Abstract
The ETS family of proteins is a large group of transcription factors implicated in many aspects of normal hematopoietic development, as well as oncogenesis. For example, the TEL1/ETV6 (TEL1) gene is required for normal yolk sac angiogenesis, adult bone marrow hematopoiesis, and is rearranged or deleted in numerous leukemias. This report describes the cloning and characterization of a novelETS gene that is highly related to TEL1 and is therefore called TEL2. The TEL2 gene consists of 8 exons spanning approximately 21 kilobases (kb) in human chromosome 6p21. Unlike the ubiquitously expressed TEL1 gene, however,TEL2 appears to be expressed predominantly in hematopoietic tissues. Antibodies raised against the C-terminus of the TEL2 protein were used to show that TEL2 localizes to the nucleus. All ETS proteins can bind DNA via the highly conserved ETS domain, which recognizes a purine-rich DNA sequence with a GGAA core motif. DNA binding assays show that TEL2 can bind the same consensus DNA binding sequence recognized by TEL1/ETV6. Additionally, the TEL2 protein is capable of associating with itself and with TEL1 in doubly transfected Hela cells, and this interaction is mediated through the pointed (PNT) domain of TEL1. The striking similarities ofTEL2 to the oncogenic TEL1, its expression in hematopoietic tissues, and its ability to associate withTEL1 suggest that TEL2 may be an important hematopoietic regulatory protein.
The ETS (E26-transformation specific) family of transcription factors is a large group of evolutionarily conserved transcriptional regulators that play an important role in a variety of cellular processes throughout development and differentiation (reviewed in Dittmer and Nordheim,1 Ghysdael and Boureux,2 and Sharrocks et al3). All ETS proteins bind DNA via a highly conserved approximately 85 amino acid (aa) region, the ETS domain, that recognizes a purine-rich GGAA/T core motif within the promoters and enhancers of various target genes (reviewed in Graves and Petersen4 and Janknecht and Nordheim5). In addition to sequence recognition, DNA binding may also be regulated through phosphorylation of ETS proteins and by protein-protein interactions mediated via other domains (eg, the “pointed” [PNT] domain) within ETS proteins, as well as the ETS domain itself.6 7
Although expressed in a variety of tissues, most currently knownETS genes are expressed predominantly in hematopoietic cells and many are key regulators of blood cell development and differentiation.8 Indeed, transgenic and gene-targeting experiments in the mouse have uncovered essential roles for several ETS factors in the regulation of various cell lineages within the hematopoietic system (reviewed in Dittmer and Nordheim1 and Sharrocks et al3). Additionally, ETS protein binding sites have been identified in numerous genes involved in the regulation of the hematopoietic system.2 8
In addition to regulating normal blood cell functions, several ETS proteins have shown oncogenic potential. For example,v-ets, the original member of the ETS gene family, induces avian erythroblastosis when fused to the viralgag gene and the v-myb oncogene.9 Specific integration of the Friend murine leukemia virus results in overexpression of either spi1/pu.1 or fli1 in murine erythroleukemias.10,11 Translocations involving theETS gene TEL1/ETV6 (henceforth referred to asTEL1) are associated with many different human leukemias.12 Indeed, TEL1 is fused with several different proteins in both myeloid and lymphoid leukemias and can contribute to cellular transformation by diverse molecular mechanisms. Given the extensive involvement of ETS factors in normal hematopoiesis and in hematopoietic malignancies, a complete understanding of these complex processes relies on the continued characterization of knownETS genes, as well as the identification and characterization of new ETS genes that may also regulate normal and/or aberrant hematopoiesis.
This report describes the identification and initial characterization of a new human ETS-family gene called TEL2. Analysis of the TEL2 complementary DNA (cDNA) and the genomic locus shows that TEL2 is closely related to TEL1 in both sequence and structure. However, in contrast to the ubiquitously expressedTEL1, TEL2 appears to be expressed predominantly in hematopoietic tissues. Like a subset of other ETS proteins, including TEL1, TEL2 contains an amino- (N-) terminal PNT domain and this domain is thought to mediate protein-protein interactions. We show that TEL2 can associate with itself and with TEL1, and that the PNT domain of TEL1 is crucial for this interaction. Additionally, we show that TEL2 can bind the TEL1 consensus DNA binding sequence (CDBS). Together, these data describe a new ETS transcription factor closely related to TEL1 and, in light of the essential role of TEL1 in both normal and aberrant hematopoiesis, suggest that it likely plays an important role in hematopoietic regulation.
Materials and methods
Cloning and sequence analysis
TEL2 was identified by sequencing expressed sequence-tagged clones (ESTs) from Human Genome Systems (Rockville, MD) libraries as previously described.13 The TEL2 cDNA sequence was used for searches of the nonredundant GenBank, EMBL, DDBJ, and PDB sequence databases and the nonredundant databases of the EST divisions of GenBank, EMBL, and DDBJ, via the Baylor College of Medicine (BCM) “BLASTN” search engine.14 The TEL2 peptide sequence was also searched against the GenBank CDS translations, PDB, SwissProt, PIR, and PRF databases through the BCM “BLASTN” search engine. Sequence comparisons with other ETS factors were performed with the CLUSTAL W Multiple Sequence Alignment Program15 and the Align program.16 The genomic organization of TEL2was determined by directly comparing the cDNA sequence to the genomic sequence of PAC clone 50J22 (accession number [acc #] Z84484). The sequence data of PAC 50J22 were produced by the Chr. 6 Sequencing Group at the Sanger Centre (Hinxton, Cambridge, UK) and can be obtained fromhttp://www.ncbi.nlm.nih.gov. PAC clone 50J22 was mapped by the Chr. 6 Mapping Group at the Sanger Centre, and the mapping data were obtained from the World Wide Web at http://www.sanger.ac.uk/HGP/Chr6.
Fluorescent in situ hybridization
The TEL2 cDNA was labeled with digoxigenin-11-dUTP (Boehringer Mannheim, Indianapolis, IN) by nick translation. The labeled probe was combined with sheared human DNA and hybridized to normal metaphase chromosomes derived from PHA-stimulated peripheral blood lymphocytes in a solution containing 50% formamide, 10% dextran sulfate, and 2 × SSC. Specific hybridization signals were detected by incubating the hybridized slides in fluorescein-conjugated sheep antibodies against digoxigenin (Boehringer Mannheim). The chromosomes were then counterstained with 4, 6-diamidino-2-phenylindole (DAPI) and analyzed. Band assignment was made by conducting fractional length measurements of 10 specifically hybridized chromosomes 6.
Northern analysis
A human Multiple Tissue Northern (MTN) blot (Clontech, Palo Alto, CA) containing 2 μg of purified messenger RNA (mRNA) per lane was hybridized with a randomly primed, α-32P-dATP-radiolabeled (Amersham Pharmacia Biotech, Arlington Heights, IL) full-length TEL2 cDNA fragment, and a similarly labeled human α-actin control probe. Hybridization and washing conditions were according to the manufacturer's specifications. Blots were exposed on a phosphor screen and imaged on a Molecular Dynamics Storm 860 phosphorimager (Amersham Pharmacia Biotech, Sunnyvale, CA).
Antibodies
A decapeptide corresponding to the 10 carboxy-(C)-terminal amino acids (aa) of TEL2 was synthesized, conjugated to keyhole limpet hemocyanin (KLH), and injected into New Zealand White rabbits (Rockland, Gilbertsville, PA). The KLH-decapeptide used for antibody production contained a 1 amino acid change. The sequence of the KLH-decapeptide, KDKRQEISP, should have been KDKRPEISP. However, all subsequent assays of the TEL2-specific antibody showed that it was specific for the TEL2 protein. TEL2-specific antibodies were purified using an AminoLink Plus Immobilization Kit column (Pierce, Rockford, IL), coupled with glutathione-S-transferase (GST)-TEL2 fusion proteins. A monoclonal antibody, 12CA5 (Boehringer Mannheim), was used to recognize influenza hemagglutinin (HA) tagged–TEL2.
Plasmids
Polymerase chain reaction (PCR) products encompassing theTEL2 full-length cDNA and the TEL2 ETS domain were inserted in-frame into the EcoRI site of the GST fusion pGEX-2T vector (Pharmacia, Piscataway, NJ). Primer sequences, including an introduced-EcoRI site at each 5′ end, are as follows: (full-length TEL2) (sense) 5′-CGGAATTCATATGCAGGAGGGAGAATTG-3′; (antisense) 5′-CGGAATTCCCTCACGGAGAGATTTCTGG-3′; (ETS domain) (sense) 5′-ATCGGAATTCTCTGTTCCTTCCCCGCGATGCC-3′; (antisense) the same antisense oligomer listed above was used. GST protein preparations were made by electroporating these pGEX-2T constructs, or the pGEX-2T vector only, into Escherichia coli BL21 cells. Protein expression and purification were performed according to manufacturer's instructions (Pharmacia). The full-length TEL2 cDNA, and a triple HA-tagged TEL2, were inserted into the cytomegalovirus (CMV) promoter-driven expression vector pSCTOP.17 All TEL1 constructs used have been previously described.18 Ten micrograms of each construct were transfected into Hela cells, 50% to 80% confluent on 10 cmol/L dishes, using the FuGene 6 Transfection Reagent (Boehringer Mannheim).
Western blotting and immunoprecipitation
For Western blotting, protein lysates were separated on 8% SDS-polyacrylamide gels (PAGE) and electroblotted onto PVDF membranes (Millipore, Bedford, MA). Kaleidescope Prestained Molecular Weight Standards (Bio-Rad, Hercules, CA) were used as molecular weight markers. Immunoprecipitations were performed as previously described.19 Blots were blocked overnight in phosphate-buffered saline (PBS) containing 3% bovine serum albumin (BSA) and incubated with specific antibodies overnight at 4°C, or 1 hour at room temperature. Bound antibody was detected using alkaline phosphatase-conjugated antirabbit (for α-TEL2) or antimouse (for 12CA5) IgG secondary antibodies and colorimetry (Renaissance Western Blot Chemiluminescence Reagent; NEN Life Science Products, Boston, MA).
Indirect immunofluorescence analysis
Cytospins were prepared from transfected Hela cells. Cells were fixed and stained as previously described.18Images were obtained using confocal microscopy (Olympus B × 50 laser confocal microscope; Olympus, Lake Success, NY).
Electrophoretic mobility shift assay
The DNA binding assay was performed as previously described.18
Results
Identification and sequence analysis of TEL2
TEL2 was identified by sequence comparisons of EST clones obtained from a human activated T-cell cDNA library. Ten overlapping clones were found that showed homology to ETS transcription factors. Sequences matching these ESTs were also found among clones sequenced from a primary human breast tissue cDNA library and a human meningioma cDNA library (1 clone from each). Alignment of the clones revealed overlapping sequences of 1550 nucleotides (Figure1). Database searches with the full-length cDNA sequence showed that it was highly similar to that of the humanTEL1 ETS gene. Therefore, we have named this geneTEL2.
Nucleotide sequence and predicted amino acid sequence ofTEL2.
Nucleotides are numbered beginning with +1 at the ATG start codon. Numbering of the aa sequence is italicized. A putative MAPK phosphorylation site (aa 10-13) is in bold type and italicized. Both the nucleotide and aa sequences of the PNT (aa 51-117) and ETS (aa 225-304) domains are in bold type and underlined. The last 10 aa of TEL2, used as a KLH-conjugated peptide for antibody production, are underlined. The stop codon used and additional in-frame stop codons in the 3′ UTR are italicized and marked with an asterisk. Within the 3′ UTR, an attta motif, associated with mRNA turnover, is in bold type and the putative polyadenylation signal, aataaa, is in bold type and underlined. This sequence has been assigned the GenBank accession number AF175387.
Nucleotide sequence and predicted amino acid sequence ofTEL2.
Nucleotides are numbered beginning with +1 at the ATG start codon. Numbering of the aa sequence is italicized. A putative MAPK phosphorylation site (aa 10-13) is in bold type and italicized. Both the nucleotide and aa sequences of the PNT (aa 51-117) and ETS (aa 225-304) domains are in bold type and underlined. The last 10 aa of TEL2, used as a KLH-conjugated peptide for antibody production, are underlined. The stop codon used and additional in-frame stop codons in the 3′ UTR are italicized and marked with an asterisk. Within the 3′ UTR, an attta motif, associated with mRNA turnover, is in bold type and the putative polyadenylation signal, aataaa, is in bold type and underlined. This sequence has been assigned the GenBank accession number AF175387.
Analysis of this sequence identified an open reading frame (orf) of 341 aa, initiating from an ATG codon 25 base pairs (bp) from the 5′ end of the aligned cDNA sequence. After a stop codon beginning at position +1027 is a 3′ untranslated region (UTR) of 499 bp. The 3′ UTR contains additional stop codons, as well as an ATTTA motif that may be involved in mRNA turnover,20 and a putative polyadenylation signal 14 bp upstream of a poly (A) sequence.
Sequence comparison of the deduced aa sequence of our cDNA clone to the aa sequences of over 30 other known ETS genes revealed that it is most closely related to human TEL1 (data not shown). Indeed, the deduced 341 aa sequence is 38% identical to the 452 aa sequence of the TEL1 protein. Although all ETS domains show significant aa sequence conservation,21 the ETS domains of TEL2 andTEL1 are highly conserved (85.4% aa identity) (Figure2A). Comparison of all other ETS domains to that of TEL2 shows that the next highest level of homology is with the ETS domains of the Drosophila (D)-YAN (47.6% aa identity), human ELF1 (44.6% aa identity), and D-E74 proteins (44.6% aa identity) (Figure 2A; data not shown). Sequence identity of TEL2 to ETS domains from all other ETSfamily members examined ranges from approximately 33% to 43% (data not shown).
ETS and PNT domains.
Comparison of the aa sequence of the ETS domain (A) and the PNT domain (B) of human (hu) TEL2 with those of hu-TEL1 and D-YAN. Identical aa are shaded in black and similar aa are shaded gray. Dashes represent spaces introduced for optimal alignment. The percentage of aa identity with TEL2 is shown on the left of each aa sequence. Comparisons were made using the CLUSTALW program15 and percentage identity was determined with the ALIGN program.16
ETS and PNT domains.
Comparison of the aa sequence of the ETS domain (A) and the PNT domain (B) of human (hu) TEL2 with those of hu-TEL1 and D-YAN. Identical aa are shaded in black and similar aa are shaded gray. Dashes represent spaces introduced for optimal alignment. The percentage of aa identity with TEL2 is shown on the left of each aa sequence. Comparisons were made using the CLUSTALW program15 and percentage identity was determined with the ALIGN program.16
A subset of ETS transcription factors, including ETS1, ETS2, PNT, GABP, ERG, FLI1, TEL1, YAN, and ESE1, contains a second highly conserved domain called the PNT domain (reviewed in Graves and Petersen4). This domain is thought to mediate protein-protein interactions.22 A region of approximately 67 aa near the N-terminus of TEL2 is homologous to the PNT domains from other ETS proteins. Comparison of the TEL2 PNT domain with those of other ETS factors shows the highest aa identity with TEL1 (62.5%) (Figure 2B). The next closest match is the D-YAN PNT domain, which is 48.6% identical to TEL2. Sequence comparisons to all other ETS-factor PNT domains show an aa identity with TEL2 ranging from approximately 23% to 39% (data not shown).
The transcriptional activity of several ETS proteins is regulated by Ras-induced phosphorylation by mitogen-activated protein kinases (MAPK).7 The sequence of TEL1 contains at least 2 putative MAPK phosphorylation sites, and the protein is phosphorylated in vivo.23 Analysis of the TEL2 aa sequence reveals the presence of at least 1 potential MAPK phosphorylation site (Figure 1). This site is located at aa positions 10 to 13, approximately 37 aa upstream of the PNT domain. No other putative MAPK phosphorylation sites were detected in the TEL2 sequence.
Mapping TEL2 and genomic organization
The chromosomal map position of TEL2 was determined by fluorescent in situ hybridization (FISH), using a digoxigenin-11-dUTP–labeled TEL2 cDNA probe hybridized to normal metaphase human chromosomes (chr) derived from phytohemaglutanin (PHA)- stimulated peripheral blood lymphocytes. Hybridization signals were detected on the short arm of chr 6, corresponding to a position of 6p21 (data not shown).
A subsequent BLAST search of GenBank sequences14 using theTEL2 cDNA sequence identified a perfect match with sequences from PI-derived artificial chromosome (PAC) clone HS50J22 on human chr 6p21, thus confirming our FISH result above. Clone HS50J22 is part of a bacterial clone contig of human chr 6 generated by the Sanger Centre Chr 6 Mapping Group (UK), and it has been entirely sequenced (see “Materials and methods”). The completeTEL2 cDNA sequence is found within this clone; thus, access to this genomic sequence permitted the deduction of the structural organization of the TEL2 locus.
The TEL2 cDNA is encoded by at least 8 exons that span approximately 21 kb of DNA (Table 1). Exon 3 and part of 4 encode the PNT domain and exons 6, 7, and part of 8 encode the ETS domain. Sequences at the borders of the exons and the sizes of introns and exons are shown in Table 1; however, because the exact transcriptional start site has not been determined, the exact size of exon 1 is unknown. As shown in Table 1, all splice junctions show canonical GT/AG dinucleotides.
Exon/intron boundaries of TEL2
| Exon no. . | Size (bp) . | 5′ splice donor . | Intron length (kb) . | 3′ splice acceptor . | TEL1/ETV6 exon size (bp)* . |
|---|---|---|---|---|---|
| 1 | >31† | ATGCAG gtaaga | 1.7 | taggag CGAGAA | 57 |
| 2 | 134 | GACTCC gtaagt | 9.5 | ttgcag GCATCC | 130 |
| 3 | 166 | GCTCAG gtcagg | 2.3 | ctccag GTGACG | 164 |
| 4 | 125 | CGGAAG gtgagg | 1.9 | ttgcag AGGTGA | 134 |
| 5 | 230 | TCGCTG gtgagt | 2.3 | ttgcag ACTGCC | 545 |
| 6 | 142 | CACAAG gtgagc | 2.0 | ttccag AACCGG | 142 |
| 7 | 100 | GTTCAG gtgagt | 0.1 | ttctag ATTTCT | 100 |
| 8 | 117 | — | — | — |
| Exon no. . | Size (bp) . | 5′ splice donor . | Intron length (kb) . | 3′ splice acceptor . | TEL1/ETV6 exon size (bp)* . |
|---|---|---|---|---|---|
| 1 | >31† | ATGCAG gtaaga | 1.7 | taggag CGAGAA | 57 |
| 2 | 134 | GACTCC gtaagt | 9.5 | ttgcag GCATCC | 130 |
| 3 | 166 | GCTCAG gtcagg | 2.3 | ctccag GTGACG | 164 |
| 4 | 125 | CGGAAG gtgagg | 1.9 | ttgcag AGGTGA | 134 |
| 5 | 230 | TCGCTG gtgagt | 2.3 | ttgcag ACTGCC | 545 |
| 6 | 142 | CACAAG gtgagc | 2.0 | ttccag AACCGG | 142 |
| 7 | 100 | GTTCAG gtgagt | 0.1 | ttctag ATTTCT | 100 |
| 8 | 117 | — | — | — |
TEL2 expressed in hematopoietic tissues
Because several TEL2 cDNA clones were sequenced from an activated T-cell cDNA library (in addition to a breast tissue and meningioma cDNA library), we anticipated that this gene may be expressed predominantly in hematopoietic tissues. To determine the expression pattern of TEL2, several human multitissue Northern blots (Clontech) containing poly (A)+ mRNAs from various human tissues were hybridized with the full-length TEL2 cDNA (Figure3; data not shown). Expression was seen only in hematopoietic tissues; 1 predominant band, approximately 1.2 kb, was detected in fetal liver and bone marrow (Figure 3), which is slightly smaller than the transcript size predicted from theTEL2 cDNA sequence (approximately 1.5 kb). The same band is also faintly present in mRNAs isolated from peripheral blood leukocytes, which may represent expression in activated T cells (Figure3). Smith and Ostrowski24 have reported a sequence to Genbank (acc # AF147782), excluding exon 5, identical to TEL2. A truncated transcript, omitting exon 5 (230 bp) might account for the smaller band detected in Figure 3. However, a reverse transcriptase–polymerase chain reaction (RT-PCR) using RNA from human fetal liver and primers flanking exon 5 yielded only 1 band equal to a size that includes exon 5 (data not shown). Additionally, Shin et al26 have reported 3 sequences to Genbank corresponding toTEL2 (acc # AF116508, AF116509, AF11610), including 2 isoforms that are 70 to 80 aa shorter than the full-length TEL2.Therefore, the difference in size of the detected band in Figure 3 and the TEL2 cDNA may be due to alternative splicing, an unknown modification(s) of the transcript, or altered migration of theTEL2 transcript in the denaturing gel.
Expression of TEL2 in human tissues.
A Northern blot containing mRNA from the indicated human tissues was hybridized sequentially with the full-length TEL2 cDNA and a control hu α-actin probe. The approximate molecular weight (MW) of the band detected by TEL2 is indicated on the right. The 2.0-kb band identified by α-actin band is also shown.
Expression of TEL2 in human tissues.
A Northern blot containing mRNA from the indicated human tissues was hybridized sequentially with the full-length TEL2 cDNA and a control hu α-actin probe. The approximate molecular weight (MW) of the band detected by TEL2 is indicated on the right. The 2.0-kb band identified by α-actin band is also shown.
Generation of antibodies specific for TEL2
To facilitate further characterization of the TEL2 protein, rabbit polyclonal antibodies (abys) were generated against a KLH-conjugated decapeptide corresponding to the C-terminal 10 aa of TEL2 (Figure 1). These 10 aa show significant sequence divergence (8 of 10 different) from the corresponding 10 aa of TEL1 and, therefore, polyclonal abys directed against this decapeptide were not expected to cross-react with the TEL1 protein.
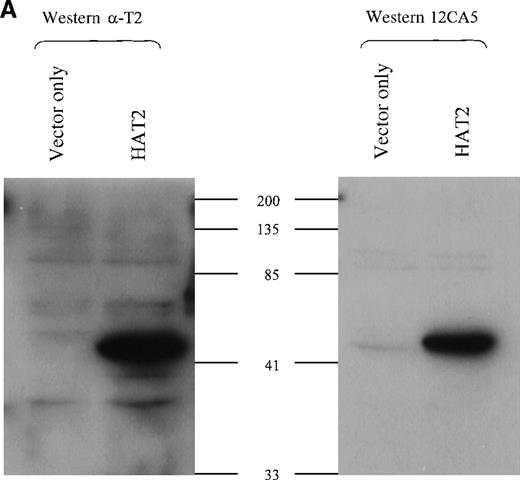
To test the specificity of the TEL2 antisera, we collected lysates from Hela cells transiently transfected with an HA-tagged TEL2expression construct (HATEL2). Western blots were made with this lysate, and lysates from vector-only transfected Hela-cells, and these blots were screened with either the α-TEL2 aby (α-T2) or the HA-specific monoclonal aby 12CA5 (Figure4A). As shown in Figure 4A, α-T2 recognizes the same protein as 12CA5; that is, it specifically binds to the HATEL2 protein, which is near the predicted size, based on the predicted 38 kd size of TEL2 plus the 3-kd HA tag.
TEL2-specific antibodies.
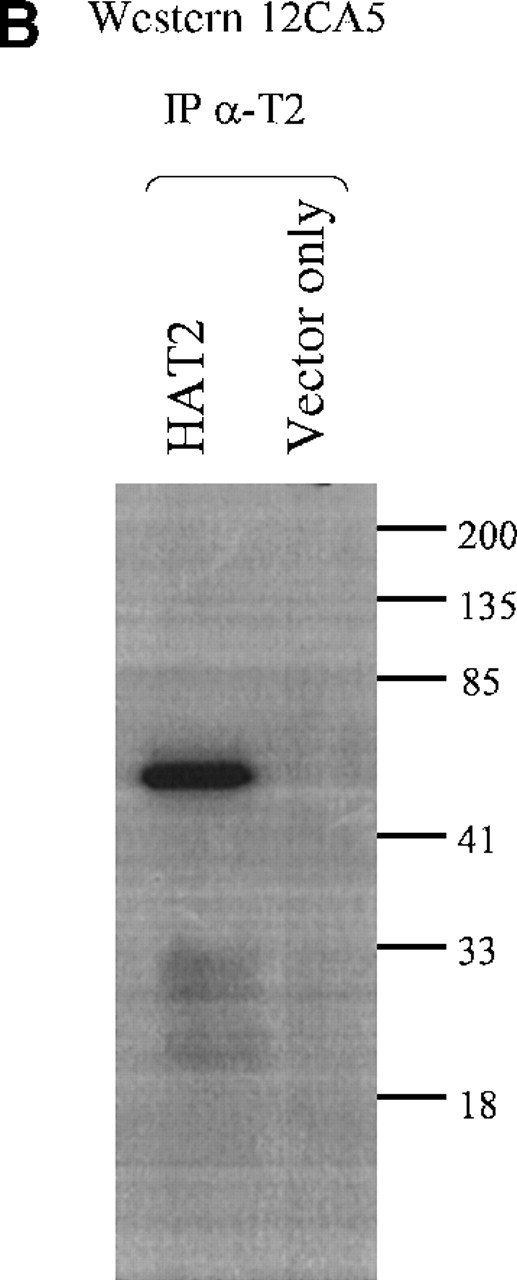
(A) Western blot analysis of TEL2-specific antibodies (α-T2). Western blots containing proteins from the indicated Hela-cell lysates were made and incubated with either anti-TEL2 antisera (α-T2) (left) or an anti-HA antibody (12CA5) (right). MW standards are indicated in kilodaltons (kd). (B) Immunoprecipitation of TEL2 with α-T2. Indicated lysates isolated from Hela cells were incubated with α-T2. IP products were separated by SDS-PAGE, Western blotted, and probed with the 12CA5 aby. MW standards are shown on the right.
TEL2-specific antibodies.
(A) Western blot analysis of TEL2-specific antibodies (α-T2). Western blots containing proteins from the indicated Hela-cell lysates were made and incubated with either anti-TEL2 antisera (α-T2) (left) or an anti-HA antibody (12CA5) (right). MW standards are indicated in kilodaltons (kd). (B) Immunoprecipitation of TEL2 with α-T2. Indicated lysates isolated from Hela cells were incubated with α-T2. IP products were separated by SDS-PAGE, Western blotted, and probed with the 12CA5 aby. MW standards are shown on the right.
The α-TEL2 abys were then tested for their ability to immunoprecipitate (IP) the TEL2 protein from cellular lysates. The α-T2 aby was incubated with either HATEL2 only or vector only–transfected Hela-cell lysates. Figure 4B shows that α-T2 effectively immunoprecipitates HATEL2 without immunoprecipitating a significant amount of extraneous proteins. No significant protein is immunoprecipitated from the vector only–transfected Hela cells.
TEL2 is localized within the nucleus
One mechanism of regulating the activity of transcription factors, including some ETS factors, is based on subcellular localization of the protein. For example, the ETS protein D-YAN is regulated by shuttling between the nucleus and cytoplasm.27 Because TEL2 is closely related to the D-YAN protein, as well as TEL1, indirect immunofluorescence (IF) was used to study the subcellular localization of TEL2 in transiently transfected Hela cells.
Hela cells transfected with the HATEL2 expression construct were fixed to slides by cytospin centrifugation 24 to 48 hours after transfection. These cells were then incubated with either α-T2 or 12CA5. As shown in Figure 5A, the HATEL2 protein is localized within the nucleus, excluding the nucleoli. The specificity of α-T2 for TEL2 protein via IF is demonstrated by the identical signal found in cells stained with 12CA5 (Figure 5C), and by the lack of signal in vector-only transfected cells stained with α-T2 or 12CA5 (Figure 5B and 5D, respectively). Thus, the TEL2 protein is capable of localizing within the nucleus.
Subcellular localization of TEL2 transiently expressed in Hela cells.
Nuclear staining of TEL2, detected with α-T2 and a FITC-conjugated goat-α-rabbit secondary aby (A), or with 12CA5 and a FITC-conjugated goat-α-mouse secondary aby (C), is shown in cytospin preparations of transiently transfected Hela cells. Hela cells transfected with an empty vector expression construct, stained with the same antibodies, are also shown ([B] and [D], respectively). Original magnification, ×40.
Subcellular localization of TEL2 transiently expressed in Hela cells.
Nuclear staining of TEL2, detected with α-T2 and a FITC-conjugated goat-α-rabbit secondary aby (A), or with 12CA5 and a FITC-conjugated goat-α-mouse secondary aby (C), is shown in cytospin preparations of transiently transfected Hela cells. Hela cells transfected with an empty vector expression construct, stained with the same antibodies, are also shown ([B] and [D], respectively). Original magnification, ×40.
TEL2 binds to the TEL1 consensus recognition sequence
The CDBS has been determined for several ETS transcription factors and it is thought that all ETS proteins bind a DNA sequence with a purine-rich GGAA/T core motif (reviewed in Graves and Petersen4 and Janknecht and Nordheim5). The sequences flanking this core are more variable, but not random, and may confer some binding specificity among ETS proteins. The CDBS for TEL1 has been determined to be ccGGAAgt.18 Because of the high sequence identity between TEL2 and TEL1, particularly within the ETS domain, we sought to determine whether the TEL2 protein could bind the TEL1 CDBS.
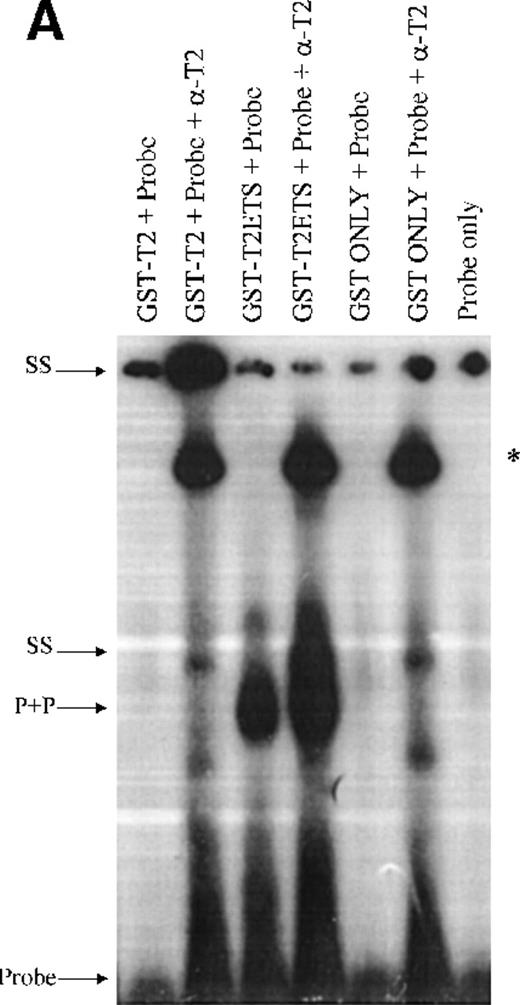
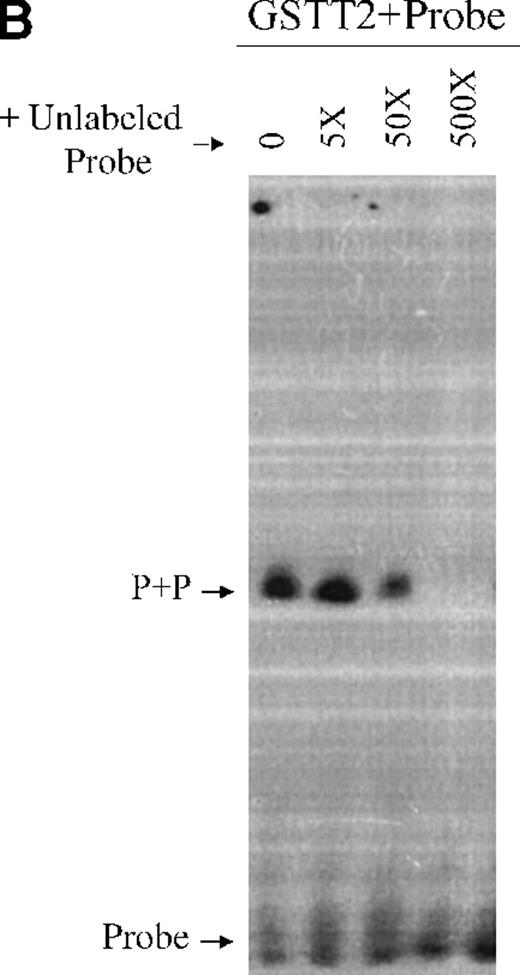
Double-stranded oligomers containing the TEL1 CDBS were used in electrophoretic mobility shift assays (EMSA) to analyze the DNA binding activities of a GST-fusion TEL2 protein (GST-T2). Because many full-length ETS proteins do not bind DNA efficiently, due to intramolecular regulatory mechanisms,28,29 we also generated a GST fusion of the ETS domain only of TEL2 (GST-T2ETS). As shown in Figure 6A, GST-T2 does not bind the TEL1 CDBS. However, the addition of α-T2 causes a GST-T2 + DNA + aby complex to shift to the well (Figure 6A). Binding of α-T2 likely changes the intramolecular conformation of GST-T2, allowing the ETS domain to bind, and a similar phenomenon has been observed in DNA binding experiments with TEL1.18 Figure 6A also shows that GST-T2ETS can efficiently bind the TEL1 CDBS, and the addition of α-T2 demonstrates the specificity of this interaction, resulting in a supershift of the protein-DNA-aby complex. Additionally, the specificity of the GST-T2ETS+CDBS interaction is demonstrated in Figure6B. Binding of GST-T2ETS to the 32P-labeled CDBS is competed away by a molar excess of unlabeled CDBS. Therefore, these results show that the TEL2 ETS domain is capable of recognizing the same CDBS as TEL1.
EMSA analysis of bacterially expressed GST-TEL2 proteins.
(A) Radiolabeled oligomers encoding the TEL1 CDBS were incubated with eluates of either GST only, GST-T2, or GST-T2ETS proteins. DNA-protein complexes were supershifted with α-T2. Combinations of DNA, protein, and aby are indicated above each lane. The GST-T2+DNA supershift (top of figure) and the GST-T2ETS supershift are denoted by arrows labeled “SS.” A complex associated with the addition of α-T2 antisera is denoted with an “*.” (B) GST-T2 protein was incubated with radiolabeled-TEL1 CDBS and increasing amounts of unlabeled TEL1 CDBS. The molar excess of unlabeled probe added to each binding reaction is shown above each lane. Complexed and free probe were separated by PAGE and detected by autoradiography. The GST-T2ETS protein + CDBS probe complex is indicated with an arrow labeled “P+P.” Uncomplexed probe is shown at the bottom of the figure by the “Probe”-labeled arrow.
EMSA analysis of bacterially expressed GST-TEL2 proteins.
(A) Radiolabeled oligomers encoding the TEL1 CDBS were incubated with eluates of either GST only, GST-T2, or GST-T2ETS proteins. DNA-protein complexes were supershifted with α-T2. Combinations of DNA, protein, and aby are indicated above each lane. The GST-T2+DNA supershift (top of figure) and the GST-T2ETS supershift are denoted by arrows labeled “SS.” A complex associated with the addition of α-T2 antisera is denoted with an “*.” (B) GST-T2 protein was incubated with radiolabeled-TEL1 CDBS and increasing amounts of unlabeled TEL1 CDBS. The molar excess of unlabeled probe added to each binding reaction is shown above each lane. Complexed and free probe were separated by PAGE and detected by autoradiography. The GST-T2ETS protein + CDBS probe complex is indicated with an arrow labeled “P+P.” Uncomplexed probe is shown at the bottom of the figure by the “Probe”-labeled arrow.
TEL2 can associate with itself and TEL1.
ETS proteins have been shown to form homodimers and heterodimers with other ETS proteins, as well as complexes with other proteins outside the ETS family.22,30-35 The TEL1 protein, for example, can associate with itself22 and with the ETS protein FLI1,32 via the PNT domain. Because TEL2 contains a PNT domain, it may also associate with itself and/or other proteins via this domain. To determine whether TEL2 can self-associate and/or associate with TEL1, co-IP experiments were performed.
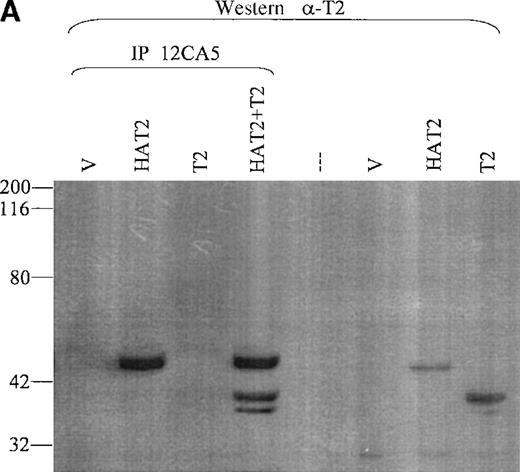
Hela cells were cotransfected with a TEL2 expression construct and either an HA-TEL2 construct (HAT2), an HA-TEL1 construct (HAT1), or an HA-TEL1 minus the PNT domain construct (HAT1ΔPNT). Lysates from cells transfected with TEL2 + HATEL2 were incubated with 12CA5, and IP products were separated by SDS-PAGE, Western blotted, and incubated with α-T2. If TEL2 can form homodimers and/or oligomers, then immunoprecipitation with 12CA5 should IP both HATEL2 and the smaller, untagged TEL2. Figure 7A shows that TEL2 is immunoprecipitated with the 12CA5 antibody; therefore TEL2 can associate with itself. It should be noted that a second band specific for TEL2 is also immunoprecipitated (Figure 7A). This band may represent a second isoform of the TEL2 protein, initiating from a secondary ATG codon. Indeed, TEL1 is expressed in several cell types as 2 protein isoforms, initiating from the ATG codons at position 1 and at position 43.23
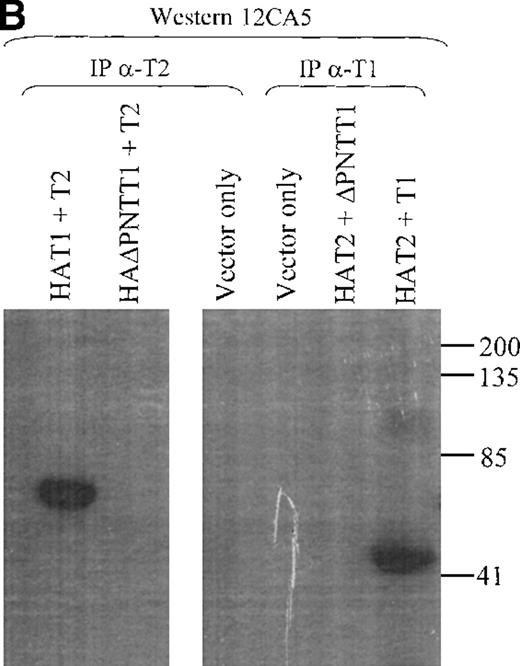
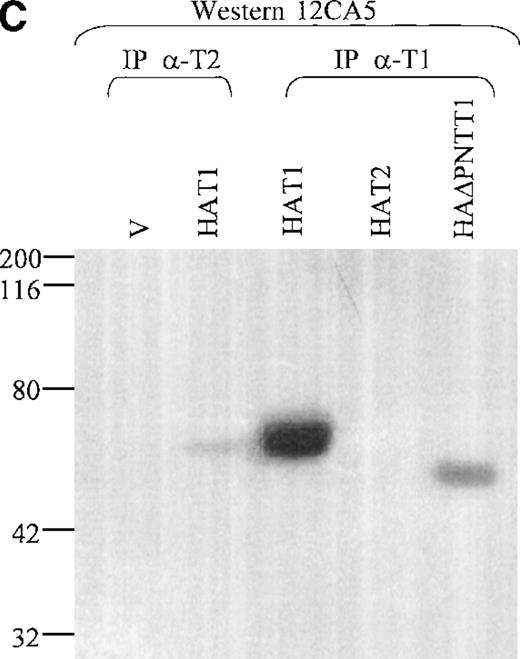
Coimmunoprecipitation of TEL2.
(A) Coimmunoprecipitation of TEL2 with HATEL2. Human fibroblast cells (293T) were transiently transfected with empty vector, HAT2, T2, or HAT2 + T2, and lysates were immunoprecipitated with 12CA5. Immunocomplexes were separated by SDS-PAGE, Western blotted, and incubated with α-T2. Lysates from cells transfected with the indicated constructs were also Western blotted with α-T2 (right 3 lanes). MW standards are indicated on the left. (B) Coimmunoprecipitation of TEL2 and TEL1. Lysates of Hela cells transiently transfected with the indicated expression constructs were incubated with either α-T2 (left 3 lanes) or α-T1 antibodies (right 3 lanes). Immunoprecipitates were separated by SDS-PAGE, Western blotted, and incubated with the 12CA5 aby. MW standards are indicated on the right. (C) 293T cells were transiently transfected with the constructs indicated above each lane and lysates were immunoprecipitated with either α-T2 or α-T1. Immunoprecipitates were separated by SDS-PAGE, Western blotted, and incubated with 12CA5. MW standards are shown on the left.
Coimmunoprecipitation of TEL2.
(A) Coimmunoprecipitation of TEL2 with HATEL2. Human fibroblast cells (293T) were transiently transfected with empty vector, HAT2, T2, or HAT2 + T2, and lysates were immunoprecipitated with 12CA5. Immunocomplexes were separated by SDS-PAGE, Western blotted, and incubated with α-T2. Lysates from cells transfected with the indicated constructs were also Western blotted with α-T2 (right 3 lanes). MW standards are indicated on the left. (B) Coimmunoprecipitation of TEL2 and TEL1. Lysates of Hela cells transiently transfected with the indicated expression constructs were incubated with either α-T2 (left 3 lanes) or α-T1 antibodies (right 3 lanes). Immunoprecipitates were separated by SDS-PAGE, Western blotted, and incubated with the 12CA5 aby. MW standards are indicated on the right. (C) 293T cells were transiently transfected with the constructs indicated above each lane and lysates were immunoprecipitated with either α-T2 or α-T1. Immunoprecipitates were separated by SDS-PAGE, Western blotted, and incubated with 12CA5. MW standards are shown on the left.
Lysates from cells transfected with either TEL2 + HATEL1 or TEL2 + HAΔTEL1 were incubated with α-T2, and IP products were subjected to SDS-PAGE, Western blotted, and incubated with 12CA5. Figure 7B shows that HAT1 does associate with TEL2. Additionally, Figure 7B shows that this interaction is mediated through the PNT domain of TEL1. To ensure that the TEL1 proteins were not specifically being immunoprecipitated by α-T2, this experiment was also performed with Hela cell lysates from cells cotransfected with HAT2 construct and either a TEL1 or TEL1 minus the PNT domain construct (T1 or T1ΔPNT, respectively). These lysates were immunoprecipitated with an α-TEL1 (α-T1) immunopurified antibody, subjected to SDS-PAGE, and Western blotted with 12CA5. These results, in Figure 7B, confirm the previous results (above); that is, the TEL2 protein can interact with the TEL1 protein via the PNT domain of TEL1.
Discussion
The data reported here represent the identification and initial characterization of a new member of the ETS gene family. Because of its similarity to the ETS gene TEL1, we have called this gene TEL2. The ETS family of proteins is a large group of eukaryotic transcription factors that regulate the expression of target genes involved in a wide variety of cellular functions. The biologic significance of this group of transcription factors is underscored by their evolutionary conservation, and by evidence of disrupted or aberrant expression of ETS proteins in numerous disease conditions, including cancer.
The ETS family of proteins have been subgrouped on the basis of sequence comparisons, the position of the ETS domain within each protein, and the presence of additional protein domains, such as the PNT domain.2,3,21 Sequence analysis shows that theTEL2 cDNA encodes a protein of 341 aa that contains a C-terminal ETS domain, as well as an N-terminal PNT domain. Comparison of the TEL2 sequence with the sequences of other ETS factors showed that TEL2 is most closely related to TEL1(38.2% aa identity). Furthermore, sequence comparisons of the ETS and PNT domains of TEL2 revealed that they are highly homologous to those of TEL1 (85.4% and 62.5% aa identity, respectively). Phylogenetic studies most often group TEL1 with the D-YAN protein and occasionally the D-ets4 protein.2,3 21 Therefore, the highly related TEL2 protein described here represents only the second human ETS sequence in this subgroup.
Further examination of the TEL2 aa sequence reveals a potential MAPK phosphorylation site located near the N-terminal end of the protein, approximately 37 aa upstream of the PNT domain. The activity of several ETS proteins may be regulated by Ras-induced phosphorylation.7 TEL1 contains at least 2 candidate MAPK sites, and it has been shown to be phosphorylated in vivo.23 In this context, it is interesting to note that functional MAPK sites in D-YAN, D-PNT P2, ETS1, and ETS2 proteins are located in a similar position as the potential TEL2 MAPK site.3,4 7 Additionally, 1 of the putative TEL1 MAPK sites is located in the same position. Therefore, this phosphorylation site may represent a conserved regulatory mechanism among some ETS proteins that is also present in TEL2.
The TEL2 cDNA sequence was discovered to be contained within a completely sequenced PAC clone on human chr 6p21, confirming our FISH results. The TEL2 cDNA is composed of at least 8 exons that span approximately 21 kb of DNA. Comparison of the genomic organization of TEL2 with that of TEL1 shows striking similarities. Although TEL1 spans over 240 kb of DNA on human chr 12p13, it also is encoded by 8 exons.25 Exon sizes in both genes, with the exception of exon 5, are nearly identical and encode homologous regions of each protein. In light of these similarities, it should be noted that TEL2 is 111 aa shorter than TEL1 (341 aa compared with 452 aa, respectively). Of the 111 aa difference, 105 aa are accounted for in the difference in size of exon 5 of each gene. Interestingly, a putative MAPK phosphorylation site is contained within the exon 5 encoded sequences of TEL1, which is not present in TEL2. Although the function (if any) of this phosphorylation site remains to be determined, it may prove to be significant in comparing the functions of these 2 related proteins.
Our results show that TEL2 is expressed in human hematopoietic tissues. Despite regulating a variety of developmental events and being widely expressed, the majority of known ETS factors are preferentially expressed in hematopoietic cell lineages8. Gene knock-out studies in mice have shown that ETS proteins regulate key aspects of hematopoietic development and differentiation. For example, mice lacking TEL1 die early in development and have a defect in yolk sac angiogenesis.36 Additional studies withTEL1-/-embronic stem (ES) cells in chimeric mice showed thatTEL1 is essential for bone marrow hematopoiesis, and may play a role in hematopoietic stem cell homing.37 Of the tissues examined, TEL2 is expressed predominantly in fetal liver and bone marrow. Expression is also faintly detectable in mRNA from peripheral blood leukocytes. Because TEL2 sequences were discovered in cDNA libraries made from activated T cells, the band in peripheral blood leukocyte RNA may represent expression in this cell type. Thus, given that TEL2 is expressed in hematopoietic tissues, and its high homology with TEL1, it will be important to study the role of TEL2 in normal hematopoietic development.
TEL2 is likely expressed in tissues outside the hematopoietic system, because sequences were also found in cDNA libraries made from normal human breast tissue and a human meningioma, suggesting that it is also expressed in these tissues. In fact, database searches with theTEL2 sequence indicate that it is also present in several cDNA libraries used by the National Cancer Institute-Cancer Genome Anatomy Project (NCI-CGAP).38 This is interesting because dysregulation of several ETS genes has been shown to be oncogenic.1,9,10,39 40 For example, the TEL1 gene is frequently rearranged in human leukemias of myeloid and lymphoid origin. Therefore, given that TEL2 is expressed in hematopoietic tissues, the oncogenic potential of several ETS factors in general, and in particular, the role of the highly similarTEL1 in leukemias, it will be of importance to analyze the oncogenic potential of TEL2. In this light, it is interesting to note that at least 6 ETSs matching part of theTEL2 sequence were identified from the NCI-CGAP database (data not shown). These clones were sequenced from an RER+ colon tumor (acc #AI343380), pooled germ cell tumors (acc # AA729929), a B-cell chronic lymphocytic leukemia (acc # AI394727), an adenocarcinoma (acc #AI347001), tonsil tissue enriched for germinal center B cells (acc #AA748867), and an adrenal adenoma (acc # AA604808). The identification of TEL2 sequences in these tissues may simply represent normal expression of TEL2 in the tissues from which these tumors are derived, or may indicate that aberrant expression of TEL2contributes to the genesis of these malignancies.
It was determined by EMSA that the ETS domain of TEL2 can bind the CDBS of TEL1. Indeed, ETS domains among the entire family of ETS proteins are highly homologous and many ETS proteins are able to bind similar target sequences in vitro.4,5 41 Our sequence comparisons reveal that 13 of 80 aa residues within the TEL2 ETS domain differ from the those of the TEL1 ETS domain. Therefore, these aa differences may differentiate target sequences between these 2 ETS domains. Of course, differential expression patterns of individual ETS proteins provides another level of regulation of target genes. TEL1 is ubiquitously expressed and is therefore likely expressed in the same human tissues as TEL2 (including fetal liver and bone marrow). Thus, because the TEL2 protein can bind the TEL1 CDBS in vitro, these proteins may have common gene targets in vivo. However, binding site specificity is likely determined by a complex formula of criteria and not on target sequences alone. Furthermore, ETS proteins have been shown to act as both transcriptional activators and repressors; therefore the transcriptional activity of common target genes may be differentially affected by the binding of different ETS factors.
We have shown through co-IP experiments that TEL2 interacts with TEL1. Protein-protein interactions have been demonstrated with other ETS proteins and are thought to convey yet another layer of regulation on the activity of ETS proteins. For example, the ETS protein ERG and its isoforms have been shown to form homodimers, and heterodimeric complexes with other ETS proteins via the PNT and ETS domains in vitro.31 Several ETS proteins, including ERG, ETS1, and ETS2, have been shown to interact with the AP1 transcription factors through the ETS domain, and this interaction is required for transcriptional activity.30 TEL1 also associates with the ETS protein FLI1.32 The interaction of TEL1 with FLI1 prevents activation of a FLI1 specific promoter and therefore TEL1 may inhibit the transactivation activity of FLI1. Therefore, it is significant that TEL2 interacts with TEL1 because it is possible that the transcriptional activity of both TEL2- and TEL1-target genes are regulated and/or modulated by this interaction.
Additionally, we have shown by co-IP that TEL2 associates with itself. Although not shown in this report, it is assumed that the PNT domain is required for this association. TEL1 is also known to self-associate via its PNT domain.22 It is through this ability to oligomerize that many of the TEL1 translocation products form constitutively active signaling molecules. In addition, TEL1 has been shown to be a transcriptional repressor and this repression requires dimerization/oligomerization.42 Therefore, the ability of TEL2 to self-associate may have important functional consequences.
It is also interesting to note that 2 isoforms of TEL2 are immunoprecipitated in Figure 7A. TEL1 is also expressed as 2 isoforms.23 There are at least 2 additional ATG codons in TEL2, at positions 17 and 82, that might initiate a truncated protein. However, it is assumed that the PNT domain of TEL2 is necessary for oligomerization; thus, because the smaller isoform is coimmunoprecipitated with HATEL2, it likely contains the entire PNT domain. Therefore, the smaller isoform of TEL2 probably initiates from the ATG at position 17, which would encode a protein that includes the entire PNT domain.
Acknowledgments
We are grateful to Virginia Valentine for performing the FISH analysis. We also thank Andrew Hollenbach, Craig McPherson, Edwin Meintjes, Ugur Ozbek, and Colin Pritchard for many helpful discussions of the work. Additionally, we thank Sjozef van Baal, Jacqueline Bonten, and Wenkai Dou for technical help, and Charlotte Hill for secretarial assistance.
Supported in part by NCI grant CA-72996-03, the Cancer Center (CORE) support grant CA-21765, and by the American Lebanese Syrian Associated Charities (ALSAC) of St Jude Children's Research Hospital.
Reprints:Gerard C. Grosveld, Department of Genetics, St Jude's Children Research Hospital, 332 N Lauderdale, Memphis, TN.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.





![Fig. 5. Subcellular localization of TEL2 transiently expressed in Hela cells. / Nuclear staining of TEL2, detected with α-T2 and a FITC-conjugated goat-α-rabbit secondary aby (A), or with 12CA5 and a FITC-conjugated goat-α-mouse secondary aby (C), is shown in cytospin preparations of transiently transfected Hela cells. Hela cells transfected with an empty vector expression construct, stained with the same antibodies, are also shown ([B] and [D], respectively). Original magnification, ×40.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/11/10.1182_blood.v95.11.3341/6/m_bloo01144005w.jpeg?Expires=1769125850&Signature=q0yO25iYWBKZpDUmTkYE-~Ypge1eXfxkjsMd0Z2xryalK5Y8YYGwkPxbRQuzaXm46KpubWLWEz3F6cHuod-ZqWuWp1faClE8b5SxUlMdkQbgDrw1h~gyNAhvf~1HYGJnxk2GF2aS1nvMMpGS6jZVp~qiGXyYCaf-vsA8O1WGbpNywi4iSDKDzhbXuog-khgS0XzGUp4Bq0GD5QYro2EoXOEh1ojTVMiyivtf38dUQcqf3IJKea9b1SFASIqZGuVCg1TaPIbrhSYqzsija2-MztyWW7fEZu3L6hw84LJ7ZrEYgOprCKR9xgnczMPH91BJV9-wj1YqQy77Mt-i2fvuBQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal