Abstract
Recessive congenital methemoglobinemia due to nicotinamide adenine dinucleotide (NADH)-cytochrome b5 reductase (b5R) deficiency is classified into 2 clinical types: type 1 (erythrocyte type) and type 2 (generalized type). We found a Chinese family with type 1 recessive congenital methemoglobinemia, the patients from which were diagnosed according to clinical symptoms and b5R enzyme activity in the blood cells. To learn the molecular basis of type 1 recessive congenital methemoglobinemia in this Chinese family, we isolated total RNA from the peripheral leukocytes of the propositus and b5R complementary DNA (cDNA) by reverse transcription– polymerase chain reaction (RT-PCR). The coding region of the b5R cDNA was analyzed by sequencing the cloned PCR products. The results showed that the propositus was homozygous for a G→A transition at codon 203 in exon 7, changing a cysteine to a tyrosine (Cys203Tyr). To characterize the mutant enzyme, both glutathione S-transferase (GST)-fused wild-type b5R and GST-fused mutant Cys203Tyr b5R were expressed in Escherichia coli and affinity purified. The results showed that the catalytic activity of the enzyme was not much affected by this amino acid substitution, but the mutant enzyme exhibited decreased heat stability and increased susceptibility to trypsin. These properties of the mutant enzyme would account for the restricted b5R deficiency and mild clinical manifestations of these type 1 patients. The finding of this novel mutation makes codon 203 the only position within the b5R gene at which more than 1 mutation has been found.
Nicotinamide adenine dinucleotide (NADH)-cytochrome b5 reductase (b5R, EC.1.6.2.2.) deficiency leads to 2 different types of recessive congenital methemoglobinemia. In type 1, cyanosis is the only major symptom, and b5R deficiency is restricted to red blood cells.1 In type 2,2 cyanosis is associated with severe mental retardation and neurologic impairment, and the enzyme deficiency is systemic. The b5R gene is a housekeeping gene. A membrane-bound form of b5R is found in all somatic cells and plays an important role in fatty acid metabolism, cholesterol synthesis, and in vivo transformation of drugs.3 Soluble-form b5R exists mainly in erythrocytes, where it functions in the reduction of methemoglobin. It is of great interest to learn the molecular basis of recessive congenital methemoglobinemia, because identification of different mutations occurring at different positions within the b5R gene might account for the phenotypic heterogeneity of this disease. In previous reports we have described 2 point mutations (Arg57Gln and Leu72Pro) in the b5R gene of Chinese families of type 1 recessive congenital methemoglobinemia.4 5 In this paper we present analysis of the b5R gene of another Chinese pedigree by reverse transcription–polymerase chain reaction (RT-PCR), PCR–restriction fragment length polymorphism (RFLP), and dot blot hybridization, with the identification of a G→A transition at codon 203 of exon 7. We also discuss the significance of such a novel mutation.
Patients, materials, and methods
Case report and family pedigree
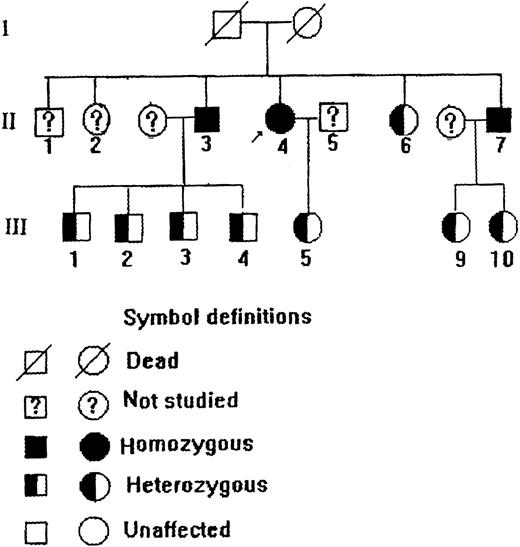
The propositus was a 75-year-old Chinese woman who was found to have persistent slate-gray cyanosis 1 year after her birth without mental retardation or any neurologic abnormalities. She could maintain normal life and labor. Laboratory studies showed (Table 1) that her methemoglobin level was elevated and the b5R activity in her erythrocytes was greatly decreased. Treatment with ascorbic acid could improve her cyanotic conditions. The propositus was diagnosed as having type 1 recessive congenital methemoglobinemia. Investigation of her family members gave such a pedigree as depicted in Figure1, in which 2 of the brothers of the propositus were also type 1 patients, and some other family members were diagnosed as heterozygous according to their methemoglobin levels and red cell b5R activities (Table 1). The diagnoses of some family members were further clarified by molecular and immunologic studies described below.
Summary of methionine hemoglobin (MetHb) level, b5R activity, and relative b5R content of the family
| Subject . | II.3 . | II.4 . | II.6 . | II.7 . | III.1 . | III.2 . | III.3 . | III.4 . | III.5 . | III.9 . | III.10 . | Control . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MetHb/total Hb (%) | 20.9 | 19.0 | 3.75 | 15.2 | 3.30 | 1.43 | 2.94 | 3.30 | 2.94 | 1.78 | 2.06 | < 2 |
| b5R activity (IU/g Hb)* | 0.84 | 0.9 | 2.5 | 2.8 | 6.5 | 8.9 | 8.5 | 10.1 | 7.4 | 10.5 | 12.1 | 12.5-22.2 |
| Relative b5R content† | 0 | 0 | NS | 0 | NS | NS | NS | NS | 0.53 | 0.56 | 0.36 | 1.0 |
| Subject . | II.3 . | II.4 . | II.6 . | II.7 . | III.1 . | III.2 . | III.3 . | III.4 . | III.5 . | III.9 . | III.10 . | Control . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MetHb/total Hb (%) | 20.9 | 19.0 | 3.75 | 15.2 | 3.30 | 1.43 | 2.94 | 3.30 | 2.94 | 1.78 | 2.06 | < 2 |
| b5R activity (IU/g Hb)* | 0.84 | 0.9 | 2.5 | 2.8 | 6.5 | 8.9 | 8.5 | 10.1 | 7.4 | 10.5 | 12.1 | 12.5-22.2 |
| Relative b5R content† | 0 | 0 | NS | 0 | NS | NS | NS | NS | 0.53 | 0.56 | 0.36 | 1.0 |
NS indicates not studied.
b5R activity was measured as NADH-ferricyanide activity.
b5R protein amount is expressed as relative to that of the normal control.
Pedigree of a Chinese family with type 1 recessive congenital methemoglobinemia.
Electrophoresis and visualization of red cell b5R on native polyacrylamide gel
A native polyacrylamide gel with a concentration gradient from 6% to 20% was prepared based on Laemmli's system without adding any sodium dodecyl sulfate (SDS) to the system and reductants and dye indicators to the loading buffer.6 For electrophoresis, red cells from various subjects were hemolyzed by adding an equal volume of 4% Triton X-100 (in 20 mmol/L Tris-HCl, pH 7.5), followed by repeated freezing and thawing. For comparison among the subjects, the hemoglobin concentrations of all hemolysates to be loaded were adjusted to the same level. After electrophoresis, the gel was stained with freshly prepared MTT-DCIP-NADH substrate solution7: 10 mmol/L phosphate buffer (pH 7.0); 0.5 mg/mL NADH (Huamei, Shanghai, China); 50 μM dichlorophenol indophenol (DCIP, Sigma, St Louis, MO); and 0.5 mg/mL 3 (4,5-dimethyl thiazoyl-2)-2,5-diphenyl tetrazolium bromide (MTT, Sigma). The staining was completed within 1 hour at room temperature. The gel was washed in distilled water to stop staining. Photographic recording of the result was best taken within 48 hours.
Determination of b5R protein in the erythrocytes by double-antibody sandwich enzyme-linked immunoabsorbent assay
b5R protein in the hemolysates was determined by double-antibody sandwich enzyme-linked immunosorbent assay (ELISA), as described in a previous report,8 using the immunoglobulin G fraction of rabbit anti-b5R serum for plate coating and horseradish peroxidase–labeled anti-b5R antibody (1E3) as the reporter. For comparison, hemolysate from a normal individual was included in the measurement, and the quantities of b5R in the hemolysates of the propositus and her family members were expressed as those relative to the b5R amount of the normal control (Table 1).
RNA isolation and RT-PCR
Total RNA was extracted from peripheral lymphocytes of the propositus and an unrelated normal control by using RNeasy Blood Mini Kit (Qiagen, Hilden, Germany). The synthesis of complementary DNA (cDNA) was performed from 1 μg of total RNA according to the instructions of the kit producer (Promega, Madison, WI). The entire b5R cDNA coding region starting with the ATG initiation site in exon 1 through the TGA stop site in exon 9 was amplified with 1 primer set (P1: 5′-GGGAATTCATGGGGGCCCAGCTCAGCACG-3′; P2: 5′-GGGGATCCCCTCAGAAGACGAAGCAGCGC-3′; synthesized on an Applied Biosystems DNA synthesizer [Perkin Elmer, Foster City, CA]) that allows the amplification of a 921-base pair (bp) fragment. The amplification reaction was performed on Gene Amp system 2400 (Perkin-Elmer Setups ) in a 50 μL reaction volume containing 5 μL of 10 × PCR buffer (500 mmol/Lol/L KCl; 100 mmol/L Tris-HCl, pH 9.0; 1% Nonodet P40), 5 μL of 25 mmol/L MgCl2, 2U of Taq polymerase (Sangon, Shangai, China), 5 μL of 2 mmol/L of each dNTP, 1 μL of 10 pmol/L of each primer, and 4 μL of cDNA. PCR reaction was started with an initial denaturation at 94°C for 5 minutes, followed by 30 cycles of denaturation at 94°C for 45 seconds, annealing at 60°C for 45 seconds, and elongation at 72°C for 60 seconds, with an additional extension at 72°C for 10 minutes after the last cycle. The PCR products were visualized by electrophoresis on 1.0% agarose gel stained with ethidium bromide.
Cloning and nucleotide sequencing of the amplified products
The RT-PCR products were purified from the agarose slice after gel electrophoresis by phenol-chloroform extraction. The purified PCR fragments were cloned into the vector pWR-4509 (kindly provided by Prof Xie Yi of Fudan University, Shanghai, China) by overnight ligation at 12°C using T4 DNA ligase (MBI Fermentas, Milan, Italy). The recombinant vector was used to transform Escherichia coliJM103. Clones of interest were screened by PCR and sequenced with the P1 and P2 primers on ABI 373A sequencer by following a dye terminator protocol.
Genomic DNA amplification and PCR/restriction analysis
The peripheral lymphocytes of the propositus, her family members, and an unrelated normal control were washed twice with phosphate-buffered saline, and genomic DNA was extracted using QiAamp Blood Kit (Qiagen). The genomic DNA was amplified with a primer set10 (5′-AGTACACCTGGGCGGGGCGG-3′ and 5′-AGAATCTAGCAGAGCTGTGCA-3′) that allows the amplification of a 262-bp fragment of exon 7. The PCR reaction volume was 25 μL, containing 2.5 μL of 10 × PCR buffer, 2.5 μL of 25 mmol/L MgCl2, 2.5 μL of 2 mmol/L of each dNTP, 1 μL of 10-mmol/L of each primer, and 2U of Taq polymerase (Sangon). Thirty cycles of denaturation at 94°C for 1 minute, annealing at 60°C for 1 minute, and elongation at 72°C for 1 minute were carried out, followed by an extended incubation at 72°C for 10 minutes. The amplified products were purified by phenol-chloroform extraction, digested with RsaI (MBI) at 37°C for 60 minutes, and subjected to electrophoresis in 12% polyacrylamide gel.
Dot blot hybridization with allele-specific oligonucleotide probes
Digoxigenin (DIG) Oligonucleotide Tailing Kit and DIG Nucleic Acid Detection Kit were purchased from Boehringer Mannheim (Mannheim, Germany). PCR products of genomic DNA from the propositus, her family members, and 3 unrelated normal controls were loaded onto nitrocellulose membranes and then baked for 2 hours at 80°C. The 1-hour prehybridization was done at 54°C for the mutant probe or at 60°C for the normal probe in prehybridization buffer: 5 × SSC (0.75 mol/L NaCl, 0.075 mol/L sodium citrate, pH 7.0), 1% blocking reagent (in 0.1 mol/L maleic acid, 0.15 mol/L NaCl, pH 7.5), 0.1% N-lauroylsarcosine, 0.02% SDS. The probe WA I (5′AGAGCAGGTGGCACACAG-3′) for normal allele (normal probe) and the probe WA II (5′-ACTGTGTACCACCTGCTCT-3′) for the mutated allele (mutant probe) were labeled by tailing incorporation of a digoxigenin-labeled nucleotide. The hybridization reaction was initiated by adding the labeled probes directly to the prehybridization mixture. After 6 hours of hybridization, the membranes were washed twice for 5 minutes with 2 × SSC, 0.1% SDS at room temperature, and twice for 15 minutes with 0.1 × SSC, 0.1% SDS at 60°C for the normal probe or at 54°C for the mutant probe. The immunologic detection was carried out as described in the product manual. Briefly, the filters were incubated at room temperature for 30 minutes in 1% blocking reagent containing 150 mU/mL anti-digoxigenin-alkaline phosphatase conjugate. The color reaction was initiated by the addition of 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium salt and was completed in 16 hours.
Expression, purification, and characterization of GST-fused b5R
To construct glutathione S-transferase (GST)-fused b5R expression vector, cDNA fragments encoding soluble-form normal and mutant b5R were isolated by PCR from the corresponding pWR-450 recombinants with 1 primer set (5′-GGGAATTCCCCTCAGAAGACGAAGCA-3′ and 5′-GGGGATCCTTCCAGCGCTCCACGCC-3′). The wild-type and mutant b5R cDNA fragments were inserted into the GST fusion gene vector, pGEX-2T (Pharmacia, Piscataway, NJ). Only 1 extra amino acid residue was introduced into the cloning (BamHI) site.E. coli BL21 harboring the expression plasmid was incubated overnight at 37°C in 3 mL of L-broth medium containing 100 μg/mL ampicillin. The culture was inoculated into 50 mL of L-broth medium and incubated at 37°C. After an initial incubation for 2.5 hours, isopropyl-β-D-thiogalactopyranoside was added to the medium, with a final concentration of 0.1 mmol/L, and incubated for another 4 hours. The bacterial cells were pelleted by centrifugation (5000g for 5 minutes), resuspended in sterile water, lysed by sonication, and spun at full speed on a microcentrifuge for 10 minutes to remove insoluble materials. The supernatant was transferred to Glutathione Sepharose 4B Microspin Column (Pharmacia) and mixed gently at room temperature for 10 minutes to ensure optimal binding of GST proteins to the Glutathione Sepharose 4B matrix. After washing the column twice with phosphate-buffered saline, elution was performed with the elution buffer containing 10 mmol/L reduced glutathione. The concentrations of fusion proteins were determined by measuring light absorbance at 280 nm. Purity of the GST-fused b5Rs was evaluated by 10% SDS-polyacrylamide gel electrophoresis.6 Western blot analysis of GST-fused b5Rs was performed according to the standard method.11 GST-fused b5Rs were separated on 10% SDS-polyacrylamide gels and blotted onto nitrocellulose filters by eletrophoretic transfer. The nitrocellulose filters were incubated with anti-b5R monoclonal antibody (1E3)12 for 1 hour at room temperature. Biotinylated goat antimouse immunoglobulin G was used as the second antibody (Zymed, San Francisco, CA). The GST-fused b5Rs were identified on the membrane by color development using 4-chloro-1-naphthol and H2O2 as the substrates of streptavidin-conjugated horseradish peroxidase. A b5R preparation (rb5R, kindly provided by Dr Yubisui, Oita University, Oita, Japan) was used as control.
Analysis of enzyme properties of GST-fused b5Rs
Kinetic parameters of the GST-fused wild-type b5R and GST-fused Cys203Tyr mutant b5R were assayed with DCIP as the electron acceptor.13 A standard reaction mixture (3 mL) contained 0.03 mmol/L NADH, 0.05 mmol/L Tris-HCl (pH 7.5), 20 μg of b5R fusion protein, 0.1 mmol/L ethylenediamine tetraacetic acid (EDTA), and 0.06 mmol/L DCIP. The Km values of each enzyme were determined by Lineweaver-Burk double-reciprocal plot. TheKcat values were calculated by dividing the maximal reaction velocity by the enzyme amount in mole. According to the method of Higasa et al,14 stability to heat was tested by incubating the b5Rs at various temperatures for 10 minutes and at 37°C for various times, and susceptibility to trypsin was examined by incubation for various times at 37°C with 1.5 U/μL trypsin. In such cases, the enzyme activity was checked by using potassium ferricyanide as an electron acceptor15 and expressed as percentages of the activity before heat or trypsin treatment for the sake of convenience. A standard reaction mixture contained 30 μL of 100 μmol/L NADH, 0.25 mL of 10 mmol/L EDTA, 0.5 mL of 50 mmol/L citrate buffer (pH 4.5), 0.8 mL of 0.2 mmol/L potassium ferricyanide, and 0.5 mL of 11.68 g/L b5R-free hemoglobin. NADH-ferricyanide activity was measured at 575 nm at 25°C with Shimadzu UV-240 spectrophotometer (Shimadzu, Kyoto, Japan).
Other methods
Total hemolysate hemoglobin concentrations were determined with an automatic blood analyzer (Coulter JT-IR, Miami, FL), the built-in protocol of which eliminates any interference of the determination by methemoglobin. Methemoglobin level was determined by the classical method of Evelyn and Malloy16 and expressed as a percentage of total hemoglobin (Table 1).
Results
Visualization of b5R activity in native gel and quantification of b5R protein
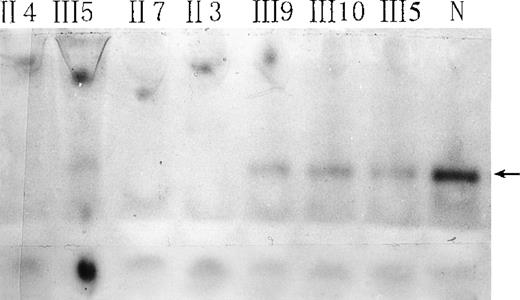
Red-cell b5R from the propositus, some of her family members, and a normal subject was studied by native gel electrophoresis. Submerging the gel in MTT-DCIP-NADH substrate solution to stain b5R in polyacrylamide gel. A diaphorase band appeared behind that of hemoglobin (Figure 2). No NADH-diaphorase activity can be observed in the hemolysates of the propositus and 2 of her brothers by this method. Her daughter and nephews had less intensive NADH-diaphorase bands than those of the normal subject. An ELISA method showed that the content of b5R protein in the erythrocytes of the daughter and nephews was much smaller than that of a normal control, and no b5R protein could be detected in the hemolysates of the propositus and 2 of her brothers (Table 1).
Native gel electrophoresis of red-cell NADH-diaphorase.
The arrow indicates the position of NADH-diaphorase bands. N indicates a normal individual. Hemoglobin concentrations in all hemolysates were adjusted to 93 g/L, and 30 μL of each sample was loaded.
Native gel electrophoresis of red-cell NADH-diaphorase.
The arrow indicates the position of NADH-diaphorase bands. N indicates a normal individual. Hemoglobin concentrations in all hemolysates were adjusted to 93 g/L, and 30 μL of each sample was loaded.
Sequence analysis of the patient's b5R gene
A fragment of 921 bp from PCR amplification of b5R cDNA was detected on 1.0% agarose gel. This fragment was purified, cloned, and sequenced. Sequencing results (Figure 3) revealed only 1 notable base change in the propositus, a G→A transition in exon 7 at the second position of codon 203, causing an amino acid change from cysteine to tyrosine.
Nucleotide sequence analysis of the b5R gene from the propositus.
The sequence shown corresponds to the nonencoding strand of the gene of a normal individual (left) or the propositus (right). The arrow indicates a homozygous G→A transition at the second position of codon 203. This mutation results in a replacement of cysteine by tyrosine.
Nucleotide sequence analysis of the b5R gene from the propositus.
The sequence shown corresponds to the nonencoding strand of the gene of a normal individual (left) or the propositus (right). The arrow indicates a homozygous G→A transition at the second position of codon 203. This mutation results in a replacement of cysteine by tyrosine.
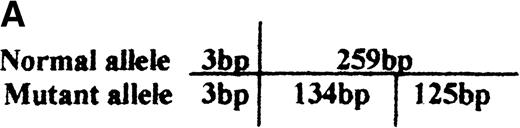
PCR/restriction enzyme analysis and dot blot hybridization
To confirm that this mutation was not an artifact derived from the misincorporation of Taq polymerase during PCR, we performed PCR-RFLP and dot blot hybridization. Because this nucleotide replacement created another RsaI recognition site within the 262-bp fragment (Figure 4A), the PCR products from genomic DNA were digested by RsaI and separated on 12% polyacrylamide gel. The 262-bp PCR product of the propositus was cleaved into 2 smaller fragments of 134 bp and 125 bp, as predicted (Figure 4B). In addition, the amplified DNA of the propositus, 5 of her family members, and 3 normal controls was allowed to interact with an oligonucleotide probe specific either for the normal (WA I) or for the mutant (WA II) allele. It was shown (Figure 5) that the mutant-specific probe was hybridized to the PCR products of the propositus and 2 of her brothers (II.3 and II.7) but not to any specimen from the normal individuals, and the normal probe was hybridized to the PCR products of the normal individuals but not to any specimen from the propositus and 2 of her brothers. The amplified DNA of her daughter (III.5) and her nephews (III.9 and III.10) could be hybridized to both the mutant and normal probes. These facts indicated that the propositus and her brothers were homozygous and her daughter and nephews were heterozygous for this G→A substitution.
Identification of the missense mutation in exon 7 of the b5R gene by PCR/restriction analysis.
(A) Diagram of genomic restriction enzyme analysis. The 262-bp genomic fragment of exon 7 was amplified. The mutated allele possesses an additional RsaI site and produces 3 fragments (3 bp, 125 bp, 134 bp) upon digestion. (B) Restriction enzyme analysis. The 262-bp genomic PCR fragments from the propositus, some of her family members, and a normal control were digested with RsaI and electrophoresed on 12% polyacrylamide gel. The 2 visible products of digestion are indicated by arrows. The DNA markers used areBsuRI digests of pBR322 (MBI). N1 and N2 indicate the PCR products of the normal control after and before digestion, respectively.
Identification of the missense mutation in exon 7 of the b5R gene by PCR/restriction analysis.
(A) Diagram of genomic restriction enzyme analysis. The 262-bp genomic fragment of exon 7 was amplified. The mutated allele possesses an additional RsaI site and produces 3 fragments (3 bp, 125 bp, 134 bp) upon digestion. (B) Restriction enzyme analysis. The 262-bp genomic PCR fragments from the propositus, some of her family members, and a normal control were digested with RsaI and electrophoresed on 12% polyacrylamide gel. The 2 visible products of digestion are indicated by arrows. The DNA markers used areBsuRI digests of pBR322 (MBI). N1 and N2 indicate the PCR products of the normal control after and before digestion, respectively.
Dot blot hybridization of PCR-amplified genomic fragment spanning exon 7.
PCR products from the propositus, 5 of her family members, and 3 healthy individuals were hybridized with the normal allele-specific oligonucleotide probe (A) or mutant allele-specific oligonucleotide probe (B) as described in “Materials and methods”; 1: three normal controls; 2: three homozygotes from the family (from top to bottom: II.3, II.4, and II.7); 3: three heterozygotes (from top to bottom: III.5, III.9, and III.10).
Dot blot hybridization of PCR-amplified genomic fragment spanning exon 7.
PCR products from the propositus, 5 of her family members, and 3 healthy individuals were hybridized with the normal allele-specific oligonucleotide probe (A) or mutant allele-specific oligonucleotide probe (B) as described in “Materials and methods”; 1: three normal controls; 2: three homozygotes from the family (from top to bottom: II.3, II.4, and II.7); 3: three heterozygotes (from top to bottom: III.5, III.9, and III.10).
Preparation and identification of fusion-expressed b5R proteins
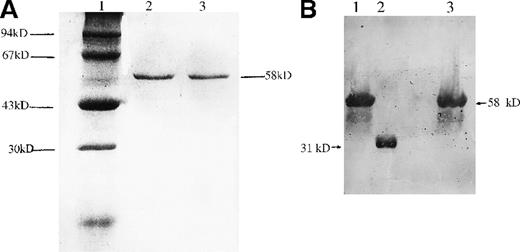
To characterize the effect of the Cys203Tyr missense mutation on the biochemical properties of b5R, the normal and mutant b5R cDNAs were inserted into GST fusion gene expression vector separately. GST-fused wild-type b5R and GST-fused Cys203Tyr b5R were expressed in E. coliBL21and purified as described in “Materials and methods.” A single band of 58 kd was observed after purification (Figure6A). To determine whether GST-fusion proteins expressed in E. coli were b5R proteins, Western blot analysis was carried out with a monoclonal antibody against b5R. As shown in Figure 6B, a single band with molecular mass of 58 kd was revealed. These results confirmed that GST-fusion proteins expressed inE. coli were indeed b5R proteins.
Gel electrophoresis and Western blotting of GST-fused b5R and GST-fused Cys203Tyr b5R.
(A) Gel electrophoresis of GST-fused b5Rs expressed in E. coliBL21. A total of 5 μg of GST-fused b5R or GST-fused Cys203Tyr b5R purified with glutathione Sepharose 4B was subjected to electrophoresis on 10% SDS-polyacrylamide gel. Lane 1, molecular weight markers used are phosphorylase B (94 kd), bovine serum albumin (67 kd), actin (43 kd), and bovine carbonic anhydrase (31 kd). Lane 2, GST-fused Cys203Tyr b5R. Lane 3, GST-fused wild-type b5R. (B) Western blotting of GST-fused b5R and GST-fused Cys203Tyr b5R purified with glutathione Sepharose 4B. Lane 1, GST-fused wild-type b5R. Lane 2, known rb5R as control (see text). Lane 3, GST-fused Cys203Tyr b5R.
Gel electrophoresis and Western blotting of GST-fused b5R and GST-fused Cys203Tyr b5R.
(A) Gel electrophoresis of GST-fused b5Rs expressed in E. coliBL21. A total of 5 μg of GST-fused b5R or GST-fused Cys203Tyr b5R purified with glutathione Sepharose 4B was subjected to electrophoresis on 10% SDS-polyacrylamide gel. Lane 1, molecular weight markers used are phosphorylase B (94 kd), bovine serum albumin (67 kd), actin (43 kd), and bovine carbonic anhydrase (31 kd). Lane 2, GST-fused Cys203Tyr b5R. Lane 3, GST-fused wild-type b5R. (B) Western blotting of GST-fused b5R and GST-fused Cys203Tyr b5R purified with glutathione Sepharose 4B. Lane 1, GST-fused wild-type b5R. Lane 2, known rb5R as control (see text). Lane 3, GST-fused Cys203Tyr b5R.
Kinetic properties of the expressed enzymes
The enzyme activity of mutant b5R was almost the same as that of the wild-type enzyme when measured at the same concentration. Kinetic properties of the wild-type and mutant enzymes are summarized in Table2. The Km values of the GST-fused wild-type b5R for DCIP and NADH were 26 μmol/L and 68 μmol/L, respectively. The Km values of the mutant b5R were close to those of the wild type. Little difference between the wild-type and mutant b5Rs was noted in Kcatvalues, as well as in the Kcat/Km values. These results indicated that the kinetic properties of mutant enzyme b5R were essentially the same as those of the wild-type b5R, suggesting that the cysteine to tyrosine substitution at residue 203 does not affect the affinity for NADH and DCIP and the catalytic activity of b5R.
Kinetic properties of the GST-fused wild-type and mutant (Cys203Tyr) b5Rs
| Enzyme . | Km (NADH) (μmol/L) . | Km (DCIP) (μmol/L) . | Kcat (NADH) (s−1) . | Kcat (DCIP) (s−1) . | Kcat/Km (NADH) (s−1) · (mol/L)−1 . | Kcat/Km (DCIP) (s−1) · (mol/L)−1 . |
|---|---|---|---|---|---|---|
| Wild type | 26 | 68 | 376 | 673 | 1.4 × 107 | 9.9 × 106 |
| Mutant type | 22 | 66 | 368 | 645 | 1.7 × 107 | 9.8 × 106 |
| Enzyme . | Km (NADH) (μmol/L) . | Km (DCIP) (μmol/L) . | Kcat (NADH) (s−1) . | Kcat (DCIP) (s−1) . | Kcat/Km (NADH) (s−1) · (mol/L)−1 . | Kcat/Km (DCIP) (s−1) · (mol/L)−1 . |
|---|---|---|---|---|---|---|
| Wild type | 26 | 68 | 376 | 673 | 1.4 × 107 | 9.9 × 106 |
| Mutant type | 22 | 66 | 368 | 645 | 1.7 × 107 | 9.8 × 106 |
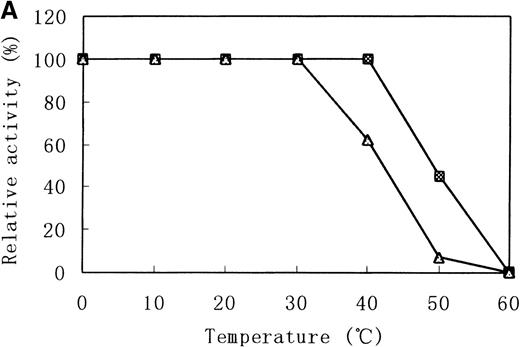
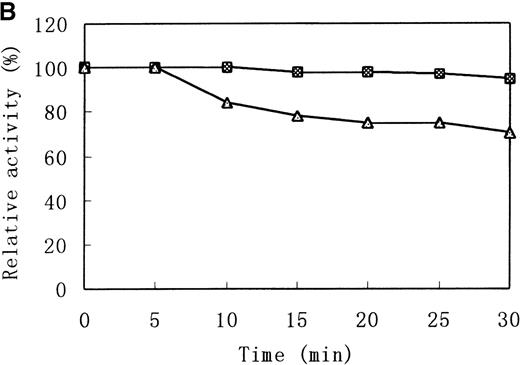
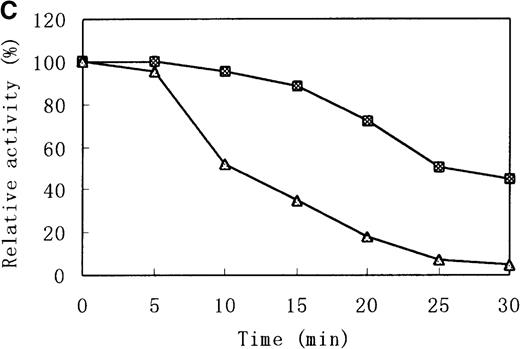
Heat stability and protease susceptibility of GST-fused b5Rs
There were distinct differences in heat stability between the mutant enzyme and the wild-type enzyme. Residual activities of the mutant enzyme after 10 minutes at 37°C and 50°C were about 70% and less than 5% of the initial activity, respectively, whereas the wild-type b5R retained 50% of the initial activity after 10 minutes at 50°C (Figure 7A). After a 30-minute incubation at 37°C, enzyme activity of the wild type dropped only slightly (more than 95% initial activity), but mutant Cys203Tyr b5R was heat-inactivated to less than 70% of the initial activity under the same conditions (Figure 7B). We made use of trypsin to reveal the protease sensitivity of the mutant Cys203Tyr b5R. The mutant b5R showed only 5% of the initial activity after being incubated with 3 μmol/L trypsin for 30 minutes at 37°C, whereas the wild-type b5R showed much higher resistance to the action of trypsin, retaining 47% of the initial activity under the same conditions (Figure 7C).
Heat stability and trypsin susceptibility of the wild-type and mutant b5R.
b5R was diluted to 2 μg/μL with 50 mmol/L Tris-HCl (pH 8.0) containing 1 mmol/L EDTA and 0.1 mmol/L DTT. □, wild type; ▵, mutant. (A) Samples were incubated for 10 minutes at various temperatures. (B) Incubation at 37°C for various times. (C) Incubation for various times at 37°C with 1.5 U/μL trypsin.
Heat stability and trypsin susceptibility of the wild-type and mutant b5R.
b5R was diluted to 2 μg/μL with 50 mmol/L Tris-HCl (pH 8.0) containing 1 mmol/L EDTA and 0.1 mmol/L DTT. □, wild type; ▵, mutant. (A) Samples were incubated for 10 minutes at various temperatures. (B) Incubation at 37°C for various times. (C) Incubation for various times at 37°C with 1.5 U/μL trypsin.
Discussion
The organization of the NADH-cytochrome b5 reductase gene has been determined in rats and humans. The human gene is about 31 kilobases long with 9 exons and 8 introns. The translational start site is in exon 1. Human liver b5R cDNA has a 903-bp open reading frame, predicting a protein of 301 residues.17 Identification of different mutations occurring at different positions within the b5R gene might account for the phenotypic heterogeneity of this disease. Meanwhile, the characterization of mutant b5R protein might provide insight into the understanding of the molecular basis of 2 types of recessive congenital methemoglobinemia and shed light on the correlation between protein structure and activity of mutant enzymes.
Mutations of the b5R gene are highly heterogeneous. Seventeen mutations in the b5R gene have been reported, including 6 missense mutations (Arg57Gln,4,18 Val105Met,19Leu148Pro,18 Glu212Lys,20Leu72Pro,5 Ala178Val14) in the case of type 1 and, in the case of type 2, 3 missense mutations (Ser127Pro,21 Pro95His,22Cys203Arg10), 3 nonsense mutations (Thr42stop,22 Arg218stop,10Arg83stop14), 2 in-frame 3-bp deletions (del phe-298,23 del Met-27210), 1 exon 6 deletion,24 and 2 splicing mutations (5′ splice site of the exon 510 and 3′ splice site of exon 925). The Arg57Gln mutation has been found in 3 unrelated Japanese and 2 unrelated Chinese families, and no other mutations have been found in more than 1 family. Interestingly, no mutations in the upstream regulatory regions have been reported.
In this paper, we present the study on the molecular basis of type 1 recessive congenital methemoglobinemia in another Chinese family. The results showed that the propositus and 2 of her brothers were homozygous for a Cys203Tyr (TGC→TAC) missense mutation, whereas her daughter and nephews were heterozygous for this mutation. In 1996, Vieira et al reported a compound heterozygote in a Spanish–French family with type 2 recessive congenital methemoglobinemia, 1 allele of which was a Cys→Arg (TGC→CGC) mutation at codon 203, the other a 3-bp deletion eliminating the methionine residue at codon 272.10 The present finding of the Cys203Tyr mutation in a Chinese family makes codon 203 the only codon within the b5R gene where more than 1 mutation has been pinpointed.
To clarify the effects of Cys203Tyr missense mutation on the enzyme properties, GST-fused Cys203Tyr b5R and wild-type b5R were expressed inE. coli BL21 and purified to homogeneity for kinetic analysis. The Km and Kcat values of the mutant enzyme for DCIP and NADH were essentially the same as those of the wild type, indicating that the mutant enzyme had roughly normal catalytic activity. However, there were remarkable differences in heat stability and susceptibility to trypsin between the mutant and wild-type enzymes, ie, the mutant enzyme was less thermostable than the wild type and more susceptible to trypsin. In mature erythrocytes, where protein-synthesizing machinery is lost, such a mutation would greatly shorten the half-life of b5R molecules, whereas in somatic cells, where b5R protein is constantly replenished, gene mutations resulting in chemophysically less stable but catalytically normal b5R would not cause much damage to the total cellular b5R activity. In other words, properties of the mutant enzyme revealed in the present study would well explain why b5R deficiency was restricted to erythrocytes of the patients and why the patients' symptoms were so mild. Similar results have been documented by Higasa et al.14
According to a prediction on the secondary structure of b5R,26 Cys203 residue is located in the β2 sheet of the NADH-binding domain and is 1 of the 4 cysteine residues (Cys203, Cys273, Cys283, and Cys297) found in b5R protein. Our finding that the mutant Cys203Tyr b5R had roughly normal catalytic properties implies that the Cys203 residue would not play a key role in NADH binding. This is in accordance with a previous report indicating that the Cys203, together with the other 3 cysteine residues, is not essential in the catalytic reaction27; nevertheless, replacement of the Cys203 residue by tyrosine would influence either the formation of disulfide bond or conformation of the enzyme, resulting in instability of b5R enzyme.
Acknowlegments
We thank Mr Dezhu Zheng for determining hemoglobin concentration and Professor Yi Xie of Fudan University for providing pWR-450 plasmid and useful comments.
The sequence reported in this paper has been submitted to GenBank. The accession number is AJ0110118.
Reprints:Yao Wang, Research Laboratories, Center for Laboratory Medicine, Fuzhou General Hospital, 156 Xihuanbei Road, Fuzhou City, Fujian Province, 350025, People's Republic of China; e-mail: SZQ@public.fz.fj.cn or yctang@pub2.fz.fj.cn.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal