Abstract
Twenty-five patients (22 adults and 3 infants) withALL1/AF4-positive acute lymphoblastic leukemia (ALL) were prospectively monitored by reverse transcriptase-polymerase chain reaction (RT-PCR) between January 1992 and July 1999. After high-dose induction and consolidation chemotherapy without bone marrow transplantation, all patients had a complete hematologic remission. Using nested RT-PCR (sensitivity 10−4), we observed conversion to PCR negativity in 11 (44%) of the patients. Thirteen of the 14 patients who did not have a molecular remission had a relapse at a median time of 4 months (range, 1 - 20 months). Of the 11 patients who had a conversion to PCR negativity, 5 reconverted to PCR positivity within 1 to 14 months. These 5 patients all progressed to hematologic relapse after 2, 3, 4, 4, and 7 months, respectively. Of the remaining 6 patients, 4 are in persistent hematologic and molecular remission at 12, 14, 88, and 96 months, whereas 2 are early in their follow-up. Actuarial probabilies of relapse and overall survival were 100% and 0% at 14 and 24 months and 67% and 43% at 96 and 100 months, respectively, in patients who had persistent RT-PCR positivity and in those who had a molecular remission. For both relapse and survival, the differences observed between the two groups were significant (P = .003 andP < .005, respectively). This study, which represents the first prospective analysis of residual-disease monitoring carried out in a substantial series of patients with t(4;11)-positive ALL, emphasizes the clinical relevance of RT-PCR-based methods to monitor minimal residual disease in this leukemia subset. (Blood. 2000;95:96-101)
In recent years, cloning of recurrent chromosome translocations associated with hematologic malignancies has led to a better understanding of the mechanisms underlying the pathogenesis of both leukemia and lymphoma.1-3 The characterization of genes involved in these karyotypic aberrations has also allowed the use of molecular strategies for a rapid recognition of distinct genetic abnormalities of diagnostic relevance and the sensitive monitoring of minimal residual disease (MRD).4 In particular, reverse transcriptase-polymerase chain reaction (RT-PCR) assays, which permit the identification of a single leukemic cell among 103 to 106 normal cells, have been used extensively to better assess response to therapy and to identify, during hematologic remission, patients with the highest risk of relapse.5-10
For the acute leukemias, RT-PCR studies have proved to be particularly useful in acute promyelocytic leukemia, in which RT-PCR detection of the specific PML-RARα fusion transcript during remission was found to correlate with subsequent relapse, suggesting that information provided by these studies may be used for early implementation of salvage treatment modalities.7
With regard to acute lymphoblastic leukemia (ALL), chromosomal translocations have been demonstrated in more than 50% of cases. The t(4;11)(q21;q23), one of the most frequent abnormalities, characterizes a subset of ALL with aggressive clinical features, such as hyperleukocytosis, organomegaly, frequent central nervous system involvement, and poor prognostic outcome. The prevalence of this aberration in ALL varies among age groups, being extremely high in infants aged < 12 months (up to 70% of cases) and more uncommon in children and adults (5% and 10% of cases, respectively).11-15 At the molecular level, the t(4;11) fuses the ALL1 gene (MLL, HRX, Hrtx1) on chromosome 11 band q23 to the AF4 gene (FEL) on chromosome 4 band q21, resulting in an ALL1/AF4 chimeric gene.16-21Because this gene is transcribed into a hybrid mRNA, RT-PCR assays have been developed to amplify the t(4;11)-associated transcript and aid rapid diagnosis and monitoring of MRD.8,23 Using a 2-round (nested) RT-PCR with a sensitivity level of 104, Janssen et al9 showed that patients in long-term complete remission (CR) had no detectable ALL1/AF4 transcripts in their bone marrow. This finding was confirmed by our retrospective study in 12 patients with ALL1/AF4-positive ALL.10 Moreover, we showed that the inability to achieve a molecular remission (ie, the persistence of PCR positivity after treatment) was invariably associated with a subsequent hematologic relapse.10
It was recently reported that, by using 2-round RT-PCR, low levels ofALL1/AF4 transcript may be found in hematopoietic tissues from healthy individuals, as well as in a proportion of children with ALL without cytogenetically detectable t(4;11) or an unfavorable clinical outcome.24 These data raised concerns about the use of RT-PCR for detecting MRD in patients withALL1/AF4-positive ALL.24,25
We here report a prospective RT-PCR study in 25 patients withALL1/AF4-positive ALL. In all cases the leukemic clone was characterized at diagnosis by cytogenetic or Southern blot evidence of t(4;11) or ALL1 gene rearrangement. Our results indicate that PCR status during remission is predictive of the clinical outcome, thereby supporting the value of molecular monitoring studies in this subgroup of patients with ALL.
Materials and methods
Patient samples
Sequential bone marrow samples from 25 patients withALL1/AF4-positive ALL were obtained at diagnosis and during the clinical follow-up. The diagnosis of ALL was established according to standard morphocytochemical and immunophenotypic criteria.26 Eight of the patients were diagnosed and treated at the Department of Cellular Biotechnologies and Hematology, University La Sapienza, Rome, between 1992 and 1996 (patients 1 to 8 on Table 1). One additional infant patient (patient 24 on Table 1) was diagnosed and treated at the Hematologic Unit, Bambino Gesù Pediatric Hospital, Vatican City. The remaining 16 patients, in whom ALL was diagnosed after 1996, were enrolled in the Italian GIMEMA 0496 multicenter study (ie, patients 9 to 23 and patient 25 on Table 2). This study includes a centralized program of cell-sample handling at diagnosis and at predetermined intervals during the clinical follow-up of patients. According to this program, cytogenetic and molecular analyses in the 16 patients were performed at diagnosis in the laboratories of the Department of Cellular Biotechnologies and Hematology, University La Sapienza; the Department of Biomedical Sciences, University of Turin; the Department of Hematology, University of Perugia; and the Department of Biomedical Sciences, Hematology Unit, University of Ferrara. In all cases, the molecular monitoring of MRD by RT-PCR was done at the laboratory of the Department of Cellular Biotechnologies and Hematology, University of Rome. Patients 1, 2, and 3 were also included in our previous retrospective study.10Their follow-up has now been extended by 3 years.
Clinical and biologic features, treatment, and clinical outcome in 25 patients with ALL1/AF4-positive acute lymphoblastic leukemia
| Patient . | Age (y)/ Sex . | WBC (109/L) . | Karyotype . | ALL1 Gene Status . | Immunophenotype . | Treatment . | Treatment Resp HR/MR . | Clinical Outcome . |
|---|---|---|---|---|---|---|---|---|
| 1 | 29/F | 380 | 46,XX,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0288 | Yes/yes | 1st CCR 96+ mo |
| 2 | 52/F | 328 | 46 XX,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0288 | Yes/yes | 1st CCR 90+ mo |
| 3 | 33/F | 17 | 46,XX,t(4;11)(q21;q23) | R | Pre-pre B* | GIMEMA 0288 | Yes/no | Rel 10 mo; died 12 mo |
| 4 | .3/F | 17 | 46;XX,t(4;11)(q21;q23) | R | Pre-pre B* | AIEOP 9503 | Yes/no | Rel 9 mo; died 16 mo |
| 5 | 71/F | 126 | NE | R | Common | VCR + IDA + PDN† | Yes/no | Rel 4 mo; died 11 mo |
| 6 | 21/F | 290 | 46,XX,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0394 | Yes/yes | Rel 5 mo; died 15 mo |
| 7 | 26/F | 270 | NE | R | Pre-pre B | GIMEMA 0394 | Yes/no | Rel 18 mo; died 19 mo |
| 8 | .8/M | 34 | 46,XY,t(4;11)(q21;q23) | R | Pre-pre B | AIEOP 9503 | Yes/yes | Rel 14 mo; died 20 mo |
| 9 | 38/M | 22.4 | 46,XY,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0496 | Yes/no | Rel 6 mo; died 8 mo |
| 10 | 44/F | 93 | 46,XX,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0496 | Yes/no | Rel 7 mo; died 8 mo |
| 11 | 16/M | 21.6 | 46,XY,17p- | R | Pre-pre B | GIMEMA 0496 | Yes/yes | Rel 9 mo; 2nd CCR 28+ mo |
| 12 | 42/F | 8.6 | 46,XX,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0496 | Yes/no | Rel 6 mo; 2nd CCR 12+ mo |
| 13 | 21/M | 131.5 | 46,XY,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0496 | Yes/yes | Rel 22 mo; died 25 mo |
| 14 | 58/F | 227 | 46,XX,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0496 | Yes/no | Rel 8 mo; died 12 mo |
| 15 | 39/M | 274 | 46,XX,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0496 | Yes/no | Rel 4 mo; died 9 mo |
| 16 | 49/M | 19.6 | NE | R | Pre-pre B | GIMEMA 0496 | Yes/no | Rel 11 mo; died 13 mo |
| 17 | 25/M | 41.2 | 46,XY | R | Pre-pre B | GIMEMA 0496 | Yes/yes | Rel 14 mo; died 17 mo |
| 18 | 49/F | 41.4 | NE | R | Pre-pre B | GIMEMA 0496 | Yes/no | Rel 14 mo; died 15 mo |
| 19 | 28/M | 281 | NE | R | Pre-pre B | GIMEMA 0496 | Yes/no | Rel 3 mo; died 4 mo |
| 20 | 47/M | 60 | 46,XY,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0496 | Yes/yes | 1st CCR 14+ mo |
| 21 | 47/M | 135 | 46,XY,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0496 | Yes/yes | 1st CCR 12+ mo |
| 22 | 60/F | 112 | 46,XX,t(4;11)(q21;q23) | ND | Pre-pre B | GIMEMA 0496 | Yes/no | Rel 6 mo; died 9 mo |
| 23 | 53/F | 75 | 46,XX,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0496 | Yes/yes | 1st CCR 7+ mo |
| 24 | .1/M | 128 | 46,XY,t(4;11)(q21;q23) | R | Pre-pre B | INTERFANT 99 | Yes/yes | 1st CCR 3+ mo |
| 25 | 40/F | 41 | ND | R | Pre-pre B | GIMEMA 0496 | Yes/no | 1st CCR 3+ mo |
| Patient . | Age (y)/ Sex . | WBC (109/L) . | Karyotype . | ALL1 Gene Status . | Immunophenotype . | Treatment . | Treatment Resp HR/MR . | Clinical Outcome . |
|---|---|---|---|---|---|---|---|---|
| 1 | 29/F | 380 | 46,XX,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0288 | Yes/yes | 1st CCR 96+ mo |
| 2 | 52/F | 328 | 46 XX,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0288 | Yes/yes | 1st CCR 90+ mo |
| 3 | 33/F | 17 | 46,XX,t(4;11)(q21;q23) | R | Pre-pre B* | GIMEMA 0288 | Yes/no | Rel 10 mo; died 12 mo |
| 4 | .3/F | 17 | 46;XX,t(4;11)(q21;q23) | R | Pre-pre B* | AIEOP 9503 | Yes/no | Rel 9 mo; died 16 mo |
| 5 | 71/F | 126 | NE | R | Common | VCR + IDA + PDN† | Yes/no | Rel 4 mo; died 11 mo |
| 6 | 21/F | 290 | 46,XX,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0394 | Yes/yes | Rel 5 mo; died 15 mo |
| 7 | 26/F | 270 | NE | R | Pre-pre B | GIMEMA 0394 | Yes/no | Rel 18 mo; died 19 mo |
| 8 | .8/M | 34 | 46,XY,t(4;11)(q21;q23) | R | Pre-pre B | AIEOP 9503 | Yes/yes | Rel 14 mo; died 20 mo |
| 9 | 38/M | 22.4 | 46,XY,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0496 | Yes/no | Rel 6 mo; died 8 mo |
| 10 | 44/F | 93 | 46,XX,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0496 | Yes/no | Rel 7 mo; died 8 mo |
| 11 | 16/M | 21.6 | 46,XY,17p- | R | Pre-pre B | GIMEMA 0496 | Yes/yes | Rel 9 mo; 2nd CCR 28+ mo |
| 12 | 42/F | 8.6 | 46,XX,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0496 | Yes/no | Rel 6 mo; 2nd CCR 12+ mo |
| 13 | 21/M | 131.5 | 46,XY,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0496 | Yes/yes | Rel 22 mo; died 25 mo |
| 14 | 58/F | 227 | 46,XX,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0496 | Yes/no | Rel 8 mo; died 12 mo |
| 15 | 39/M | 274 | 46,XX,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0496 | Yes/no | Rel 4 mo; died 9 mo |
| 16 | 49/M | 19.6 | NE | R | Pre-pre B | GIMEMA 0496 | Yes/no | Rel 11 mo; died 13 mo |
| 17 | 25/M | 41.2 | 46,XY | R | Pre-pre B | GIMEMA 0496 | Yes/yes | Rel 14 mo; died 17 mo |
| 18 | 49/F | 41.4 | NE | R | Pre-pre B | GIMEMA 0496 | Yes/no | Rel 14 mo; died 15 mo |
| 19 | 28/M | 281 | NE | R | Pre-pre B | GIMEMA 0496 | Yes/no | Rel 3 mo; died 4 mo |
| 20 | 47/M | 60 | 46,XY,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0496 | Yes/yes | 1st CCR 14+ mo |
| 21 | 47/M | 135 | 46,XY,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0496 | Yes/yes | 1st CCR 12+ mo |
| 22 | 60/F | 112 | 46,XX,t(4;11)(q21;q23) | ND | Pre-pre B | GIMEMA 0496 | Yes/no | Rel 6 mo; died 9 mo |
| 23 | 53/F | 75 | 46,XX,t(4;11)(q21;q23) | R | Pre-pre B | GIMEMA 0496 | Yes/yes | 1st CCR 7+ mo |
| 24 | .1/M | 128 | 46,XY,t(4;11)(q21;q23) | R | Pre-pre B | INTERFANT 99 | Yes/yes | 1st CCR 3+ mo |
| 25 | 40/F | 41 | ND | R | Pre-pre B | GIMEMA 0496 | Yes/no | 1st CCR 3+ mo |
WBC indicates white blood cells; Resp, response; HR, hematologic remission; MR, molecular remission; R, rearranged; Pre-pre B, HLDR+, CD19+, CD10−; CCR, continuous complete remission; Rel, relapse; NE, not evaluable; common, HLA-DR+, CD19+, CD10+; and ND, karyotyping not done.
Coexpression of myeloid antigen(s).
Vincristine, idarubicin, and prednisone.
Mononuclear cells obtained from bone marrow samples after centrifugation on a Ficoll-Hypaque gradient were washed and cryopreserved in 4 mol of guanidinium isothiocyanate per liter at −20°C for total RNA extraction or as dry pellets for DNA extraction. At diagnosis, 24 of the 25 patients were assessed by Southern blot study and all had a genomic rearrangement of theALL1 locus. Seventeen of the 19 evaluable patients had the t(4;11) abnormality at karyotypic analysis, including one for whom no DNA studies were available.
Cytogenetic analyses
Cytogenetic analyses were performed on bone marrow cells after 24 hours of unstimulated culture. GTG bands with trypsin were obtained. Karyotypes were reviewed and defined according to criteria of the International System for Human Cytogenetic Nomenclature.
DNA analysis
High-molecular-weight DNA was obtained from cell pellets after proteinase K digestion and phenol-chloroform extraction, digested to completion with BamHI and BglII, sized, fractionated by electrophoresis through a 0.8% agarose gel, and transferred to nitrocellulose membranes. Filters were prehybridized for 16 to 24 hours at 42°C in a solution containing 50% deionized salmon sperm, 5 × SSC, 5 × Denhart's solution, and 100 μg of denatured salmon sperm DNA per milliliter, and then hybridized in the same solution with the addition of a denatured B859 probe previously labeled with the phosphorus 32 random priming technique. Filters were washed twice for 30 minutes in 0.2 × SSC and 0.1% sodium dodecyl sulfate at 65°C and exposed with an intensifying screen at −70°C for 24 to 72 hours. The B859 probe is a complementary DNA (cDNA) insert that explores the entire ALL1 breakpoint cluster region.20
RNA preparation
Total RNA was extracted from cells cryopreserved in guanidinium isothiocyanate, according to the method of Chomczynski and Sacchi.27 The quality of RNA was assessed on an ethidium bromide-stained 1% agarose gel containing 2.2 mol of formaldehyde per liter.
RT-PCR amplification
With use of a commercial kit (Gene Amp RNA PCR kit; Perkin Elmer-Cetus, Norwalk, CT), in vitro reverse transcription of 1 μg of total RNA to cDNA was performed at 42°C for 20 minutes in a 20 μL reaction volume containing 2.5 U of cloned Moloney murine leukemia virus, reverse transcriptase, and random examers as primers. A volume of 5 μL was then diluted with 95 μL of a PCR mixture containing 1.5 mmol of Mg2Cl per liter, 50 mmol of KCl per liter, 10 mmol of Tris HCl per liter (pH 8.3), 200 μmol of dNTPs per liter, 2.5 U of TAQ DNA polymerase (Perkin Elmer-Cetus), and 15 pmol of primers Ex 5 and AF4.1. After initial denaturation at 94°C for 2 minutes, 30 cycles of amplification were done on a DNA thermal cycler (Perkin Elmer-Cetus). One cycle of denaturation, annealing, and extension consisted of processing at 94°C for 1 minute, 56°C for 1 minute, and 72°C for 1 minute, respectively. At the end, 1 μL of a 1:10 dilution of the first PCR product was used for a second round of amplification for 30 additional cycles by using the primers AF4.1 and E × 6 (half-nested PCR). Finally, 1/10 of the PCR products were run on a 2% agarose gel stained with ethidium bromide and visualized under a UV lamp. The sequences of the primers used were as follows: Ex 5: 5′-GAGGATCCTGCCCCAAAGAAAAG-3′ (sense); Ex 6: 5′-CGCCCAAGTATCCCT GTAAAAC-3′ (sense); AF4.1: 5′-TGAGCTGAAGGTCGTCTT CGAGCAT-3′ (antisense).
Amplification of normal ALL1 gene mRNA was done with the same cDNA preparation and the same conditions used to identify theALL1/AF4 junctions by using Ex 5 and Ex 6 as sense primers and the Ex 9 antisense primer (Ex 9: 5′-TTGTAGCCTGATGTTGCCTTCCACA-3′).
Negative controls (all reagents and water) were included in all PCR experiments. To rule out the possibility of a cDNA contamination of RNA samples, all tests with positive results were repeated, mixing all reagents without RNA.
As previously reported,8 serial dilution experiments established that our RT-PCR assay allows detection of 1ALL1/AF4-positive cell in a background of 104normal cells. Dilution experiments that showed the same sensitivity level have been serially performed.
Treatment modalities
All adult patients were enrolled in the GIMEMA trials for adult ALL, which include a conventional 4-drug induction treatment without consolidation with bone marrow transplantation. In particular, 3 patients were treated according to the GIMEMA ALL 0288 regimen28 and 2 patients received the GIMEMA 0394 protocol.29 Sixteen patients underwent the GIMEMA ALL 0496 protocol, which is currently open to enrollment and is derived from the ALLVR589 regimen, which includes high-dose daunorubicin in induction followed by high-dose cytosine arabinoside as consolidation.30 The remaining adult patient (who was > 70 years old) received vincristine, daunorubicin, and prednisone as induction treatment. Two of the 3 infant patients were treated according to the AIEOP 9503 regimen, which is a modification of the German BFM protocol,31 whereas the third infant is being treated according to the INTERFANT 99 pilot protocol, which includes both anti-ALL agents and agents active against acute myeloblastic leukemia (AML). The treatment protocol is based on an AML-like schedule and both low- and high-dose cytarabine.
Criteria of response and statistical methods
Hematologic complete remission (HCR) was defined as normal bone marrow cellularity with < 5% undifferentiated cells and normalization of peripheral blood counts. Molecular remission was characterized by the absence, on ethidium bromide-stained electrophoresis gel, of the specific ALL1/AF4 amplification band detected at diagnosis in the presence of RNA integrity, as evaluated by minigel visualization and successful amplification of the control gene. Hematologic and molecular monitoring of MRD were performed simultaneously at predetermined intervals during the clinical follow-up of patients (ie, postinduction, postconsolidation, every 3 months during maintenance therapy, and every 6 months for the first 2 years of clinical follow-up and once a year thereafter).
The probabilities of hematologic relapse and overall survival were estimated with the Kaplan-Meier method and compared with use of the log-rank test.
Results
Clinical and biologic features of the 25 patients at presentation, treatment modalities, and overall outcome are shown in Table1. Three patients were infants aged 1.5, 4, and 8 months, respectively. The median age of the 22 adults was 40.5 years (range, 16-71). Twelve patients had hyperleukocytosis (white blood cell count [WBC] > 100 × 109/L). The immunophenotypic characterization showed a pre-pre B phenotype (CD19+, CD10−) in 24 patients and a common B-lineage ALL (CD10+) in 1 patient.
All 25 patients had a HCR (ie, no leukemic blasts in the bone marrow at the morphologic level and normalization of peripheral blood cell count) after induction treatment. Molecular analyses showed a conversion to PCR negativity in 8 (32%) patients after induction and in 11 (44%) patients after consolidation. As shown in Table 1, biologic and clinical features of patients, such as age, sex, WBC, karyotype, and immunophenotype, did not differ between patients who did and those who did not convert to a PCR-negative status. During the clinical follow-up, 18 patients had a hematologic relapse at a median time of 5 months (range, 1-20) after achieving HCR, whereas 7 patients are in hematologic and molecular remission at 3, 3, 7, 12, 14, 90, and 96 months, respectively. Three of the 18 patients who had a relapse have received an allogeneic bone marrow transplantation during their second HCR from HLA-identical (2 patients) or haploidentical (1 patient) donors. Two of these 3 patients are in their second HCR at 12 and 28 months, respectively. One of these 2 patients, who was tested for theALL1/AF4 transcript, had a conversion to ALL1/AF4-PCR negativity after transplantation. Sixteen of the 25 patients died of ALL between 3 and 25 months after diagnosis.
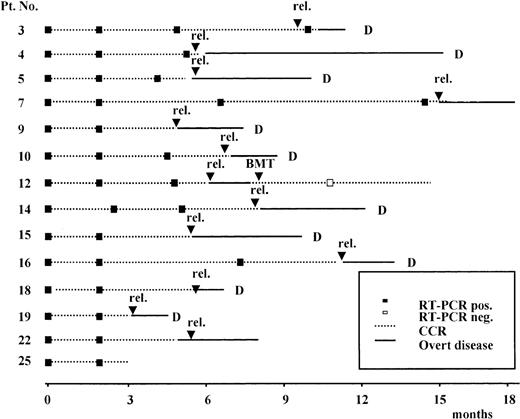
Results of the prospective RT-PCR analysis are illustrated in Figure1 and Figure 2. Thirteen of the 14 patients who never had a conversion to PCR negativity after consolidation (Figure 1) had an hematologic relapse at a median time of 5 months after HCR (range, 1-20 months), whereas the remaining patient currently in HCR is early in her follow-up. Of the 11 patients who had a conversion to PCR negativity after consolidation (Figure 2), 5 had a molecular relapse at a median time of 4 months after achieving molecular remission (range, 1-14 months). All 5 patients subsequently had a hematologic relapse at a median time of 3.5 months (range, 2-7 months) after conversion to PCR positivity (Figure2). Of the remaining 6 patients, 2 (patients 24 and 25) are early in their follow-up, whereas 4 (patients 1, 2, 20, and 21) are in persistent hematologic and molecular remission at 12, 14, 90, and 96 months, respectively, after diagnosis.
Prospective longitudinal monitoring by RT-PCR analysis of the ALL1/AF4 transcript in 14 patients who never had conversion to PCR-negative status.
Time 0 corresponds to diagnosis; rel indicates hematologic relapse; BMT, allogeneic bone marrow transplantation; and D, dead.
Prospective longitudinal monitoring by RT-PCR analysis of the ALL1/AF4 transcript in 14 patients who never had conversion to PCR-negative status.
Time 0 corresponds to diagnosis; rel indicates hematologic relapse; BMT, allogeneic bone marrow transplantation; and D, dead.
Prospective longitudinal monitoring of residual disease by RT-PCR analysis of the ALL1/AF4 transcript in 11 patients who had conversion to PCR- negative status after consolidation.
Time 0 corresponds to diagnosis; rel indicates hematologic relapse; BMT, allogeneic bone marrow transplantation; and D, dead.
Prospective longitudinal monitoring of residual disease by RT-PCR analysis of the ALL1/AF4 transcript in 11 patients who had conversion to PCR- negative status after consolidation.
Time 0 corresponds to diagnosis; rel indicates hematologic relapse; BMT, allogeneic bone marrow transplantation; and D, dead.
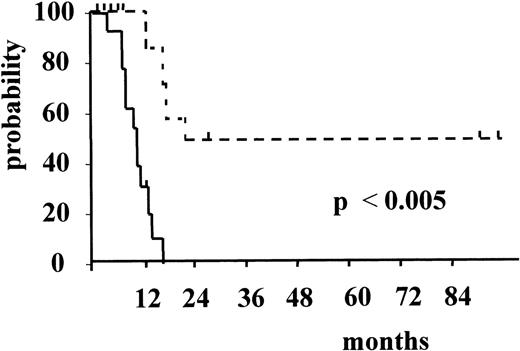
Figure 3 and Figure4 illustrate the actuarial probabilities of hematologic relapse and overall survival according to the RT-PCR status after consolidation. The actuarial probabilities of hematologic relapse and survival in patients who never had a molecular remission were 100% and 0% at 14 and 24 months, respectively. In the 11 patients who had a conversion to PCR negativity after consolidation, the actuarial probabilities of relapse and survival were 67% and 43% at 96 and 100 months, respectively. The differences observed between the 2 groups of patients with respect to both relapse and survival were significant (P = .003 and P < .005, respectively).
Actuarial probability of hematologic relapse according to RT-PCR status after consolidation.
The solid line indicates patients who had persistent PCR-positive status, and the dotted line, patients who had conversion to PCR-negative status.
Actuarial probability of hematologic relapse according to RT-PCR status after consolidation.
The solid line indicates patients who had persistent PCR-positive status, and the dotted line, patients who had conversion to PCR-negative status.
Actuarial probability of overall survival according to RT-PCR status after consolidation.
The solid line indicates patients who had persistent PCR-positive status, and the dotted line, patients who had conversion to PCR negative status.
Actuarial probability of overall survival according to RT-PCR status after consolidation.
The solid line indicates patients who had persistent PCR-positive status, and the dotted line, patients who had conversion to PCR negative status.
Discussion
To our knowledge, this study is the first RT-PCR prospective analysis of MRD in patients with t(4;11) positive ALL. After conventional intensive chemotherapy, 44% of our patients had a PCR-negative remission, and this was associated with a lower actuarial risk of relapse and a better actuarial probability of overall survival than in patients with persistent PCR-positive status. In addition, our results support our previous observation that persistent PCR positivity or conversion to PCR positivity after remission induction is strongly predictive of a subsequent hematologic relapse.10 In fact, all 18 patients (100%) who had persistence of or reconversion to PCR-positive status subsequently had a full hematologic relapse. These findings, together with the notion that t(4;11) ALL is a potentially eradicable disease, even with current chemotherapy programs, strengthen the value of molecular monitoring studies in this leukemia subtype. Although the results obtained were related to the treatment given to our patients, it is likely that their clinical impact will, over time, parallel the increasing efficacy of future therapeutic strategies for patients with t(4;11) positive ALL. In fact, RT-PCR monitoring of MRD has already been proved to have great clinical validity in different subgroups of acute leukemia, such as acute premyelocytic leukemia and childhood ALL, in which current treatment protocols are associated with a high cure rate.7 32
These considerations are particularly pertinent in light of recent concerns about the clinical utility of RT-PCR monitoring studies of MRD in t(4;11) ALL. Uckun et al,24 using a 2-round (nested) RT-PCR approach, detected the ALL1/AF4 fusion gene in the hematopoietic tissues of healthy individuals and in 12% of pediatric ALL patients with no cytogenetic or genomic evidence of 11q23 alterations. With respect to other leukemia-specific hybrid genes, low levels of BCR/ABL have been detected in a sizable proportion of healthy individuals,33 and the chimeric genes CBFβ/MHY11and AML1/ETO, originated by t(8;21)(q22;q22) and inv16, respectively, were found in patients with long-term disease-free intervals after chemotherapy.34 35
It is worth noting that the patients in the current study had cytogenetic or Southern blot evidence of an ALL1 gene rearrangement at diagnosis, indicating that the entire leukemic clone harbored this aberration. Moreover, the clinical outcome in our patients was extremely poor, as it was in previously reported studies in this leukemia subset.11,36-39 In contrast, the PCR-positive patients with ALL1/AF4 in the study of Uckun et al24 had an ALL1 germline configuration at presentation and clinicoprognostic features of pediatric standard-risk ALL, leading the authors to suggest that the percentage ofALL1/AF4-positive cells was presumably very low (< 5%) and not representative of the main leukemic clone. In our opinion, the findings of Uckun et al highlight the importance of the routine use of Southern blot study to identify ALL1 gene alterations at diagnosis of leukemia. This idea is supported by research reporting variability in gene recombinatory events participating in ALL1alterations, including lesions not identified by karyotyping and fluorescence in situ hybridization, such as insertions and tandem duplications.14,40 41
Recently, quantitative RT-PCR analyses have provided a useful tool for measuring chimeric transcripts at diagnosis and during the follow-up of leukemic patients.42 These studies will allow researchers to draw correlations among variations of chimeric transcripts, biologic and clinical features of patients at diagnosis, and the clinical outcome. However, with regard to ALL1-positive acute leukemia, there are experimental data that might raise concerns about the existence of a linear relation between the amount of hybrid transcript, tumor burden, and patients' clinical outcome. In fact, in vitro cultured cells transfected with ALL1 fusion genes did not show a stable expression of fusion transcripts and proteins and, more interestingly, Lavau et al43 showed that experimental immortalization of primary murine bone marrow cells by ALL1/ENLwas associated with expression of very low levels of fusion transcripts and proteins. Taken together, these data demonstrate thatALL1-positive leukemic blasts may express very low amounts ofALL1 fusion transcripts, possibly because high levels of expression might be dangerous for the survival of leukemic cells. The concerns should be taken into account in future studies, and results of quantitative RT-PCR analyses must be compared carefully with those of qualitative RT-PCR analyses.
The patients in the current series had an overall dismal outcome, similar to that reported by other authors in adults with t(4;11) ALL treated with intensive treatment strategies. In particular, we did not observe any significant differences with regard to age, sex, WBC count, karyotype, or immunophenotype at diagnosis between patients who did and those who did not have a conversion to a PCR-negative status. It is worth noting, however, that the 2 long-term survivors in our series had a good response to prednisone prephase treatment. This observation is particularly relevant in light of a recent analysis of prognostic factors in patients with infant ALL by Dördelmann et al,44 who showed that prednisone response was the best predictor of treatment outcome in those patients.
In a recent study of pro-B ALL that included t(4;11), Ludwig et al45 reported an improved outcome after intensification of postremission treatment. Using high-dose cytarabine, allogeneic bone marrow transplantation, or both, as consolidation, these authors found that 16 of 30 patients were in continuous complete remission for between 883 and 2886 days, with an actuarial probability of remaining in continuous complete remission of 52% at 8 years. Although these data require confirmation, it is worth noting that, in the current study, a conversion to PCR negativity was observed in 3 patients (patients 17, 21, and 23; Figure 3) after the administration of high-dose cytarabine as consolidation and that the patient who was tested after allogeneic bone marrow transplantation had a conversion to PCR negativity. These observations support the strong recommendation of Ludwig et al45 that early postremission intensification, including allogeneic bone marrow transplantation, should be used. Because all the 18 patients in our series who had persistent PCR positivity or a conversion to PCR positivity after induction and consolidation treatments subsequently had a full hematologic relapse, we believe that such t(4;11)-positive ALL patients should be treated with aggressive early postremission intensification programs. The optimal postinduction treatment strategy for patients who have a conversion to a PCR-negative status remains to be clarified.
In conclusion, our results support the value of PCR monitoring studies in ALL1/AF4-positive patients with ALL who have cytogenetically detectable t(4;11) or Southern blot ALL1 rearrangements at diagnosis. Analyses of larger series of patients in multicenter studies that include centralized handling of biologic materials at diagnosis and during the clinical follow-up, as in the current GIMEMA 0496 trial, will allow conclusive definition of the impact of PCR monitoring and the efficacy of new therapeutic strategies for this highly aggressive ALL subset.
Supported in part by Associazione Italiana contro le Leucemie-Sezione diRoma (ROMAIL); Istituto Superiore di Sanità, Italy-USA project on Therapy of Tumors; AIRC(Associazione Italiana Ricerca sl Cancro; and Ospedale Bambino Gesù—IRCCS grant 98/02/P/480.
M.C.R. is in receipt of a fellowship from FIRC (Federazione Italiana Ricerca sul Cancro).
Reprints:Giuseppe Cimino, Dipartimento di Biotecnologie Cellulari ed Ematologia, Via Benevento 6, 00161 Rome, Italy; e-mail: cimino@bce.med.uniroma1.it.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal