Abstract
Genetic modification of hemopoietic progenitor cells ex vivo, followed by the infusion of the genetically modified cells into the human immunodeficiency virus-1 (HIV-1) infected donor, has been proposed as a treatment for HIV-1 infection. The current study was undertaken to evaluate the effect of hemopoietic stem cell mobilization and harvesting on HIV-1 replication in persons with HIV-1 infection. Eighteen HIV-1–infected persons received recombinant granulocyte colony-stimulating factor (G-CSF; Filgrastim) 10 μg/kg per day, for 7 days. On days 4 and 5, peripheral blood mononuclear cells were harvested by leukapheresis. The CD4+ lymphocyte count at entry was >500/μL for 6 subjects, 200 to 500/μL for 6 subjects, and <200/μL for 6 subjects. For 9 of 18 subjects, plasma HIV-1 RNA levels increased 4- to 100-fold (>0.6 log10) above baseline between days 4 and 7 and returned to baseline by day 27. Significant increases of plasma HIV-1 RNA levels occurred in 5 subjects despite 3-drug antiretroviral therapy. Changes in CD4+ and CD34+ cells during mobilization and harvesting were similar in all subjects whether they had or did not have increased plasma HIV-1 RNA levels. Thus, mobilization and harvesting of bone marrow progenitor cells from persons infected with HIV-1 induced a transient increase in viral replication in some patients but was not associated with adverse effects. (Blood. 2000;95: 48-55)
Therapies that inhibit human immunodeficiency virus-1 (HIV-1) protease provide potent inhibition of viral replication in HIV-1–infected persons1,2 and, when used in combination with inhibitors of HIV-1 reverse transcriptase, provide effective treatment of HIV-1 infection.3 Although treatment of HIV-1 infection with combinations of protease inhibitors and reverse transcriptase inhibitors often leads to effective suppression of viral replication and at least partial reversal of HIV-1–related immunosuppression,4 available treatments do not result in HIV-1 eradication.5,6 Because long-term therapy with antiretroviral regimens may be limited by toxicities and by the eventual emergence of resistant viruses7 and because the long-term benefits of available antiretroviral treatments are unknown, additional strategies for the treatment of HIV-1 infection are needed.
It has been proposed that HIV-1 infection could be treated by the genetic modification of HIV-1 host cells to confer resistance to infection by HIV-1.8 One proposed strategy for genetic therapy for HIV-1 infection is the delivery of a gene that confers resistance to HIV-1 infection to hemopoietic progenitor cells. This approach requires 3 basic steps. First, hemopoietic stem cells are harvested from the HIV-1 infected person. Second, the gene that confers resistance to HIV-1 infection is delivered to hemopoietic stem cells ex vivo. Third, the genetically modified stem cells are infused into the autologous HIV-1–infected person. Expansion and differentiation of the progeny of the genetically modified stem cells in the HIV-1–infected person would theoretically provide a population of CD4+ lymphocytes resistant to HIV-1 infection, thereby inhibiting viral replication and providing immune reconstitution.
For stem cell–based gene therapy of HIV-1 infection to be feasible, it is first necessary to harvest safely adequate numbers of hemopoietic progenitor cells from HIV-1 infected–persons. Mobilizing hemopoietic progenitor cells with recombinant granulocyte colony-stimulating factor (G-CSF; Filgrastim; Amgen, Thousand Oaks, CA) and harvesting the mobilized cells by leukapheresis in persons with malignant diseases are not associated with significant adverse events, and they provide adequate quantities of progenitor cells for bone marrow reconstitution after ablation chemotherapy.9-11 Preliminary studies have found that hemopoietic progenitor cells can be harvested from HIV-1 infected–persons and that HIV-1–resistant genes can be effectively delivered to the purified progenitor cells.12-14 Although adverse effects of stem cell harvesting and mobilization on HIV-1 virus load have not been described, studies reported to date have included only small numbers of HIV-1–infected persons and have not included persons with advanced HIV-1 infection or persons with AIDS-related illnesses.
The Adult AIDS Clinical Trials Group (ACTG) Protocol 285 was undertaken to determine the safety and efficacy of stem cell mobilization with G-CSF and harvesting by leukapheresis in persons with various stages of HIV-1 infection. Detailed descriptions of the efficacy of stem cell mobilization and harvesting from this group of research subjects are forthcoming.44 In this article we describe the effects of mobilizing and harvesting hemopoietic progenitor cells on HIV-1 virus load in this research group of 18 infected persons.
Methods
Subject selection
To be eligible for participation in this study, subjects were required to meet all of the following criteria: (1) HIV-1 infection documented by positive HIV-1 enzyme-linked immunosorbent assay and confirmed by Western blot analysis, positive serum p24 antigen, positive HIV-1 culture, or second antibody test other than an enzyme-linked immunosorbent assay; (2) no antiretroviral therapy within 30 days before study entry for cohort 1 or stable antiretroviral therapy for at least 60 days before study entry for cohorts 2 and 3; (3) minimum age of 18 years; (4) Karnofsky performance score of at least 70; (5) venous access suitable for leukapheresis; (6) hemoglobin count of at least 9.1 g/dL for men or at least 8.8 g/dL for women; (7) absolute neutrophil count of at least 1000 cells/μL without the use of G-CSF; (8) platelet count of at least 75 000/μL; (9) serum aspartate transaminase level no more than 5× upper normal limit; (10) serum creatinine level no more than 1.5× upper normal limit; (11) willingness and ability to give informed consent; (12) agreement to use barrier methods of birth control; and (13) negative result of urine human chorionic gonadotropin pregnancy test within 14 days of study entry for women of childbearing potential. Subjects on antiretroviral therapy at study entry were encouraged not to change their antiretroviral regimen during the first 4 weeks after treatment. Subjects who were not on antiretroviral therapy at study entry were encouraged not to begin it during the first 4 weeks after treatment.
Potential subjects were excluded from participation in this study if they met any of the following criteria: (1) malignant neoplastic disease, past or present; (2) pregnancy; (3) known sensitivity to proteins derived from Escherichia coli; (4) leukapheresis or lymphapheresis within 180 days before study entry; (5) opportunistic infection within 14 days before study entry; (6) active alcohol or substance abuse; (7) seizures within 1 year before study entry or clinically significant central nervous system disease; (8) investigational antiretroviral therapy within 30 days before study entry; or (9) any medical condition that would interfere with evaluation of the subject.
The protocol was approved by the institutional review boards of the University of Colorado Health Sciences Center, the Thomas Jefferson University Hospital, and the University of California Los Angeles Medical Center. Informed consent was obtained from all subjects before their participation in this study.
Study design
At study entry, subjects were stratified into 1 of 3 cohorts by baseline CD4+ lymphocyte count. Cohort 1 consisted of 6 persons with >500 CD4+ lymphocytes/μL and without symptoms of HIV-1 infection. Cohort 2 consisted of 6 persons with ≥200 and ≤500 CD4+ lymphocytes/μL, with or without HIV-1–related symptoms but without prior AIDS-defining illness. Cohort 3 consisted of 6 persons with <200 CD4+ lymphocytes/μL, with or without AIDS-defining illness. All subjects received daily subcutaneous injections of 10 μg/kg G-CSF (Neupogen; Amgen) on study days 1 through 7. On days 4 and 5, peripheral blood mononuclear cells (PBMC) were harvested by leukapheresis. Complete blood counts and lymphocyte subset analyses were performed on days −3, 0, 4, 5, 6, 7, 10, and 27.
Quantitation of plasma HIV-1 RNA
Plasma specimens were obtained by venipuncture on days −3, 0, 4, 5, 6, 7, and 27 and stored at −70°C. The amount of HIV-1 RNA in thawed plasma specimens was determined by the Amplicor HIV-1 Monitor assay (Roche Molecular Systems, Indianapolis, IN), a reverse transcription–polymerase chain reaction (RT-PCR) assay with a lower limit of quantitation of 400 copies/mL (2.6 log10). Except for subject 620 733 (see Table1), all plasma specimens obtained at different time points from the patients were assayed together in a batch. All plasma samples from subjects with <400 copies/mL by the standard HIV-1 monitor assay at study entry were assayed in a batch with the ultrasensitive HIV-1 Monitor assay (Roche Molecular Systems) with a lower limit of quantitation of 50 copies/mL (1.7 log10). Because the 95% confidence interval for interassay variation of plasma HIV-1 RNA quantitation by RT-PCR is ±0.5 log10,15 16 a significant change in HIV-1 RNA level for each subject was defined as ≥0.6-log10difference from baseline.
Subject characteristics at study entry
| Cohort . | Subject* . | Age (y) . | CD4+ Count† (cells/μL) . | HIV-1 RNA‡ (log10 copies/mL) . | HIV-1 DNA1-153 (log10 copies/ 105cells) . | Antiretroviral therapy1-155 . |
|---|---|---|---|---|---|---|
| 1 | 610 219 | 36 | 499 | 4.06 | 3.00 | None |
| 620 733 | 48 | 943 | 4.08 | 2.52 | None | |
| 620 745 | 37 | 798 | 3.66 | 1.99 | None | |
| 61 145 | 58 | 495 | <1.70 | 1.44 | None | |
| 61 146 | 58 | 1270 | <1.70 | <0.9 | None | |
| 610 354 | 31 | 884 | 3.00 | 2.14 | None | |
| Median | 43 | 841 | 3.33 | 2.07 | — | |
| 2 | 610 051 | 38 | 338 | 2.70 | 2.78 | ZDV/3TC/IDV |
| 620 495 | 38 | 230 | <1.70 | 3.40 | ZDV/3TC/IDV | |
| 620 738 | 45 | 321 | 3.05 | 2.69 | DDI | |
| 61 149 | 49 | 368 | 3.51 | 1.36 | None | |
| 61 147 | 43 | 307 | 3.72 | 2.41 | ZDV/3TC/SQV | |
| 610 020 | 38 | 278 | <1.70 | 2.69 | ZDV/3TC/IDV | |
| Median | 41 | 314 | 2.88 | 2.69 | — | |
| 3 | 61 144 | 40 | 9 | 5.73 | 4.75 | DEL/SQV/RTV |
| 620 743 | 42 | 33 | 4.06 | 3.63 | ZDV/3TC/SQV | |
| 61 148 | 34 | 150 | 4.18 | 3.04 | ZDV/3TC/SQV | |
| 620 736 | 53 | 162 | <2.60 | 2.97 | ZDV/3TC/IDV | |
| 61 150 | 48 | 153 | <1.70 | 2.91 | D4T/3TC/SQV | |
| 61 821 | 33 | 154 | 3.89 | 2.63 | DDI/D4T/SQV | |
| Median | 41 | 152 | 3.98 | 3.00 | — |
| Cohort . | Subject* . | Age (y) . | CD4+ Count† (cells/μL) . | HIV-1 RNA‡ (log10 copies/mL) . | HIV-1 DNA1-153 (log10 copies/ 105cells) . | Antiretroviral therapy1-155 . |
|---|---|---|---|---|---|---|
| 1 | 610 219 | 36 | 499 | 4.06 | 3.00 | None |
| 620 733 | 48 | 943 | 4.08 | 2.52 | None | |
| 620 745 | 37 | 798 | 3.66 | 1.99 | None | |
| 61 145 | 58 | 495 | <1.70 | 1.44 | None | |
| 61 146 | 58 | 1270 | <1.70 | <0.9 | None | |
| 610 354 | 31 | 884 | 3.00 | 2.14 | None | |
| Median | 43 | 841 | 3.33 | 2.07 | — | |
| 2 | 610 051 | 38 | 338 | 2.70 | 2.78 | ZDV/3TC/IDV |
| 620 495 | 38 | 230 | <1.70 | 3.40 | ZDV/3TC/IDV | |
| 620 738 | 45 | 321 | 3.05 | 2.69 | DDI | |
| 61 149 | 49 | 368 | 3.51 | 1.36 | None | |
| 61 147 | 43 | 307 | 3.72 | 2.41 | ZDV/3TC/SQV | |
| 610 020 | 38 | 278 | <1.70 | 2.69 | ZDV/3TC/IDV | |
| Median | 41 | 314 | 2.88 | 2.69 | — | |
| 3 | 61 144 | 40 | 9 | 5.73 | 4.75 | DEL/SQV/RTV |
| 620 743 | 42 | 33 | 4.06 | 3.63 | ZDV/3TC/SQV | |
| 61 148 | 34 | 150 | 4.18 | 3.04 | ZDV/3TC/SQV | |
| 620 736 | 53 | 162 | <2.60 | 2.97 | ZDV/3TC/IDV | |
| 61 150 | 48 | 153 | <1.70 | 2.91 | D4T/3TC/SQV | |
| 61 821 | 33 | 154 | 3.89 | 2.63 | DDI/D4T/SQV | |
| Median | 41 | 152 | 3.98 | 3.00 | — |
ZDV, zidovudine; 3TC, lamivudine; IDV, indinavir; DDI, didanosine; SQV, saquinavir; DEL, delavirdine; RTV, ritonavir.
All subjects were male.
Baseline values for CD4+ lymphocyte counts are averages of values obtained on days −3 and 0.
Baseline plasma HIV-1 RNA values are the average of log10 values obtained on days −3 and 0 for all subjects except subject 620 733, for whom the baseline value is from a single determination on day −18.
Baseline copies HIV-1 DNA per 105 CD4+ lymphocytes are averages of log10 values obtained on days −3 and 0 for all subjects.
Antiretroviral treatment regimen for days −60 through 27.
Culture of HIV-1 from leukapheresis products
HIV-1 was isolated from leukapheresis products obtained on study days 4 and 5 according to the ACTG consensus protocol.17Approximately 106 fresh PBMCs harvested by leukapheresis of the HIV-1–infected patients were cocultured with an equal number of 3- to 4-day-old phytohemagglutinin-stimulated lymphoblasts obtained from random HIV-1 seronegative donors. Fresh 3- to 4-day-old phytohemagglutinin-stimulated lymphoblasts were added weekly, and culture supernatants were monitored for p24 antigen production (Coulter Diagnostics, Hialeah, FL) twice weekly. Cultures were considered negative if p24 antigen was not detected after 30 days.
Quantitation of peripheral blood mononuclear cell HIV-1 DNA
An internal quantitation standard (IQS) plasmid (pIQSGAG) was constructed by inserting a chimeric 251-nucleotide bp HIV-1 gag gene fragment (position 1291–1542) into the plasmid pCR-Script Amp Sk+ (Stratagene, La Jolla, CA). The chimeric fragment contained a unique, conserved probe region that consisted of a 33-bp sequence derived from the Drosophila “white” locus18 and was designed to replace the SK102 (position 1403–1435) probe region of the HIV-1 gag gene. The chimeric fragment was generated by first amplifying 2 separate products from the gag gene using gag primers gag 04 and gag 0619 and primers FLYINTA (5′-GCCGGATTGTAGTTGGTAGGACACTGGTTTTAA CATTTGCATGGCTGCTTG) and FLYINTB (5′-TCCTACCAACTACAATCCGGCGGACTTAGATTGCATCCAGTGCATGCAG, which contain both Drosophila and HIV-1 gag sequence. Product 1 (generated from gag 04 and FLYINTB) and product 2 (generated from gag 06 and FLYINTA) were then gel purified, mixed, and simultaneously extended and amplified to generate the full-length chimeric fragment that was used to generate pIQSGAG. The plasmid was linearized withXho I and diluted to appropriate concentrations. This plasmid served as a coamplified control and allowed for the amplification of a product identical in size to that generated from proviral DNA, except that it hybridized with the Fly-C probe (5′-GTCCTACCAA CTACAATCCGG) but not with the gag-specific GAGP1 probe (5′-GAGGAAGCTGCAGAATG GGA).20
The PBMC was purified by Ficoll-Hypaque gradient sedimentation of whole blood specimens obtained by venipuncture on days −3, 0, 4, 5, 6, 7, and 27. Purified PBMC was counted, and aliquots were stored as either dry-cell pellets at 70°C or as viable cells in dimethyl sulfoxide and liquid nitrogen. Dry pellets were used directly, and viable cells were quickly thawed at 37°C and washed with phosphate-buffered saline (PBS). Dry and viable cell pellets were resuspended in 200 μL PBS. Total specimen DNA was extracted using Qiagen Blood Kits (Qiagen, Santa Clara, CA). The amount of total DNA was quantified using a DyNA Quant 200 fluorometer (Hoeffer Pharmacia Biotech, San Francisco, CA), calibrated with a 100 ng/μL calf-thymus DNA standard (Hoeffer Pharmacia Biotech). DNA was diluted to 20 ng/μL, and 50 μL (1 μg) was used in each PCR reaction. Samples from individual subjects were run in batch to minimize intrasubject variability.
For each amplification reaction, 1 μg specimen DNA was used. For each specimen 5 reactions were performed with varying amounts (0–10 000 copies) of the IQS. Biotin-labeled primers SK462 and SK43121 were custom synthesized by Gibco BRL (Gaithersburg, MD) and used to amplify a portion of both the specimen-associated HIV-1 gag gene and the IQS. Each 100-μL reaction contained 10 mmol/L Tris-HCl, pH 8.3, 50 mmol/L KCl, 1.5 mmol/L MgCl2, 0.001% gelatin, 150 μmol/L dNTPS, 200 μmol/L dUTP, 2.5 U Taq Polymerase (Perkin Elmer, Foster City, CA), 1 U heat-labile uracil-N-glycoslyase (Boehringer Mannheim, Indianapolis, IN), and 1 μmol/L each of SK431 and SK462. Samples were amplified in a Perkin Elmer 9600 thermocycler using the following conditions (modified from Kwok and Sninsky21): 10 minutes at 25°C; 5 cycles consisting of 10 seconds at 95°C, 10 seconds at 55°C, and 10 seconds at 72°C; 35 cycles consisting of 10 seconds at 90°C, 10 seconds at 60°C, and 10 seconds at 72°C; 5 minutes at 72°C; and 2.5 minutes at 95°C.
The detection method was modified after Hockett et al.22Briefly, 10-μL aliquots of the amplified product were transferred to 4 wells of a streptavidin-coated microtiter plate (Boehringer Mannheim) containing 150 μL PBS and 0.01% Tween 20 and incubated at 42°C for 1 hour. The plates were washed once with PBS containing 0.01% Tween 20, denatured 2 minutes with 160 μL of denaturing solution, 50 mmol/L NaOH, 0.15 NaCl, 2 mmol/L EDTA, pH 8, followed by 2 more washes with the PBS-Tween 20 solution. The contents of 2 wells were then hybridized at 42°C for 1 hour with 160 μL 7.5 nmol/L GAGP1 (3′ digoxigenin; custom synthesized by Genosys Biotechnologies, Woodlands, TX) in hybridization buffer (20% formamide, 0.9 mol/L NaCl, 1.2 mol/L NaH2PO4, 6 mmol/L EDTA, and 50 ng/mL sheared herring sperm DNA) to detect amplified specimen, and 2 wells were hybridized with 160 μL 7.5 nmol/L FLY-C (3′ digoxigenin) in hybridization buffer to detect amplified IQS. The well-plates were washed 3 times with PBS-Tween and were incubated at 37°C for 1 hour with anti-digoxigenin Fab conjugated with a diluted alkaline phosphatase solution (3 U/20 mL in PBS, 1% bovine serum albumin, and 0.01% NaN3), then washed 4 times with PBS-Tween and incubated with 1 mg/mL p-nitrophenyl phosphate solution (Kierkegaard and Perry, Gaithersburg, MD) for 15 minutes; the absorbance was read at 405 nm. Copy number (HIV-1 DNA copies/μg PBMC DNA) was determined by linear regression from a plot of the log of absorbance of HIV-1 gag/IQS versus the log of the nominal (input) IQS copy number. The measured HIV-1 proviral DNA copy number was determined at the equivalency intercept (ie, log absorbance HIV-1 gag/IQS = 0). The copies of HIV-1 DNA/μg PBMC DNA were converted to copies of HIV-1 DNA/105 PBMC using 6.6 × 109 bp as an estimate of the size of the human genome. To adjust for effects of fluctuations in the numbers of circulating CD4+ lymphocytes, copies of HIV-1 DNA/105 PBMC were divided by the fraction of whole blood lymphocytes that were CD4+ to give copies of HIV-1 DNA/105 CD4+ lymphocytes. Use of this calculation assumed that the fraction of CD4+ lymphocytes in purified PBMC was equal to the fraction of whole blood lymphocytes that were CD4+. Because the 95% confidence interval for the interassay variation of the PCR method used to quantify HIV-1 DNA is ±0.3 log10,20a significant change in HIV-1 DNA was defined as ≥0.4 log10 change from baseline.
Results
Subjects
Eighteen HIV-1-infected–persons were enrolled in this study. Six subjects were stratified into each cohort; their characteristics at study entry are summarized in Table 1. All subjects were men, and the median age of subjects in each cohort was similar. None of the subjects in cohort 1 received antiretroviral therapy during the first 27 study days. Although the entry criteria specified that subjects in cohort 2 be on stable antiretroviral therapy, an exception was made for subject 61 149 who did not receive antiretroviral therapy. Four subjects in cohort 2 were administered 3-drug antiretroviral regimens that included at least 1 inhibitor of HIV-1 protease, whereas subject 620 738 was administered monotherapy with didanosine. All subjects in cohort 3 were administered 3-drug antiretroviral regimens that included at least 1 protease inhibitor. All subjects received 10 μg/kg G-CSF on days 1 through 7 and underwent leukapheresis on days 4 and 5. Subject 61 148 (cohort 3) reported bone pain on days 3 and 4. The other subjects reported no treatment-related side effects. After stem cell mobilization and harvesting, CD4+ lymphocyte counts decreased transiently on day 27 but returned to baseline by day 83.44
Stem cell mobilization and harvesting affects plasma HIV-1 RNA levels
Plasma HIV-1 RNA levels were determined for all 18 subjects. Sixteen of them had quantifiable HIV-1 RNA in at least 2 plasma samples obtained during days 4 through 27. Viral RNA was not detected in any of the specimens obtained from subject 610 020 by the ultrasensitive PCR method (<1.7 log10 copies/mL), nor was it detected in 8 of 9 specimens from subject 620 736 by the standard PCR method (<2.6 log10 copies/mL; sufficient amounts of plasma for repeat analysis of this subject's specimens with the ultrasensitive PCR assay were not available).
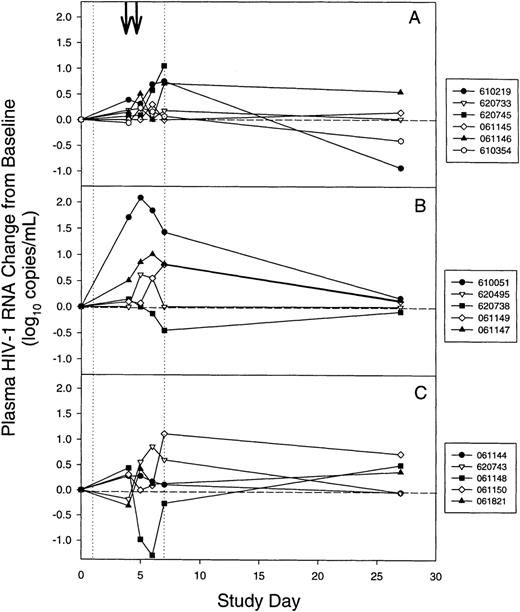
The change from baseline for plasma HIV-1 RNA levels (Figure1) was determined for 16 subjects for whom virus RNA could be detected on at least 2 occasions by subtracting the baseline level of HIV-1 RNA (Table 1) from the level on days 4, 5, 6, 7, and 27. To avoid the effects of changes in antiretroviral therapy on HIV-1 virus load, this analysis was limited to the first 27 study days. For the 16 subjects with measurable plasma HIV-1 RNA levels, the median viral RNA increased to 0.3 log10 copies/mL above baseline on day 5. Increases above baseline were statistically significant on days 5 and 7 (P = .038 and .0495, respectively, using the Bonferroni adjustment for performing 5 separate tests; 2-tailed Wilcoxon signed rank test for paired data). On day 6 the increase above baseline approached significance (P = .0565). Plasma HIV-1 RNA levels were not significantly different from baseline on days 4 or 27 (P > .1). When the data were analyzed for each cohort, similar trends were noted but statistically significant differences were not found.
Changes in plasma HIV-1 virus load during mobilization and harvesting of stem cells for individual subjects.
(A) Cohort 1. (B) Cohort 2. (C) Cohort 3. (vertical dotted lines) Granulocyte colony-stimulating factor was administered on days 1 through 7. (arrows) Leukapheresis was performed on days 4 and 5. For subject 620 745 in cohort 1, a day-27 plasma sample was unavailable. Levels of plasma HIV-1 RNA for subjects 610 020 and 620 736 were below the limit of detection and are not shown.
Changes in plasma HIV-1 virus load during mobilization and harvesting of stem cells for individual subjects.
(A) Cohort 1. (B) Cohort 2. (C) Cohort 3. (vertical dotted lines) Granulocyte colony-stimulating factor was administered on days 1 through 7. (arrows) Leukapheresis was performed on days 4 and 5. For subject 620 745 in cohort 1, a day-27 plasma sample was unavailable. Levels of plasma HIV-1 RNA for subjects 610 020 and 620 736 were below the limit of detection and are not shown.
For 9 of 18 subjects in the 3 cohorts, HIV-1 RNA levels increased to >0.6 log10 copies/mL (4-fold) above baseline at least once during study days 4 through 7 (Figure 1). On day 6 or 7, 3 subjects (620 745, 61 147, and 61 150; Figures 1A, 1B, 1C) had a 1.0 to 1.1 log10 copies/mL (10-fold) rise above baseline. The most striking changes in plasma HIV-1 RNA levels occurred for subject 610 051 (Figure 1B) who, while receiving antiretroviral therapy with zidovudine, lamivudine, and indinavir, experienced a 2.1 log10 (126-fold) rise above baseline on study day 5 that was followed by a return to baseline by day 27. Four subjects (620 495, 610 020, 620 736, and 061 150) had undetectable HIV-1 RNA levels at study entry while on 3-drug antiretroviral therapy (Table 1). Plasma HIV-1 RNA levels remained at baseline during G-CSF treatment for subjects 610 020 and 620 736. Subject 620 495 had a maximum 0.6 log10 copies/mL above baseline in plasma RNA on day 5 but returned to baseline on day 27 (Figure 1B). Despite effective antiretroviral therapy at study entry, subject 61 150 experienced a 1.1 log10 rise in plasma HIV-1 RNA level on day 7 and remained above baseline on days 27 and 55 (0.72 and 0.69 log10 above baseline, respectively). For 1 subject (61 148) plasma viral RNA levels fell 1.3 log10 copies/mL below baseline on day 6, transiently increased to 0.5 log10above baseline on day 27 (Figure 1C), and fell below baseline on day 55. None of the subjects with elevated plasma HIV-1 levels reported symptoms associated with increased HIV-1 replication (eg, fever, night sweats, lymphadenopathy).
Comparison of subjects with and without increased HIV-1 RNA levels during stem cell harvesting and mobilization
To evaluate the factors that led to increased HIV-1 RNA levels during stem cell mobilization and harvesting, the characteristics of 9 subjects who had significant increases in HIV-1 RNA level (≥0.6 log10) during days 4 through 7 were compared with 9 subjects who did not have significant increases in HIV-1 RNA level (<0.6 log10) during this period (Table2). Subjects in whom plasma HIV-1 RNA levels increased to >0.6 log10 above baseline were not more likely to be in either of the 3 cohorts. Characteristics at study entry (age, baseline CD4 cell count, baseline HIV-1 RNA level, and concurrent antiretroviral therapy) were similar for subjects with or without increased HIV-1 RNA levels. The response of subjects to G-CSF mobilization (maximum peripheral CD4+ lymphocyte count, relative change of CD4+ lymphocytes from baseline, and the maximum peripheral CD34+ cell count) was also similar in both groups. To assess whether subjects with increased plasma HIV-1 RNA levels had increased numbers of productively infected cells in the peripheral circulation, the PBMC collected by leukapheresis on days 4 and 5 was cultured for infectious HIV-1. The frequency of recovery of infectious HIV-1 from PBMC harvested by leukapheresis on either day 4 or day 5 was similar for both groups of subjects (4 of 9 vs. 5 of 9; Table 2).
Comparison of subjects with and without increases in HIV-1 virus load during stem cell mobilization and harvesting
| Variable . | HIV-1 RNA Maximum Increase* . | ||
|---|---|---|---|
| <0.6 log10 (n = 9) . | ≥0.6 log10 (n = 9) . | P . | |
| Number of subjects in cohort 1 | 3 | 3 | |
| Number of subjects in cohort 2 | 4 | 2 | .84† |
| Number of subjects in cohort 3 | 2 | 4 | |
| Age (years)‡ | 40 | 42 | >.99 |
| Baseline CD4+ lymphocytes (cells/μL)‡ | 278 | 338 | .23 |
| Baseline HIV-1 RNA (log10 copies/mL)‡ | 3.11 | 3.5 | .53 |
| Number of subjects on ARV | 6 | 5 | .632-153 |
| Maximum peripheral CD4+ lymphocytes (cells/μL)‡ | 557 | 608 | .44 |
| CD4+ lymphocyte count increase from baseline (%)‡,2-154 | 79 | 86 | .44 |
| CD4+ lymphocyte count change from baseline, day 27 (%)‡,2-154 | −25 | −25 | .86 |
| Maximum peripheral CD34+ count (cells/μL)‡,2-155 | 63 | 39 | .93 |
| Number of subjects with positive HIV-1 culture findings# | 4 | 5 | .642-153 |
| Variable . | HIV-1 RNA Maximum Increase* . | ||
|---|---|---|---|
| <0.6 log10 (n = 9) . | ≥0.6 log10 (n = 9) . | P . | |
| Number of subjects in cohort 1 | 3 | 3 | |
| Number of subjects in cohort 2 | 4 | 2 | .84† |
| Number of subjects in cohort 3 | 2 | 4 | |
| Age (years)‡ | 40 | 42 | >.99 |
| Baseline CD4+ lymphocytes (cells/μL)‡ | 278 | 338 | .23 |
| Baseline HIV-1 RNA (log10 copies/mL)‡ | 3.11 | 3.5 | .53 |
| Number of subjects on ARV | 6 | 5 | .632-153 |
| Maximum peripheral CD4+ lymphocytes (cells/μL)‡ | 557 | 608 | .44 |
| CD4+ lymphocyte count increase from baseline (%)‡,2-154 | 79 | 86 | .44 |
| CD4+ lymphocyte count change from baseline, day 27 (%)‡,2-154 | −25 | −25 | .86 |
| Maximum peripheral CD34+ count (cells/μL)‡,2-155 | 63 | 39 | .93 |
| Number of subjects with positive HIV-1 culture findings# | 4 | 5 | .642-153 |
ARV, antiretroviral therapy; PBMC, peripheral blood mononuclear cells.
Subjects were divided into 2 groups based on the maximum increase of HIV-1 RNA levels above baseline during study days 5 through 7. Each of the indicated variables was compared for the 2 groups by a 2-tailed Wilcoxon rank sum test except where indicated.
Fisher exact test for combined cohorts 1, 2, and 3.
Values are the median.
z-test for comparing 2 binomial proportions.
Maximum value during days 4 through 7.
((CD4+ count − baseline CD4+ count) / (baseline CD4+ count)) × 100.
#Ability to propagate HIV-1 by coculture of cells in leukapheresis product obtained on day 4 or 5. HIV-1 culture was taken for 8 subjects with <0.6 log10 increase and for 8 subjects with ≥0.6 log10 increase in plasma HIV-1 RNA level.
Stem cell mobilization and harvesting affects the levels of HIV-1 DNA in CD4+ lymphocytes
Levels of HIV-1 DNA were quantified for all 18 subjects from PBMC collected during stem cell mobilization and harvesting. Microscopic examination of Wright's stained purified PBMC obtained from 4 subjects (610 021, 610 051, 610 219, and 610 354) revealed that at baseline and after stem cell mobilization 95% to 98% and 80% to 94%, respectively, of the cells in PBMC preparations were mononuclear cells. The levels of HIV-1 DNA were adjusted for the CD4+ lymphocyte count at each time point. In at least 2 PBMC samples obtained during days 4 through 27 (Table 1), 17 of the 18 subjects had quantifiable HIV-1 DNA. It was not detected in any of the specimens obtained from subject 61 146 (<0.9 log10copies/105 CD4+ lymphocytes). For the other 17 patients, HIV-1 DNA levels at baseline had inverse linear relationships to the baseline CD4+ lymphocyte counts (Spearman's ρ = −0.71;P = .0047).
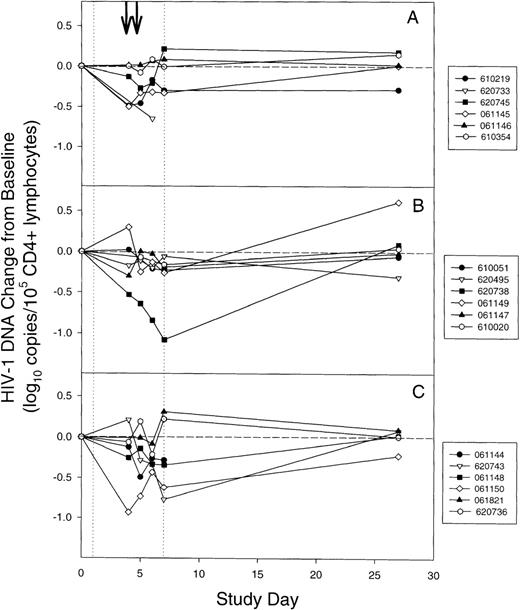
The HIV-1 DNA change from baseline was calculated by subtracting the baseline level of HIV-1 DNA (Table 1) from the level on days 4, 5, 6, 7, and 27. To avoid confounding effects from changes in antiretroviral therapy, this analysis was limited to the first 27 study days. In contrast to the effects of stem cell harvesting and mobilization on plasma HIV-1 RNA, the levels of HIV-1 DNA did not significantly increase from baseline (≥0.4 log10 increase) at any time during study days 4 through 7 (Figure 2). However, 6 subjects (61 144, 61 145, 061 150, 610 219, 620 733, and 620 743) had 0.5- to 0.9-log10 decreases from baseline CD4+ lymphocyte HIV-1 DNA levels, and 1 subject (620 738) had more than a 1.0-log10 decrease during study days 4 through 7. Overall, significant decreases in the median level of viral DNA of 0.19 and 0.21 log10 copies/105 CD4+ lymphocytes below baseline occurred on days 5 and 6, respectively (P = .01 for day 5 and P = .001 for day 6 using a Bonferroni adjustment for multiple tests; 2-tailed Wilcoxon signed-rank test for paired data). Changes in viral DNA levels from baseline on days 4, 7, and 27 were not statistically significant (Bonferroni-adjusted P = .24, P = .06 , andP > .99, respectively).
Changes in CD4+ lymphocyte HIV-1 DNA during mobilization and harvesting of stem cells for individual subjects.
(A) Cohort 1. (B) Cohort 2. (C) Cohort 3. (vertical dotted lines) Granulocyte colony-stimulating factor was administered on days 1 through 7. (arrows) Leukapheresis was performed on days 4 and 5. For subject 620 733 in cohort 1 and subject 061 144 in cohort 3, day-27 peripheral blood mononuclear cells samples were unavailable. The levels of CD4+ lymphocyte HIV-1 DNA for subject 061 146 was below the limit of detection at all time points and are not shown.
Changes in CD4+ lymphocyte HIV-1 DNA during mobilization and harvesting of stem cells for individual subjects.
(A) Cohort 1. (B) Cohort 2. (C) Cohort 3. (vertical dotted lines) Granulocyte colony-stimulating factor was administered on days 1 through 7. (arrows) Leukapheresis was performed on days 4 and 5. For subject 620 733 in cohort 1 and subject 061 144 in cohort 3, day-27 peripheral blood mononuclear cells samples were unavailable. The levels of CD4+ lymphocyte HIV-1 DNA for subject 061 146 was below the limit of detection at all time points and are not shown.
Relationship between plasma HIV-1 RNA and CD4+ lymphocyte HIV-1 DNA during stem cell mobilization and harvesting
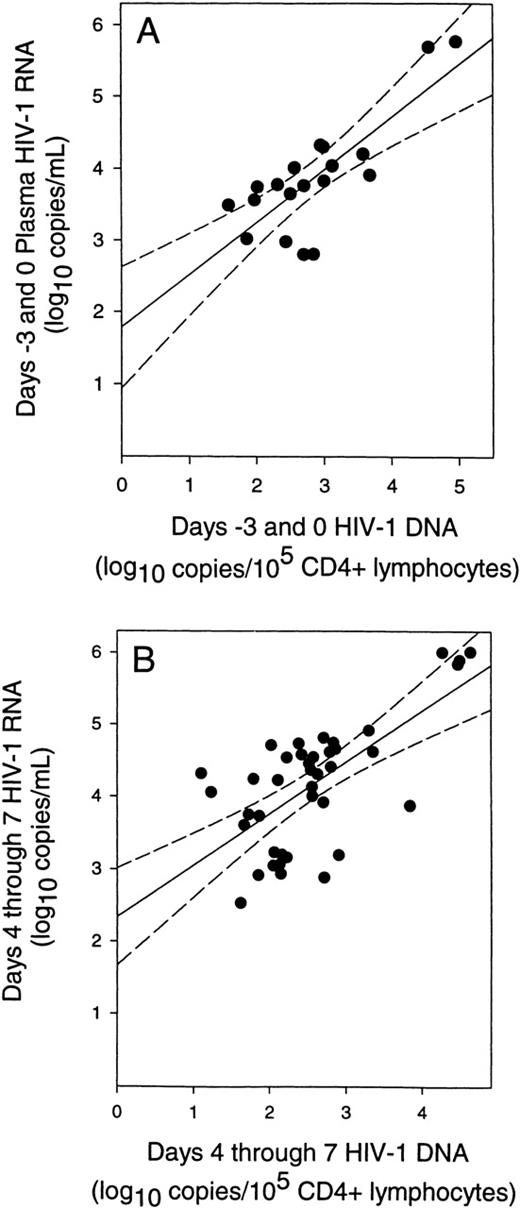
The relationship between plasma HIV-1 RNA and CD4+ lymphocyte DNA was determined for the 11 subjects who had measurable HIV-1 RNA levels on study days −3 or 0 (subjects 610 219, 620 745, 610 345, 610 051, 620 738, 61 149, 61 147, 61 144, 620 743, 61 148, and 61 821; Table 1). At study entry, HIV-1 RNA levels in plasma before stem cell harvesting and mobilization were directly related to HIV-1 DNA levels (Figure 3A). This relationship existed despite a broad range of CD4+ lymphocyte counts and viral loads for these 11 subjects. We therefore examined the relationship between plasma HIV-1 RNA and HIV-1 DNA during study days 4 through 7. Fit of these data also found a linear relationship (Figure 3B) with a slope (0.69) that was not significantly different from the slope observed at baseline (0.73; P = .83). However, the y-intercept (2.4 log10 copies/mL) was 0.6 log10 greater than at baseline (1.8 log10 copies/mL; P = .004) (compare Figures 3A and 3B). The slopes of both regression analyses were heavily dependent on the data from a single subject (61 144). Removal of this subject from both regression analyses did not change the conclusion that the slopes of both regression analyses were not significantly different (P = .95) and that there was a significant increase in the y-intercept (+0.4 log10;P = .029) for study days 4 through 7.
Correlation of plasma HIV-1 RNA levels with CD4+ lymphocyte HIV-1 DNA levels.
(A) Relationship of plasma HIV-1 RNA to CD4+ lymphocyte HIV-1 DNA levels before stem cell mobilization and harvesting. (solid line) Fit of data by linear regression (P < .0001;r = 0.79). (B) Relationship of plasma HIV-1 RNA levels to the level of CD4+ lymphocyte HIV-1 DNA during stem cell mobilization and harvesting (days 4 through 7) for each subject. (solid line) Fit of data by linear regression (P < .0001; r = 0.68). (dashed lines) 95% confidence intervals for the regressions. This analysis used only data pairs for which quantifiable values of both HIV-1 RNA and DNA levels were available.
Correlation of plasma HIV-1 RNA levels with CD4+ lymphocyte HIV-1 DNA levels.
(A) Relationship of plasma HIV-1 RNA to CD4+ lymphocyte HIV-1 DNA levels before stem cell mobilization and harvesting. (solid line) Fit of data by linear regression (P < .0001;r = 0.79). (B) Relationship of plasma HIV-1 RNA levels to the level of CD4+ lymphocyte HIV-1 DNA during stem cell mobilization and harvesting (days 4 through 7) for each subject. (solid line) Fit of data by linear regression (P < .0001; r = 0.68). (dashed lines) 95% confidence intervals for the regressions. This analysis used only data pairs for which quantifiable values of both HIV-1 RNA and DNA levels were available.
Discussion
The plasma HIV-1 RNA level is a sensitive marker of HIV-1 replication that predicts disease progression and response to antiviral therapy in infected persons.23-29 Half the subjects who underwent stem cell mobilization by G-CSF and harvesting by leukapheresis had transient increases in plasma HIV-1 RNA levels that exceeded the variability of this assay. Thus, these subjects had significant increases in HIV-1 replication. This finding has not been reported previously, and it contradicts a previous report that stem cell mobilization and harvesting did not activate HIV-1 replication.12 In the previous study, treatment of 7 HIV-1–infected subjects with G-CSF, 10 μg/kg per day for 6 consecutive days, followed by leukapheresis on day 6, did not affect serum p24 antigen levels. However, the serum p24 antigen level is an insensitive marker of HIV-1 replication and disease progression,30-32 and increases in HIV-1 replication that produce changes in plasma HIV-1 RNA do not always produce measurable changes in serum p24 antigen.33 Thus, it is likely that the use of a sensitive marker of HIV-1 replication in the current study allowed for the detection of an effect on viral replication that was previously unnoticed.
The magnitude of changes in HIV-1 RNA levels that occurred in the subjects in the current study are comparable to those observed after immunization with recall antigen,34 influenza vaccine,35 and IL-2 injection33 and during AIDS-related opportunistic infection.36 The results of these studies suggest that immune stimulation leads to enhanced HIV-1 replication. Treatment of HIV-1 infection with 3-drug combinations that include an HIV-1 protease inhibitor effectively suppresses viral replication by preventing de novo infection. However, populations of cells that are latently infected with HIV-1 persist despite effective therapy,5 6 and available antiretroviral therapies do not prevent the activation of proviral gene expression in latently infected cells.
In the current study, transient increases in plasma HIV-1 RNA levels occurred in 5 subjects despite treatment with potent 3-drug regimens. The finding that HIV-1 RNA levels promptly returned to baseline by day 27 for 4 of these 5 subjects suggests that stem cell mobilization and harvesting stimulated viral gene expression in a population of latently infected cells and that the “breakthrough” virus remained sensitive to the antiretroviral agents in use at the time. It is unlikely that stem cell harvesting and mobilization fostered the replication of antiretroviral, drug-resistant virus in these 4 subjects. However, 1 subject (61 150) who had undetectable HIV-1 RNA at study entry experienced a sustained and unresolved increase in HIV-1 RNA level. It is possible that the breakthrough virus acquired mutations that confer resistance to the antiretroviral agents in use. Analyses are planned to determine whether the virus that emerged during stem cell mobilization and harvesting had reduced susceptibility to antiviral agents.
The factors associated with increased HIV-1 replication during stem cell mobilization and harvesting are not evident. Subjects who experienced increased HIV-1 RNA levels were similar to those subjects in whom plasma RNA levels did not increase from baseline by all comparisons. However, the relatively small number of subjects in our study limits the strength of these comparisons. In 2 subjects (610 051, 61 147) (Figure 1), significant increases in plasma HIV-1 RNA were first apparent on day 4, before the first leukapheresis. This suggests that mobilization with G-CSF, not harvesting with leukapheresis, caused increased viral replication in these subjects. Because treatment of neutropenia in persons with HIV-1 infection with G-CSF at doses of 5 μg/kg per day or less does not affect plasma HIV-1 RNA levels,37 38 G-CSF may have a dose-dependent effect on HIV-1 replication. Given that all subjects in the current study underwent stem cell harvesting and mobilization, we cannot exclude the possibility that the combination of treatment with G-CSF and leukapheresis activated HIV-1 replication.
At study entry the median level of HIV-1 DNA in circulating CD4+ lymphocytes ranged from 1 copy/1000 cells for subjects with early-stage HIV-1 infection (cohort 1) to 1 copy/100 cells for subjects with advanced HIV-1 infection (cohort 3). This finding is consistent with previous estimates of the frequency of HIV-1–infected cells in the circulating CD4+ cells of HIV-1–infected persons.39-41 In contrast to the effect on plasma HIV-1 RNA levels, the levels of HIV-1 DNA in circulating CD4+ lymphocytes did not increase during stem cell mobilization and harvesting. Rather, HIV-1 DNA levels decreased during treatment and returned to baseline within 3 weeks of treatment discontinuation. The cause of decreased HIV-1 DNA levels during stem cell harvesting and leukapheresis is unknown, but possible explanations include clearance of productively infected cells by the immune system, mobilization of uninfected CD4+ lymphocytes to the circulatory compartment, removal of HIV-1–infected cells during leukapheresis, and cytopathic effects from the increased HIV-1 replication that occurred during stem cell harvesting and mobilization.
At study entry, a linear relationship between the plasma HIV-1 RNA level and the HIV-1 DNA level in circulating CD4+ lymphocytes existed. This finding is consistent with a previously reported observation that the levels of serum HIV-1 RNA are directly related to the number of HIV-1–infected cells in the circulatory compartment, regardless of disease state, and that PBMC and serum are closely related viral compartments.42 It is important to note that our analysis was limited to the 11 subjects with measurable levels of plasma HIV-1 RNA and PBMC HIV-1 DNA at baseline. Thus, the subjects in this subgroup were not on maximally effective antiretroviral regimens. During stem cell mobilization and harvesting, the linear relationship between plasma HIV-1 RNA and PBMC DNA was maintained but included a 0.6-log10 upward shift in the curve. Therefore, within the circulatory compartment, the ratio of cell-free virus to HIV-1–infected cells increased. The increased ratio of cell-free virus to infected cells is consistent with the observed overall increase in plasma level of HIV-1 RNA (+0.3 log10 copies/mL) and the concomitant overall decrease in the level of HIV-1 DNA in CD4+ lymphocytes (−0.2 log10 copies/105cells). These findings provide additional evidence that the increased plasma HIV-1 RNA levels that occurred during stem cell harvesting and mobilization resulted from stimulated production of HIV-1 from a priori infected cells and did not result from mobilization of HIV-1– infected cells to the circulatory compartment.
Recent studies have used mobilized hemopoietic progenitor cells harvested from HIV-1–infected persons to demonstrate the feasibility of gene therapy approaches to the treatment of HIV-1 infection,13 43 and studies to evaluate the safety of autologous transplantation of genetically modified progenitor cells are in progress. In the current study we observed that stem cell mobilization and harvesting caused a previously undescribed stimulation of HIV-1 replication in infected persons. It is important to note that for most subjects plasma HIV-1 RNA returned to near baseline levels within 3 weeks after treatment was discontinued and that increased HIV-1 replication was not associated with any untoward effects. The increased viral replication did not affect changes in the CD4+ lymphocyte count or in the ability to mobilize CD34+ cells. Therefore, the results of our study do not preclude further investigations that use this approach to collect hemopoietic stem cells from HIV-1–infected persons. However, investigators and HIV-1–infected subjects who participate in stem cell mobilization and harvesting studies should be aware of the possibility that HIV-1 replication could be stimulated. Careful monitoring of HIV-1 virus load should be part of future studies that use this technology.
Acknowledgments
The authors thank the other members of the ACTG 285 Study Team for their contributions: Larry Fox, Dawn Bell, Rebecca Betensky, Simon Chiu, Jim Leone, Janice Jacovini, and Scharla Estep Riley. They also thank Amgen, Inc, for supplying Filgrastim for use in this study, and they thank Monique Givens, Tiffany Kirkbride, and David Shuggarts for the HIV-1 cultures.
Supported by grants from the Department of Health and Human Services to the Adult AIDS Clinical Trials Units (AI32770), the University of Colorado General Clinical Research Center (RR0051), and the University of Colorado Cancer Center Flow Cytometry Core Laboratory (CA46934); by a Virology Advanced Technology Laboratory Award from the Adult ACTG (AI38858), University of Washington CFAR (AI-30731); and by Amgen, Inc.
Reprints:Thomas B. Campbell, Campus Box B-168, University of Colorado Health Sciences Center, 4200 East Ninth Avenue, Denver, CO 80262; e-mail: thomas.campbell@uchsc.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal