Abstract
Little is known about the natural history and the pathogenicity of the TT virus (TTV). We present our findings of a cross-sectional study based on the TTV DNA screening of 173 multiple-transfused patients and a longitudinal study based on the follow-up of TTV DNA–positive patients. Overall, 48 patients (27.7%) tested positive for TTV DNA. The influence of the number of blood donor exposures on the prevalence of blood-borne viral infection indicates that TTV, hepatitis C virus (HCV), and an RNA virus known as GB virus C/hepatitis G virus (GBV-C/HGV) share a parenteral transmission, but that TTV, in contrast to the 2 other viruses, is also transmitted by at least another efficient means. The patients having a well-defined date of TTV infection were positive for TTV DNA during a mean period of 3.1 years. A chronic infection was observed in 31 cases (86%). TTV carriage appeared clinically benign in all patients. No clinical evidence of a disease potentially linked to the TTV infection was observed in patients with TTV DNA carriage over several years. The majority of TTV carriers had no biochemical evidence of liver disease. The prevalence of elevated serum alanine aminotransferase (ALT) level was higher in the TTV DNA–positive group, even in the absence of HCV infection, but the observed peaks of ALT level were most often transient and very mild. The prevalence of TTV DNA observed in blood recipients is consistent with that of TTV infection observed in blood donors. TTV infection frequently tends to persist. (Blood. 2000;95:347-351)
By means of representational differential analysis,1 a novel unenveloped single-stranded DNA virus has recently been discovered in serum samples of posttransfusion non-A to E hepatitis.2,3 This virus has been designated TT virus (TTV), after the initials of the first patient from whom the virus was isolated. A high prevalence of TTV infection has been observed in individuals with parenteral risk exposure, such as recipients of blood products3-6 and intravenous drug users.3,7 However, as suggested by the prevalence of TTV infection in populations at low risk of parenteral exposure, such as blood donors,3,4,7,8 TTV is not solely transmitted parenterally. A vertical transmission has been documented,9and a community-acquired transmission seems probable.4,10 11 In fact, the epidemiology of TTV infection is not yet fully resolved, and other ways of transmission are possible. Large epidemiological studies are limited by the fact that in the absence of any reliable serological assay for the diagnosis of TTV infection, viral DNA detection by polymerase chain reaction (PCR) is the only available diagnostic tool indicating an ongoing infection.
Furthermore, little is known about the clinical significance and the natural history of TTV infection. If the presence of viral DNA has been detected in patients with non-A to E hepatitis, the responsibility of the virus in a specific liver disease is still debated. Recently, cases of chronic hepatitis have been linked to the presence of genomic sequences of an RNA virus termed GB virus C/hepatitis G virus (GBV-C/HGV), but this new agent was subsequently cleared in this pathology. 12-15
As follow-up studies of blood recipients were very accurate in defining the natural history and the potential outcome of infections by blood-borne viruses such as HCV16 and GBV-C/HGV,15 it seemed interesting to use such an approach with TTV infection. We present here our findings of a cross-sectional study based on the TTV DNA screening of multiple-transfused patients and a longitudinal study based on the follow-up of TTV DNA–positive multiple-transfused patients.
Materials and methods
Study population
The study population consisted of 173 immunocompetent patients multiple-transfused with leukodepleted packed red cells (PRC) and affected with hemoglobinopathy (thalassemia major or sickle cell disease) or with chronic aplastic anemia (not associated with hepatitis). All patients lived and received transfusions in the Paris area. There were 96 males and 77 females, with a mean age of 13.7 years (range, 1-89 years). They belonged to a prospective cohort initiated in 1988, and some of the patients had been followed for 11 years. The number of PRC transfusions received by each patient was recorded. During the follow-up of each patient, serum samples were collected during a routine examination preceding each transfusion. Serum samples were separated from whole blood within 2 hours after venipuncture, aliquoted, and stored frozen at -80°C until tested.
Procedures
Determination of TTV DNA in serum.
TTV DNA was extracted from 200 μL of serum (High Pure Viral Nucleic Acid Kit; Boehringer Mannheim, Frankfurt, Germany) and resuspended in 50 μL of elution buffer. We submitted 10 μL DNA (in 50 μL of final volume) to 2 rounds of PCR amplification with 2 different primer pairs, which are described by Simmonds4and located in the ORF1 region. The first round of PCR amplification was performed with 2.6 units of expanded High Fidelity enzyme mix (Boehringer Mannheim) and was carried out for 10 cycles, 30 seconds each, at 94°C, 55°C, and 68°C, and for 30 cycles, 30 seconds each, at 94°C, 55°C, and 68°C (with 5 seconds of elongation by cycle). This was followed by 68°C during 7 seconds after the last cycle (Perkin Elmer 9600 thermocycler; Perkin Elmer, Norwalk, CT).
The sequences of the selected primers for PCR amplification were A5430, 5′ CAG ACA GAG GAG AAG GCA ACA TG 3′ (sense strand), and A5427, 5′ TAC CAY TTA GCT CTC TAT TCT WA 3′ (antisense strand, degenerated primer), where Y = T or C, W = A or T.4 We transferred 2 μL (in 50 μL of final volume) of the first round of PCR, amplified the PCR for 10 cycles, and then amplified it for 10 cycles in the same conditions as the first round. The sequences of the selected primers for this second round were A8761, 5′ GGM AAY ATG YTR TGG ATA GAC TGG 3′ (sense strand, degenerated primer), and A5432, 5′ CTA CCT CTG GCA TTT TAC CA 3′ (antisense strand), where M = A or C, Y = C or T, R = A or G. The product of the first PCR round measured 328 base pairs (bp) and that of the second PCR round measured 278 bp. The amplicons were electrophoresed in 2% agarose gel, stained with ethidium bromide, and photographed under ultraviolet light. This procedure was validated by a French multicentre quality control trial.
Use of other virological markers.
The detection of serum HCV antibodies was performed through a third-generation enzyme-linked immunosorbent assay (ELISA) (EIA 3.0 HCV; Ortho Diagnostic Systems, Roissy, France), with validation of a positive result by a third-generation recombinant immunoblotting assay (Riba 3.0, Ortho Diagnostic Systems). The detection of serum human immunodeficiency virus (HIV) antibodies was performed through a specific ELISA (Sanofi-Diagnostics Pasteur, Marnes-La-Coquette, France), with confirmation of a positive ELISA by HIV-1 Western blot (Sanofi-Diagnostics Pasteur). The detection of serum human T-lymphotropic virus (HTLV-I) antibodies was performed through a specific ELISA (Abbott HTLV-I EIA; Abbott Laboratories, North Chicago, IL), with confirmation of a positive ELISA by Western blot (DB HTLV Blot 2.3; Diagnostic Biotechnology, Geneva, Switzerland).
Serum GBV-C/HGV RNA was detected using in-house reverse transcription PCR assay as previously described.15 The detection of serum anti-E2 antibody (marker of recovery to GBV-C/HGV infection) was made through an immunoassay using recombinant E2 (Anti-HGenv Enzymun-test, Boehringer Mannheim) according to the manufacturer's instructions.17 Since a majority of the patients had a serological pattern of vaccination against hepatitis B virus (HBV) infection, we did not retain the markers of HBV infection in the present study.
Determination of serum alanine aminotransferase activities.
Serum alanine aminotransferase (ALT) activities were determined by an automated method (Vitros 950, Ortho Diagnostics Systems) and expressed in IU/L. The upper limit of normal ALT activities was 40 IU/L.
Records of clinical parameters.
Any sign of hepatitis (jaundice, abdominal pain) or any unexplained clinical symptomatology potentially linked to a primary or chronic viral infection (such as cutaneous eruption, adenopathies, fever) were recorded, and/or we interviewed the patient or the parents, in the case of a minor. If a known date of TTV infection (defined as a negative viral DNA PCR followed by a positive viral DNA PCR) was evidenced through the TTV DNA screening, we carefully watched for symptoms of a primary infection, especially in the first months following contamination.
Methodology
TTV DNA screening was performed yearly, and sequential frozen serum samples were collected from each patient during the baseline visit (in 1988 for the majority of the patients, later if the patient was first seen after this date) to the most recent visit. (The study's censor date was December 1998 if the patient was still alive and followed in our center at that time, earlier if the patient was dead or did not return to follow-up).
Cross-sectional study.
We established the mean age, sex ratio, type of underlying transfusional disease (eg, thalassemia major, sickle cell disease), number of patients with a serum ALT level higher than the upper limit of normal, number of donor exposures (determined by transfusions of PRC received over entire life), and prevalence of other viral markers. These factors were included when comparing patients within TTV DNA–positive and TTV DNA–negative groups (see below).
Longitudinal study.
Per our protocol, this longitudinal study included only individuals who had a follow-up of at least 3 years among the individuals found positive through TTV DNA screening of the cross-sectional study. A chronic TTV infection was defined as a positive PCR in at least 2 successive yearly serum samples. A transient TTV infection was defined as a positive PCR sample when the yearly samples collected before and after were TTV DNA-negative. Additional samples, other than the annual samples, were available for the large majority of patients. Thus the protocol stated that all samples situated between the last annual negative TTV DNA sample and the first annual positive TTV DNA sample would be systematically tested through PCR in order to provide optimal precision as to the date of TTV DNA infection. Furthermore, irrespective of the results of the TTV DNA PCR, the serum ALT level was determined for each patient's yearly sample.
Statistical analysis.
Results were expressed as mean ±1 SD or as percentages ±95% of confidence intervals (CI). A chi-square or Fisher exact test was used to compare categorical data, and the Studentt test was used to compare quantitative data. Differences were considered significant at P < 0.05.
Results
Cross-sectional study
Among the 173 multiple-transfused patients in the study, 48 patients (27.7%; 95% CI: 21.0-34.4) tested positive for TTV DNA.
Epidemiological characteristics.
There were no significant differences between the TTV DNA–positive group and the TTV DNA–negative group with regard to age at the censor date and gender distribution (Table 1). The TTV DNA–positive group included 19 (39.6%) thalassemia major patients, 23 (47.9%) sickle cell disease patients, and 6 chronic aplastic anemia patients (12.5%), while the TTV DNA–negative group included 16 (12.8%) thalassemia major patients, 100 (80.0%) sickle cell disease patients, and 9 (7.2%) chronic aplastic anemia patients (P < .001).
Epidemiological, virological, and biochemical characteristics of TTV DNA-positive and TTV DNA-negative patients
| . | TTV DNA– Positive Individuals (n = 48) . | TTV DNA– Negative Individuals (n = 125) . | Differences . |
|---|---|---|---|
| Mean age (years) [SD] | 13.6 (13.0) | 13.8 (16.2) | NS |
| Male (%) | 29 (60.4) | 67 (53.6) | NS |
| Viral markers (%) | |||
| HCV antibody positive | 8 (16.7) | 18 (14.4) | NS |
| GBV-C/HGV RNA positive | 4 (8.3) | 12 (9.6) | NS |
| GBV-C/HGV exposed* | 11 (22.9) | 25 (20.0) | NS |
| HIV antibody positive | 0 | 3 (2.4) | NS |
| HTLV-I antibody positive | 0 | 2 (1.6) | NS |
| Number of individuals with ALT level above 40 IU/L (%) | 10 (20.8) | 10 (8.0) | 0.02 |
| Number of individuals with ALT level above 40 IU/L (%) (excluding HCV-positive individuals) | 7 (15.6) | 5 (4.2) | 0.03 |
| . | TTV DNA– Positive Individuals (n = 48) . | TTV DNA– Negative Individuals (n = 125) . | Differences . |
|---|---|---|---|
| Mean age (years) [SD] | 13.6 (13.0) | 13.8 (16.2) | NS |
| Male (%) | 29 (60.4) | 67 (53.6) | NS |
| Viral markers (%) | |||
| HCV antibody positive | 8 (16.7) | 18 (14.4) | NS |
| GBV-C/HGV RNA positive | 4 (8.3) | 12 (9.6) | NS |
| GBV-C/HGV exposed* | 11 (22.9) | 25 (20.0) | NS |
| HIV antibody positive | 0 | 3 (2.4) | NS |
| HTLV-I antibody positive | 0 | 2 (1.6) | NS |
| Number of individuals with ALT level above 40 IU/L (%) | 10 (20.8) | 10 (8.0) | 0.02 |
| Number of individuals with ALT level above 40 IU/L (%) (excluding HCV-positive individuals) | 7 (15.6) | 5 (4.2) | 0.03 |
NS indicates nonsignificant.
Indicates positive for GBV-C/HGV RNA and/or anti-E2 antibody.
Virological and biochemical characteristics.
Among the 173 multiple-transfused patients, 26 were HCV-positive (15.0%; 95% CI: 9.7-20.3); 2 were HIV-positive (1.7%; 95% CI: 0.0-3.9); 2 were HTLV-I-positive (1.2%; 95% CI: 0.0-2.2); 16 were GBV-C/HGV RNA-positive (9.2%; 95% CI: 4.9-13.5); and 36 were GBV-C/HGV–exposed (20.8%, 95% CI: 14.8-26.8). (GBV-C/HGV–exposed patients are positive for viral RNA and/or anti-E2 antibody.) Table 1 lists the number of HCV-positive and HCV-negative individuals, GBV-C/HGV RNA-positive and RNA-negative individuals, GBV-C/HGV–exposed and GBV-C/HGV–unexposed (ie, negative for viral RNA and anti-E2 antibody) individuals in both the TTV DNA–positive and TTV DNA–negative groups. There were no TTV DNA–positive individuals coinfected by HIV or HTLV-I.
The number of HCV-negative individuals with a serum ALT level above the upper limit of normal was significantly higher (P < 0.02) in the TTV DNA–positive group than in the TTV DNA–negative group (see Table 1). The mean ALT level of these 2 groups was 53 IU/L (range, 43-69 IU/L) and 52 IU/L (range, 42-73 IU/L), respectively (nonsignificant difference).
Number of blood-donor exposures.
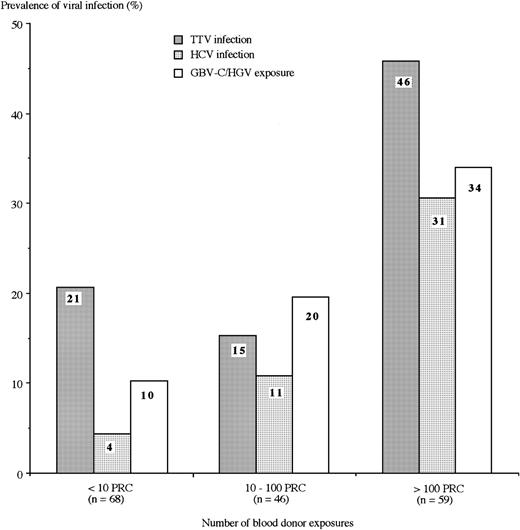
During the lifetimes of the 48 TTV DNA–positive individuals, 14 patients had received <10 PRC transfusions, 7 patients had received between 10 and 100 PRC transfusions, and 27 patients had received >100 PRC transfusions. Figure 1 shows the prevalences of TTV, HCV, and GBV-C/HGV infection in the 173 multiple-transfused patients according to the quantity of blood-donor exposures of PRC transfusions received during their lifetimes. Prevalence was significantly related to the blood-donor exposure for all 3 viruses (P < 0.001). However, the significant difference between the 3 levels of blood-donor exposure was the TTV prevalence between the highest level and each of the other 2 levels.
Prevalences of TTV and HCV infections and GBV-C/HGV exposure. Includes characteristics of 173 multiple-transfused patients (according to the number of blood-donor exposures). GBV-C/HGV exposure includes individuals positive for viral RNA and/or anti-E2 antibody. The prevalence was significantly related to the blood-donor exposure for all 3 viruses (P < .001). However, for TTV, the significant overall difference was mainly due to the difference between the group with greater than 100 PRC versus each of the 2 other groups.
Prevalences of TTV and HCV infections and GBV-C/HGV exposure. Includes characteristics of 173 multiple-transfused patients (according to the number of blood-donor exposures). GBV-C/HGV exposure includes individuals positive for viral RNA and/or anti-E2 antibody. The prevalence was significantly related to the blood-donor exposure for all 3 viruses (P < .001). However, for TTV, the significant overall difference was mainly due to the difference between the group with greater than 100 PRC versus each of the 2 other groups.
Follow-up study of TTV DNA–positive patients
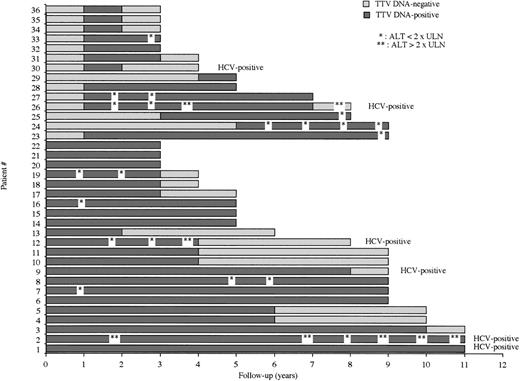
Among the 48 TTV DNA–positive patients, 36 had a follow-up of at least 3 years. The results of the TTV DNA–PCR assay in the sequential samples of these 36 multiple-transfused patients during the entire follow-up period are shown in Figure2. Prior to the beginning of the study, 22 patients had been infected with TTV. Fourteen patients were TTV-infected during the study period and thus had a well-defined date of infection. In 8 of these latter 14 individuals, the analysis of serial samples collected at each visit allowed specifying (within 1 month) the date when the PCR was negative on a sample at a given time and positive on the sample collected 1 month afterward. At the end of the study period, 19 patients were still positive for TTV DNA, whereas 17 were negative for TTV DNA PCR.
Results of the TTV DNA–PCR assay and serum ALT level during the whole follow-up period of 36 multiply-transfused patients. ULN = upper limit of normal ALT level.
Results of the TTV DNA–PCR assay and serum ALT level during the whole follow-up period of 36 multiply-transfused patients. ULN = upper limit of normal ALT level.
The 36 patients of this longitudinal study were shown to carry TTV DNA over a mean period of 4.7 years (range, 1-11 years), for a mean observational period (defined as the follow-up period since the first sample that was positive for TTV DNA) of 5.8 years (range, 2-11 years). Within the limits of this observational period, the 14 patients with a well-defined date of infection were positive for TTV DNA during a mean period of 3.1 years (range, 1-8).
Among the 36 patients, a chronic infection (defined by a PCR positive for viral DNA on at least 2 successive yearly samples) was observed in 31 cases (86%). The longest duration of TTV DNA carriage observed in the study was 11 years (ie, the longest follow-up of the study at the censor date). A transient TTV infection (defined by a PCR positive for TTV DNA on a yearly sample, when the yearly samples collected before and after were TTV DNA negative) was observed in 4 patients (11%). A patient (n°29 on Figure 2) was TTV DNA–positive only during the last year of follow-up and could not be defined as having either a transient or a chronic infection. There were no patient presenting a reappearance of viremia after the loss of TTV DNA.
The 36 patients failed to evidence a clinical symptomatology that was not attributable to the underlying transfusional disease. None of the 14 individuals with a well-defined date of TTV infection presented a clinical symptomatology evoking a primary viral infection during the first months of TTV infection; this included the 8 individuals in whom we could establish, within 1 month, the date when PCR became positive.
As shown in Figure 2, an elevated serum ALT level was observed during at least 1 visit in 12 patients (33%). The 3 individuals having the highest ALT levels were coinfected by HCV. In the 9 other patients, when retaining the highest value observed in each individual, the mean ALT level was 58 IU/L. No serum ALT level higher than twice the upper limit of normal was observed in TTV DNA–positive, HCV-negative patients. An elevated serum ALT level was observed in at least 2 successive samples in 7 TTV DNA–positive individuals (among them, 3 were coinfected by HCV). The serum ALT level was normal in the 4 patients of the study in whom a transient TTV infection was observed.
Discussion
The prevalence of individuals viremic for TTV DNA observed in the blood recipients of our study (27.7%) was higher than that of other studied blood-borne viruses. This prevalence was consistent with that of overall TTV infection observed in the blood donors of France (5.3%).7 Moreover, a higher exposure to TTV infection is likely in the blood-recipient population because of the high frequency of viremia in blood donations. Indeed, if a serological test evidencing a past and resolved TTV infection (equivalent to the assay evidencing the anti-E2 antibody detectable after the loss of GBV-C/HGV RNA15 17) was available, such an assay, combined to PCR, would reflect the total TTV exposure. Indeed, all epidemiological studies of TTV to date, including the present study, underestimated the true prevalence of TTV exposure since they were based only on the detection of viral DNA by PCR.
In the blood recipients of our series, the influence of the number of blood donor exposures on the prevalence of infection by blood-borne viruses, as shown in Figure 1, indicates that TTV, HCV, and GBV-C/HGV share a parenteral transmission, but that TTV, by contrast to the 2 others viruses, is also transmitted by at least another efficient way of transmission. The prevalences reported in populations at low risk to blood-borne viruses, such as blood donors,3,7 also indicate that parenteral transmission is not the exclusive means of TTV infection. Alternative routes of transmission, such as maternal8 or fecal-oral10 infection, are highly probable in the TTV spread, whereas sexual transmission seems inefficient.18
Our study, which provides a long-term follow-up of TTV DNA–positive individuals, shows that the TTV infection may be transient in some cases, but it frequently tends to persist. The longest duration of TTV carriage that we observed in our series was 11 years; the real duration of the viremia in this case was probably longer because the patient was already positive for TTV DNA when included in the study and was still positive at the last visit. Indeed, only a frequent chronic carriage of TTV in circulating blood of infected persons can explain the high prevalence of viremic individuals reported in the blood donor populations. Furthermore, the prevalence observed in our population was perhaps further underestimated by the characteristics of the primer pairs used in our PCR assay. A recently reported study on the prevalence of TTV infection in thalassemic patients showed a strikingly higher prevalence in this population.19
TTV infection is frequently cleared with time, but the mechanisms allowing a recovery to take place after several years of persistent carriage of the viral DNA are unknown: Host or viral characteristics or a combination of the two probably intervene. Furthermore, it is probable, but not formally established, that the patients having lost TTV DNA have totally eradicated the virus, with a definitive loss of infectivity. We failed to observe a reappearance of a viremia in the TTV DNA-negative patients of our series, but the possibility of such a phenomenon cannot be excluded, particularly in an immunocompromised host.
Generally, viruses that establish persistent infection in humans produce chronic diseases in their hosts. In our study, whether the infection was transient or chronic, TTV carriage appeared clinically benign in all patients. No clinical evidence of a disease potentially linked to the TTV infection was observed in our multiple-transfused patients who showed TTV DNA carriage over several years of follow-up. Furthermore, the majority of TTV carriers of our series had no biochemical evidence of liver disease. The prevalence of elevated serum ALT level was higher in the TTV DNA–positive group, even in the absence of HCV infection, but the observed peaks of ALT level were most often transient and very mild (lower than twice the upper limit of normal in all cases). Such mild elevations of the ALT level could be nonspecific in this multiple-transfused population. Moreover, if TTV infection was directly responsible for the slight and transient elevation of the ALT level observed in our patients, these data are insufficient to include TTV in the group of hepatitis viral agents. A mild elevation of the ALT level may be observed during the primary infection by other viruses such as B19 parvovirus, cytomegalovirus, or HIV.
Since a serological assay reflecting an ongoing TTV infection is not yet available, blood donations should be systematically screened for TTV based on the detection of the viral genome by PCR. However, in terms of transfusion safety, such a screening would be indicated only if the virus were able to generate a pathology in humans and if it were largely community-acquired by means other than fecal-oral transmission.
Acknowledgment
The authors are grateful to David Thorup for advice on preparation of this manuscript.
Reprints:Jean-Jacques Lefrère, Institut National de la Transfusion Sanguine et Faculté Saint-Antoine, UniversitéPierre et Marie Curie 53 Boulevard Diderot, 75012 Paris, France; e-mail: lefrere@worldnet.fr.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal