Abstract
The human erythropoietin gene is expressed predominantly in the kidney and liver in response to hypoxia. Although the signaling cascade for hypoxia is present in many different cell types, the expression of erythropoietin is restricted to only a few tissues. The authors show that the promoter and 5′-untranslated region (5′-UTR) of the erythropoietin gene comprise a CpG island and that methylation of the CpG island correlates inversely with expression. Methylation represses the expression of the erythropoietin gene in 2 ways: high-density methylation of the 5′-UTR recruits a methyl-CpG binding protein to the promoter, and methylation of CpGs in the proximal promoter blocks the association of nuclear proteins. (Blood. 2000;95:111-119)
CpG dinucleotides (CpGs) are underrepresented in vertebrate DNA; they occur at 0.2 to 0.25 the frequency expected from the overall nucleotide composition.1 Despite the low average abundance of CpGs, the human genome contains approximately 45,000 limited regions of high-density CpGs known as CpG islands,2,3 most of which are associated with promoters, the 5′ end of genes, or both.4 Housekeeping genes regularly contain CpG islands, as do approximately 40% of genes with tissue-specific expression patterns.5,6 In vertebrate genomic DNA, 60% to 90% of CpGs are modified by symmetric methylation of the fifth position of the cytosine ring (methyl-CpGs).7In contrast, cytosines in CpG islands in the promoters of active genes are largely unmethylated.8
Gene silencing is the legacy of CpG island methylation. Indeed the transcriptional activity of a promoter often correlates inversely with its methylation status,8,9 and agents that cause demethylation of DNA often induce the expression of previously silent genes.10,11 The best examples of gene silencing induced by CpG methylation are genomic imprinting12 and X-chromosome inactivation in females.13 More recently, DNA methylation has been implicated in the silencing of tumor-suppressor genes14-18 and other genes in cancer cells.19-21 In tissue-specific genes CpG island methylation suppresses expression in tissues that do not require those particular genes.22 Methylation of CpG islands may be epigenetic: the factors and conditions necessary to initiate methylation may not be required for the maintenance of methylation and the subsequent control of gene expression.
Two mechanisms have been proposed to explain the transcriptional repression resulting from CpG methylation. Methylation of cytosines in a DNA recognition element can block the binding of sequence-specifictrans-acting proteins.7 DNA methylation also promotes the binding of a family of proteins that interferes directly or indirectly with the transcriptional apparatus.23Methyl-CpG binding protein-1 (MeCP1) represses transcription from regions with high-density CpG methylation (> 10 methyl-CpGs),24,25 and low-density CpG methylation decreases transcription by recruiting histone deacetylase activity to chromatin through interactions with methyl-CpG-binding protein-2 (MeCP2).26-29 CpG methylation also enhances nucleosome stability by attracting histone H1 to linker regions containing methyl-CpGs.30 31
The human erythropoietin gene is expressed predominantly in kidney32,33 and liver,34,35 in limited amounts by some hematopoietic tissue,36,37 and in small amounts in other tissues.38,39 Erythropoietin expression is stimulated by hypoxia,40 and the elements that control tissue-specific, hypoxia-induced expression have been studied in detail. When expressed as a transgene in mice, the basal human erythropoietin promoter (∼400 base pair [bp]) has little tissue specificity41—tissue-specific expression requires 6 and 22 kb human DNA upstream of the transcription start site.42,43A transgene driven by a 6-kb murine erythropoietin promoter is expressed predominantly in proximal convoluted tubular cells after hypoxic stress.44 In contrast, in situ hybridization experiments suggest that expression of the native erythropoietin gene in the kidney is limited to infrequent interstitial peritubular cells.45-47 Hepatocytes and Ito cells in the adult liver contribute a small fraction to the erythropoietin pool,35,39,48,49 whereas the extent of erythropoietin production by hematopoietic cells has not been extensively characterized.36,37 Hep3B and HepG2 are human hepatoma cell lines that produce small amounts of erythropoietin under normoxic conditions but that increase their erythropoietin production up to 100-fold after exposure to hypoxia or cobalt.50-52
We hypothesize that methylation of the erythropoietin gene in nonexpressing cells contributes to tissue-specific expression. We show that the human erythropoietin gene contains a CpG island in its promoter and 5′-UTR that is differentially methylated in respect to gene expression. We demonstrate that CpG methylation of the erythropoietin gene blocks the binding of sequence-specific DNA binding proteins, recruits methyl-CpG binding proteins, and represses transcription.
Materials and methods
Cell culture
Hep3B and HeLa cells were cultured in 5% CO2 in minimum essential medium (Gibco-BRL, Gaithersburg, MD) containing 10% iron-enriched bovine calf serum (Hyclone Laboratories, Logan, UT).
Southern blot analysis
Genomic DNA was digested with Xba1 and Eag1, separated on a 1% agarose gel in 1 × TAE buffer, and transferred to Nytran Plus membrane (Schleicher & Schuell, Keene, NH) in 0.4 mol/L NaOH. The blot was incubated overnight at 42°C in 50% formamide with the radiolabeled Xba1 fragment that flanks the transcription start site (Figure 1). The blot was washed in 0.1 × SSC and 0.1% sodium dodecyl sulfate at 65°C, and a digital image was acquired on a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
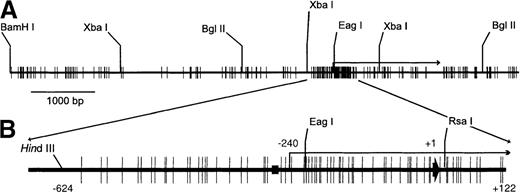
Schematic map of the erythropoietin gene.
(A) Distribution of CpG sites and a few restriction enzyme sites in an 8765-base pair (bp) fragment of the erythropoietin gene. (B) CpG island is exploded. (black rectangle) GATA box. (long bent arrow) Transcription start site. (short thick arrow) Position of the translation start site, arbitrarily numbered +1.
Schematic map of the erythropoietin gene.
(A) Distribution of CpG sites and a few restriction enzyme sites in an 8765-base pair (bp) fragment of the erythropoietin gene. (B) CpG island is exploded. (black rectangle) GATA box. (long bent arrow) Transcription start site. (short thick arrow) Position of the translation start site, arbitrarily numbered +1.
Peripheral blood mononuclear cell isolation
Peripheral blood was obtained from normal human volunteers with their informed consent. Mononuclear cells were isolated on a Ficoll step gradient (density, 1.077).
Bisulphite conversion, polymerase chain reaction amplification, and thermocycler sequencing
Bisulphite conversion of DNA was performed as described.53 DNA was denatured for 15 minutes at 37°C in freshly prepared 0.3 mol/L NaOH, then incubated with 3.1 mmol/L sodium bisulphite, pH 5, and 0.5 mmol/L hydroquinone (Sigma Chemical, St. Louis, MO) for 16 hours at 55°C. Polymerase chain reaction (PCR) amplifications were conducted with the following primers: top strand, 5410FT versus 5893R; bottom strand, 5410FB versus 5904R. In some cases, nested PCR was performed with primers 5446F versus 5893R (top strand) and 5441F versus 5904R (bottom strand), and PCR products were extracted from agarose gels with the QIAquick gel extraction kit (QIAGEN, Chatsworth, CA). The purified PCR products were sequenced using the fmol DNA cycle sequencing system (Promega, Madison, WI) and [32P]end-labeled primers. Primer sequences are listed in the Table.
Table. Primers
| Bisulfite Conversion . | . | Sequencing . | PCR . |
|---|---|---|---|
| Coding strand | |||
| 5171F | 5′-TGGGTTTTTAGATTTAGTTATTTTGTGGAA-3′ | X | |
| 5196F | 5′-TGGAATTAAGTAATTTAGGTATTTTTGAGT-3′ | X | |
| 5547R | 5′-CTCTAACCAAAAATCAAAACTATTATCTAC-3′ | X | X |
| 5410FT | 5′-GTTTTAATTTAGGTGTTTTGTTTTTGTTTT-3′ | X | |
| 5446F | 5′-GGGTGGTTTTTATTTTTGGTGATTTTTTA-3′ | X | |
| 5893R | 5′-ACCCCAACCACCTCCTAACCAATA-3′ | X | X |
| 5686R | 5′-AACCTAAAAAAAAAAACGACTATCCA-3′ | X | |
| Noncoding strand | |||
| 5202F | 5′-TCAACAACCCAAACATCTCTAAATCTC-3′ | X | |
| 5264F | 5′-CAAAAACCCAACCTTTCCCAAATAACA-3′ | X | |
| 5566R | 5′-TGGTTTAGGGATTTTGCGGTTTTGGT-3′ | X | |
| 5546R | 5′-TTTGGTTGGGGGTTGGGGTTGTTATTTG-3′ | X | |
| 5410FB | 5′-ACCTCAACCCAAACATCCTACCCCTAC-3′ | X | |
| 5441F | 5′-AACCCCAAATAACCCCTACCCCTAAC-3′ | X | X |
| 5904R | 5′-GTCGGTTTTTGAATTTAGTTATTTTTTGGT-3′ | X | X |
| EMSAs | |||
| SP1 | 5′-ATTCGATCGGGGCGGGGCGAGC-3′ | ||
| HIF1 | 5′-GGCTGGGCCCTACGTGCTGTCTCACACAGCCTG-3′ | ||
| DR2 | 5′-CTGACCTCTCGACCTACCGGCCTAGGCCACA-3′ | ||
| P56 | 5′-GTCCTGCCCCTGCTCTGACCCCGGGTGGCCCCTACCCCTGGCGACCCCTCACGCACACAGC-3′ | ||
| LMPCR | |||
| BP4 | 5′-CTAGAGGAGAGGGCGGCTGTC-3′ | ||
| BP5 | 5′-GCGGTGTCGGAGCAGAGCGG-3′ | ||
| BP6 | 5′-TCGGAGCAGAGCGGGCGCGGTG-3′ | ||
| Bisulfite Conversion . | . | Sequencing . | PCR . |
|---|---|---|---|
| Coding strand | |||
| 5171F | 5′-TGGGTTTTTAGATTTAGTTATTTTGTGGAA-3′ | X | |
| 5196F | 5′-TGGAATTAAGTAATTTAGGTATTTTTGAGT-3′ | X | |
| 5547R | 5′-CTCTAACCAAAAATCAAAACTATTATCTAC-3′ | X | X |
| 5410FT | 5′-GTTTTAATTTAGGTGTTTTGTTTTTGTTTT-3′ | X | |
| 5446F | 5′-GGGTGGTTTTTATTTTTGGTGATTTTTTA-3′ | X | |
| 5893R | 5′-ACCCCAACCACCTCCTAACCAATA-3′ | X | X |
| 5686R | 5′-AACCTAAAAAAAAAAACGACTATCCA-3′ | X | |
| Noncoding strand | |||
| 5202F | 5′-TCAACAACCCAAACATCTCTAAATCTC-3′ | X | |
| 5264F | 5′-CAAAAACCCAACCTTTCCCAAATAACA-3′ | X | |
| 5566R | 5′-TGGTTTAGGGATTTTGCGGTTTTGGT-3′ | X | |
| 5546R | 5′-TTTGGTTGGGGGTTGGGGTTGTTATTTG-3′ | X | |
| 5410FB | 5′-ACCTCAACCCAAACATCCTACCCCTAC-3′ | X | |
| 5441F | 5′-AACCCCAAATAACCCCTACCCCTAAC-3′ | X | X |
| 5904R | 5′-GTCGGTTTTTGAATTTAGTTATTTTTTGGT-3′ | X | X |
| EMSAs | |||
| SP1 | 5′-ATTCGATCGGGGCGGGGCGAGC-3′ | ||
| HIF1 | 5′-GGCTGGGCCCTACGTGCTGTCTCACACAGCCTG-3′ | ||
| DR2 | 5′-CTGACCTCTCGACCTACCGGCCTAGGCCACA-3′ | ||
| P56 | 5′-GTCCTGCCCCTGCTCTGACCCCGGGTGGCCCCTACCCCTGGCGACCCCTCACGCACACAGC-3′ | ||
| LMPCR | |||
| BP4 | 5′-CTAGAGGAGAGGGCGGCTGTC-3′ | ||
| BP5 | 5′-GCGGTGTCGGAGCAGAGCGG-3′ | ||
| BP6 | 5′-TCGGAGCAGAGCGGGCGCGGTG-3′ | ||
Primers used for bisulfite conversion polymerase chain reaction and sequencing reactions have 100% converted sequences, except for primer 5409R, which contains 1 CpG near the 5′ end that was not converted. Primers are numbered according to their distances from the BamH1 site, 5788 bp upstream of the human erythropoietin translation start site. All oligonucleotides for electrophoretic mobility shift assay reactions were double stranded; only 1 strand is represented here.
Electrophoretic mobility shift assay
Synthetic oligonucleotides and DNA fragments are listed in the Table. Probes were [32P]end-labeled with T4 polynucleotide kinase (New England BioLabs, Beverly, MA) and extracted from polyacrylamide or agarose gels with QIAEXII gel extraction or QIAquick gel extraction kits (QIAGEN). Electrophoretic mobility shift assay (EMSA) reactions (20 μL) contained nuclear extract from Hep3B and HeLa (5–10 μg protein) and 0.1 pmol probe in binding buffer: 10 mmol/L Tris, pH 8, 1 mmol/L dithiothreitol, 5% glycerol ([vol/vol], 20 mmol/L NaCl, 2 mmol/L MgCl2, 0.01 mg/mL bovine serum albumin), and either 0.1 mg/mL sonicated salmon sperm DNA (CpG 30-mer) or 0.2 mg/mL Escherichia coli DNA (CG11 and EPO236).54 Binding reactions were incubated at room temperature for 30 minutes and were analyzed on 4% nondenaturing polyacrylamide gels.
In vitro methylation, transfection, and luciferase assays
The 117-bp erythropoietin promoter (P117) was inserted immediately upstream of the luciferase reporter gene in pGL2 (Promega), with or without the 126-bp erythropoietin enhancer (E126), distal to the luciferase coding region.54 pGL2-P117 + 5′-UTR+E126 was constructed by inserting the 236-bp Eag1 to Rsa1 fragment of the erythropoietin gene into the Eag1 and Sac 2 sites of pGL2-P117+E126. The blunted erythropoietin Rsa1 site was ligated to a blunted Sac 2 site in pGL2. The P117 promoter was released from pGL2-P117 and pGL2-P117+E126 by digestion with Asp718 and Bgl2, and the 5′-UTR was released from pGL2-P117 + 5′-UTR+E126 by digestion with Eag1 and Bgl2. DNA fragments were methylated or mock-methylated with Sss 1 methylase (New England BioLabs) and then were ligated into their respective unmethylated backbones. The plasmids were introduced into Hep3B cells as calcium phosphate–DNA precipitates, and the cells were incubated under normoxia or hypoxia (1% oxygen) for 48 hours. Cell lysates were prepared, and luciferase activity was measured as previously described.54 A plasmid containing the cytomegalovirus promoter driving the lac z gene was included to correct for transfection efficiency.
In vivo digestion with restriction endonucleases
After 8 hours of hypoxia (1% oxygen), cells from three 150-mm plates were suspended in 2 mL 150 mmol/L sucrose, 35 mmol/L HEPES, pH 7.4, 80 mmol/L KCl, 5 mmol/L K2HPO4, 5 mmol/L MgCl2, and 0.5 mmol/L CaCl2. Membranes were made permeable by incubating for 1 minute with 0.05% lysolecithin (Sigma Chemical) and then were washed with digestion buffer (10 mmol/L Tris, pH 7.7, 50 mmol/L NaCl, 5 mmol/L MgCl2, and 1 mmol/L dithiothreitol).55 Aliquots containing 3 × 106 cells were treated with Msp1 for 10 minutes at 37°C. Cells were collected by centrifugation and suspended in 40 μL digestion buffer. DNA was isolated with the Genomic Prep Cells and Tissue DNA Isolation Kit (Amersham Pharmacia Biotech, Piscataway, NJ) and digested with Xba1 (−678) to provide an internal reference for digestion and amplification.
Ligation-mediated polymerase chain reaction
Ligation-mediated PCR was performed with Vent DNA polymerase (New England BioLabs) in 5% dimethyl sulfoxide with gene-specific primers BP4, BP5, and BP6 (Table) as described.56 After 24 cycles of PCR, the products were subjected to 4 cycles of linear amplification using [32P]end-labeled primers. The annealing temperatures for first-strand synthesis, PCR amplification, and labeling reactions were 65°C, 68°C, and 70°C, respectively.
Graphics
Figures were produced with Adobe Illustrator (Adobe Systems, Mountain View, CA). PhosphorImager data were analyzed with ImageQuaNT (Molecular Dynamics) without modifying the linear response curve. The CpG maps were created with VectorNTI (InforMax, Bethesda, MD).
Results
Several criteria should be met before it can be suggested that CpG methylation is involved in the tissue-specific regulation of a gene: (1) control elements of the gene should contain CpG residues, (2) methylation of the control regions of the gene should mirror gene activity in expressing and nonexpressing cells, (3) proteins that bind the control regions should be present in expressing and nonexpressing cells, (4) methylation of the control elements should affect protein binding, and (5) methylation of the control elements should affect transcriptional activity in functional assays. We provide the following experimental data in support of methylation controlling expression of the erythropoietin gene.
CpGs in the erythropoietin gene
Figure 1 is a schematic representation of 8765 bp of the human erythropoietin gene. The map begins 5788 bp upstream of the translation start site (arbitrarily defined as base pair + 1) at a BamH1 site, ends at a Pvu 2 site distal to the polyadenylation signal, and contains 203 CpGs that disproportionately cluster around the transcription start site (−240). The promoter has 25 CpGs in 320 bp, and the 5′-UTR has 31 CpGs in 240 bp, 7.8 and 12.9 CpGs per 100 bp, respectively. The highest density of CpGs is a 30-bp region in the 5′-UTR that contains 8 CpGs. The promoter and 5′-UTR comprise a CpG island based on criteria set forth by Gardiner-Garden and Frommer.6 Specifically, the GC content is 74.6%, the CpG rich region is >200 bp in length, and the observed-to-expected ratio of CpGs is 0.75.
Analysis of the erythropoietin gene with a methylation-sensitive restriction enzyme
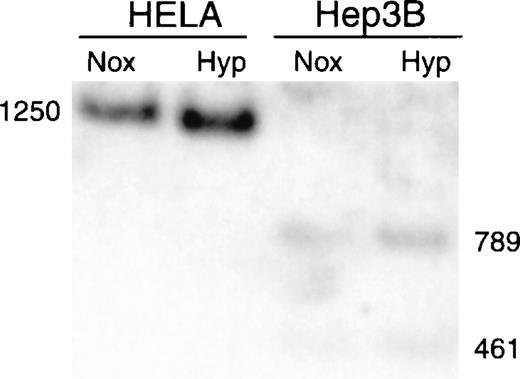
We screened the CpG island in the erythropoietin gene for methylation by exploiting the sensitivity of Eag1 to CpG methylation within its recognition site. The human erythropoietin gene (Fig. 1) has an Eag1 site 220 bp proximal to the translation start site that is flanked by sites for Xba1 (cleaves irrespective of methylation). Genomic DNA was digested with Xba1 and Eag1, and restriction fragments were identified by Southern blot analysis. Cleavage by both enzymes generates restriction fragments of 461 and 789 bp; methylation of the Eag1 site precludes Eag1 digestion, resulting in a single 1250-bp Xba1 fragment. DNA from Hep3B (expressing cells) is digested to completion with Eag1, whereas DNA from HeLa (nonexpressing cells) is resistant to Eag1 digestion (Fig. 2), consistent with methylation-sensitive expression of the erythropoietin gene.
Southern blot analysis of methylation in Hep3B and HeLa cells.
Genomic DNA from Hep3B and HeLa cells was digested with Xba1 and Eag1, fractionated by size, transferred to a nylon membrane, and hybridized to the full-length Xba1 fragment of the erythropoietin gene. The Xba1 fragment is 1250 bp long, and the combined Xba1/Eag1 fragments measure 461 and 789 bp. Nox, normoxia; Hyp, hypoxia for 8 hours.
Southern blot analysis of methylation in Hep3B and HeLa cells.
Genomic DNA from Hep3B and HeLa cells was digested with Xba1 and Eag1, fractionated by size, transferred to a nylon membrane, and hybridized to the full-length Xba1 fragment of the erythropoietin gene. The Xba1 fragment is 1250 bp long, and the combined Xba1/Eag1 fragments measure 461 and 789 bp. Nox, normoxia; Hyp, hypoxia for 8 hours.
Analysis of CpG methylation by sodium bisulfite conversion
Most CpGs in the human erythropoietin gene are not in restriction enzyme recognition sites and cannot be analyzed by methylation-sensitive cleavage assays. Sodium bisulfite conversion displays the methylation status of all cytosines.53Treatment of DNA with sodium bisulfite converts cytosine to uracil; 5-methylcytosine is unaffected because methylation blocks deamination. During PCR amplification of bisulfite-treated DNA, thymidines are incorporated in place of converted cytosines. Converted cytosines are detected by DNA sequence analysis of pooled PCR products or individual clones.
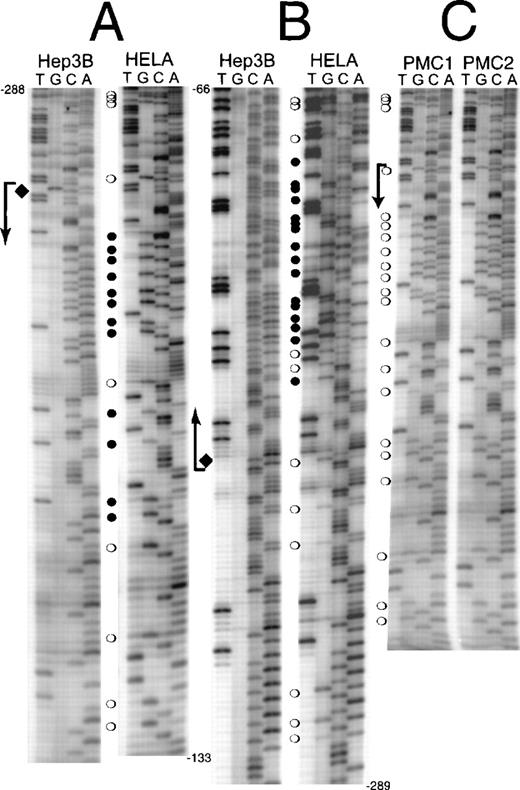
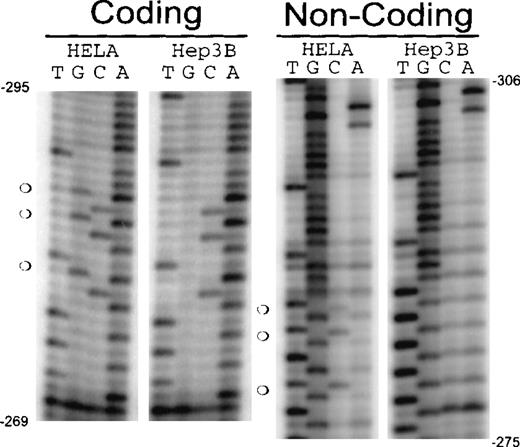
The methylation status of the human erythropoietin promoter (−288 to +81) was analyzed in Hep3B and HeLa cells by sodium bisulfite conversion (Figures 3A, 3B). In normoxic Hep3B cells, all cytosines are converted to uracil except a single CpG on the coding strand, 238 bp proximal to the translation start site that is hemimethylated (Figures 3A, 3B, diamond). In hypoxic Hep3B cells, this CpG is not methylated (data not shown). In contrast each cytosine in CpG sites from −288 to +81 of the erythropoietin gene has some resistance to sodium bisulfite conversion in HeLa (Figs. 3A,3B). The proximal 5′-UTR containing the highest density of CpGs (26.7%) is heavily methylated. The CpGs in the upstream region of the human erythropoietin promoter (−558 to −288) are only sporadically methylated in HeLa (data not shown).
CpG methylation in the erythropoietin promoter and 5′-UTR.
DNA from normoxic Hep3B, HeLa cells, and peripheral blood mononuclear cells was treated with sodium bisulfite. Polymerase chain reaction amplification was performed, and the DNA sequence was obtained from the complementary strands (guanines converted to adenines). (A) Coding strand sequence from −288 to −133. (B) Noncoding strand sequence from −66 to −289. (C) Coding strand sequence from −288 to −133 from peripheral blood mononuclear cells (PMC1 and PMC2). (bent arrows) Transcription start site. (open circles) Partially methylated CpGs. (filled circles) Fully methylated CpGs. (diamonds) Hemimethylated site present in normoxic Hep3B cells.
CpG methylation in the erythropoietin promoter and 5′-UTR.
DNA from normoxic Hep3B, HeLa cells, and peripheral blood mononuclear cells was treated with sodium bisulfite. Polymerase chain reaction amplification was performed, and the DNA sequence was obtained from the complementary strands (guanines converted to adenines). (A) Coding strand sequence from −288 to −133. (B) Noncoding strand sequence from −66 to −289. (C) Coding strand sequence from −288 to −133 from peripheral blood mononuclear cells (PMC1 and PMC2). (bent arrows) Transcription start site. (open circles) Partially methylated CpGs. (filled circles) Fully methylated CpGs. (diamonds) Hemimethylated site present in normoxic Hep3B cells.
We verified the methylation of the human erythropoietin promoter with genomic DNA from peripheral blood mononuclear cells, a heterogeneous mixture of T-lymphocytes, B-lymphocytes, natural killer cells, and monocytes. DNA from these normal human cells has a methylation pattern similar to that of HeLa DNA (Figure 3C).
Proteins bind methyl-CpGs in the erythropoietin 5′-UTR
We sought to demonstrate by EMSA that methyl-CpG binding proteins in Hep3B and HeLa extracts bind high-density CpGs in a methylation-dependent manner. The Eag1 to Rsa1 fragment of the erythropoietin gene (EPO236; see Figure 1) and a synthetic high-density CpG probe (CG11) were symmetrically methylated in vitro with a CpG methylase. Methylated CG11 is a ligand for methyl-CpG binding proteins such as MeCP1.24,57 Several protein complexes form on unmethylated CG11 (Figure 4A; lane 3) and EPO236 (Figure 4B; lane 3) in nuclear extracts from HeLa cells, but none of these complexes is methylation dependent (Figures 4A, 4B; lanes 4–8). When challenged with methylated CG11 and EPO236, new complexes are formed in both extracts (Figures 4A, 4B; lanes 2, 4; arrows). In HeLa extracts the shifted complexes form in the presence of excess unmethylated CG11 or EPO236 competitor (Figure 4; lanes 5, 6), but not when incubated with an excess of either methylated competitor (Figure4; lanes 7, 8): behavior consistent with MeCP binding proteins.23 Similar complexes form in Hep3B extracts (Fig. 4; lanes 1, 2).
Erythropoietin 5′-UTR recruits a methyl-CpG–binding protein.
Electrophoretic mobility shift assays were performed with methylated and unmethylated CG11 and EPO236 as probes and competitors. [32P]-end-labeled probes were incubated with nuclear extracts and, where indicated, with excess unlabeled competitors. (A) CG11 probe. (B) EPO236 probe. U, unmethylated; M, methylated probe or competitor. Probe identity and methylation status are noted below the panels, and competitors and sources of nuclear extracts are noted above the panels. (arrow) Methyl-CpG–binding complex. (asterisks) Complexes formed on unmethylated probes blocked by methylation. (arrowheads) Protein complexes formed on unmethylated and methylated probes, whose formation is blocked by methylated and unmethylated competitors.
Erythropoietin 5′-UTR recruits a methyl-CpG–binding protein.
Electrophoretic mobility shift assays were performed with methylated and unmethylated CG11 and EPO236 as probes and competitors. [32P]-end-labeled probes were incubated with nuclear extracts and, where indicated, with excess unlabeled competitors. (A) CG11 probe. (B) EPO236 probe. U, unmethylated; M, methylated probe or competitor. Probe identity and methylation status are noted below the panels, and competitors and sources of nuclear extracts are noted above the panels. (arrow) Methyl-CpG–binding complex. (asterisks) Complexes formed on unmethylated probes blocked by methylation. (arrowheads) Protein complexes formed on unmethylated and methylated probes, whose formation is blocked by methylated and unmethylated competitors.
In vivo protection of methyl-CpGs in HeLa cells
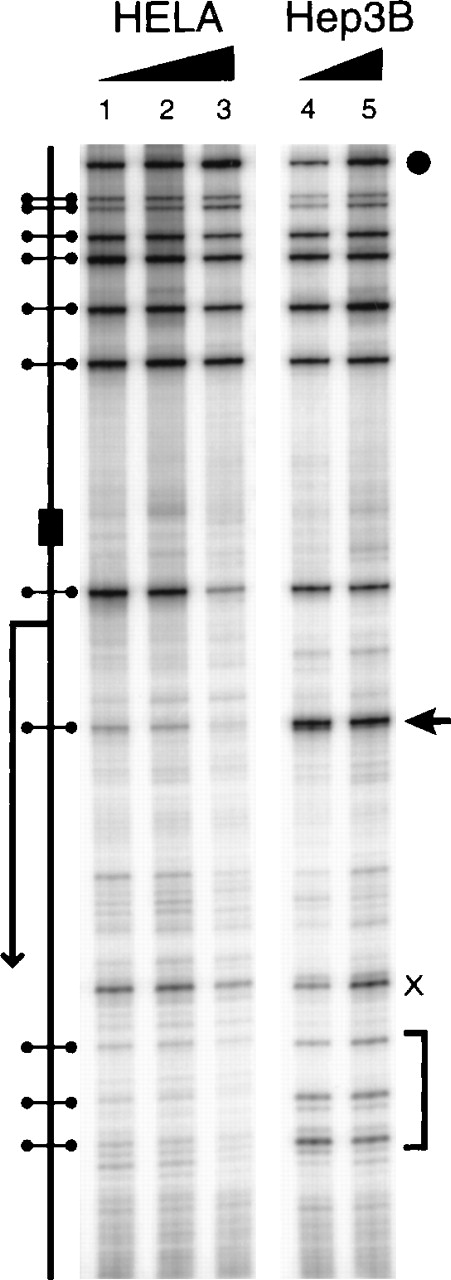
Several CpGs in the human erythropoietin promoter and 5′-UTR fall within Msp1 sites. If these CpGs recruit methyl-CpG–binding proteins when methylated, the restriction enzyme recognition sites should be protected from Msp1 cleavage in vivo. To this end we rendered the cellular and nuclear membranes of HeLa and Hep3B cells permeable and exposed the cells to Msp1, a restriction enzyme that cleaves irrespective of methylation. Cleavage sites in the DNA were detected by ligation-mediated PCR. In Hep3B cells, Msp1 cleaves the DNA at each restriction site (Figure 5; lanes 1–3); however, in HeLa cells, 4 Msp1 sites in the 5′-UTR are resistant to digestion, especially the site at −219 (Figure 5; lanes 4, 5; arrow). The in vivo resistance of HeLa DNA to Msp1 digestion suggests that methyl-CpG–binding proteins interact with a region of dense methylation in the erythropoietin 5′-UTR.
Restriction endonuclease accessibility of the erythropoietin promoter and 5′-UTR.
Hypoxic Hep3B and HeLa cells were incubated in vivo with Msp1 after a brief treatment with lysolecithin. DNA was purified and digested with Xba1 to create an amplification standard. Restriction enzyme cleavage of the DNA was detected by ligation-mediated polymerase chain reaction. X, nonspecific band. (arrow and bracket) Sites with increased resistance to Msp1 cleavage in HeLa. (filled circle) Xba1 site. The schematic diagram shows (dumbbells) Msp1 sites, (bent arrow) transcription start site, and (rectangle) GATA box. The ramp corresponds to increasing Msp1 concentration.
Restriction endonuclease accessibility of the erythropoietin promoter and 5′-UTR.
Hypoxic Hep3B and HeLa cells were incubated in vivo with Msp1 after a brief treatment with lysolecithin. DNA was purified and digested with Xba1 to create an amplification standard. Restriction enzyme cleavage of the DNA was detected by ligation-mediated polymerase chain reaction. X, nonspecific band. (arrow and bracket) Sites with increased resistance to Msp1 cleavage in HeLa. (filled circle) Xba1 site. The schematic diagram shows (dumbbells) Msp1 sites, (bent arrow) transcription start site, and (rectangle) GATA box. The ramp corresponds to increasing Msp1 concentration.
Methyl-CpGs in the proximal erythropoietin promoter
Low-density methylation inhibits transcription by recruiting a repressive protein complex to the promoter58 or interfering with the binding of essential trans-acting proteins. The promoter of the erythropoietin gene has numerous CpGs; however, the CpGs in the promoter (upstream of −240) are largely unmethylated (data not shown). A cluster of 3 CpGs immediately upstream of the GATA box (−267) in the erythropoietin proximal promoter is partially methylated in HeLa and peripheral blood mononuclear cells but is unmethylated in Hep3B cells (Figures 3,6). The differential methylation of these CpG sites and their proximity to the GATA box suggest this region may play a role in the tissue-specific regulation and basal activity of the promoter. DNA/protein interactions in this part of the promoter have been previously shown to affect erythropoietin expression.59 We next chose to evaluate the 30-bp region encompassing these CpGs (CpG 30-mer; Figure7D) for methylation-dependent DNA/protein interactions with extracts from Hep3B and HeLa cells.
CpG methylation in the erythropoietin promoter from −306 to −275.
DNA from Hep3B and HeLa cells was modified by bisulfite conversion. Converted DNA was amplified, and polymerase chain reaction products were sequenced. Coding strand: sequence from the complementary strand (guanines are converted to adenines). Noncoding strand: sequence from the original strand (cytosines converted to thymines). (open circles) Methylated CpGs.
CpG methylation in the erythropoietin promoter from −306 to −275.
DNA from Hep3B and HeLa cells was modified by bisulfite conversion. Converted DNA was amplified, and polymerase chain reaction products were sequenced. Coding strand: sequence from the complementary strand (guanines are converted to adenines). Noncoding strand: sequence from the original strand (cytosines converted to thymines). (open circles) Methylated CpGs.
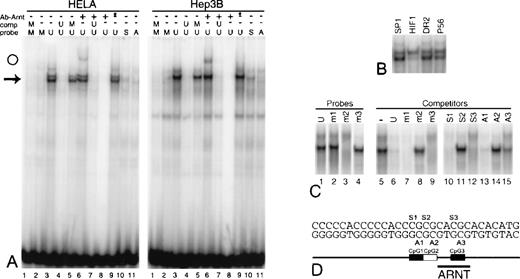
Specific DNA/protein interaction is blocked by CpG methylation.
A panel of complementary oligonucleotides comprising CpG 30-mer was synthesized with 5-methylcytosines in all 3 CpGs, in each CpG individually, and in none of the CpGs. Mixing the oligonucleotides creates probes and competitors for electrophoretic mobility shift assays with diverse methylation profiles. (A) Unmethylated CpG 30-mer (U) and CpG 30-mer symmetrically methylated on all 3 CpGs (M) were used as probes and competitors as indicated above panel A. (arrow) Position of major binding complexes. (lanes 10, 11) Probe contains 5-methylcytosine on the sense (S) or antisense (A) strands at CpG1, CpG2, and CpG3. (lanes 6–8) A murine antibody to human aryl hydrocarbon nuclear translocator, ARNT (2B10), was included in the reactions. (open circle) Position of an anti-ARNT supershift. (lane 8) No nuclear extract. (lane 9, asterisk) Addition of a murine antibody to an unrelated antibody. (B) The binding complex from HeLa extracts and unmethylated CpG 30-mer is challenged with a 50-fold excess of double-stranded competitors (see Table). (C) Shifted complexes form with Hep3B extracts and CpG 30-mer symmetrically methylated at either CpG1 (m1), CpG2 (m2), or CpG3 (m3). (lanes 5–15) Shifted complexes formed with Hep3B extracts and unmethylated CpG 30-mer are challenged with CpG 30-mer symmetrically methylated at each CpG (m1-3) or hemimethylated at each CpG on the sense (S1–S3) or antisense (A1–A3) strand. (D) Schematic map of CpG 30-mer.
Specific DNA/protein interaction is blocked by CpG methylation.
A panel of complementary oligonucleotides comprising CpG 30-mer was synthesized with 5-methylcytosines in all 3 CpGs, in each CpG individually, and in none of the CpGs. Mixing the oligonucleotides creates probes and competitors for electrophoretic mobility shift assays with diverse methylation profiles. (A) Unmethylated CpG 30-mer (U) and CpG 30-mer symmetrically methylated on all 3 CpGs (M) were used as probes and competitors as indicated above panel A. (arrow) Position of major binding complexes. (lanes 10, 11) Probe contains 5-methylcytosine on the sense (S) or antisense (A) strands at CpG1, CpG2, and CpG3. (lanes 6–8) A murine antibody to human aryl hydrocarbon nuclear translocator, ARNT (2B10), was included in the reactions. (open circle) Position of an anti-ARNT supershift. (lane 8) No nuclear extract. (lane 9, asterisk) Addition of a murine antibody to an unrelated antibody. (B) The binding complex from HeLa extracts and unmethylated CpG 30-mer is challenged with a 50-fold excess of double-stranded competitors (see Table). (C) Shifted complexes form with Hep3B extracts and CpG 30-mer symmetrically methylated at either CpG1 (m1), CpG2 (m2), or CpG3 (m3). (lanes 5–15) Shifted complexes formed with Hep3B extracts and unmethylated CpG 30-mer are challenged with CpG 30-mer symmetrically methylated at each CpG (m1-3) or hemimethylated at each CpG on the sense (S1–S3) or antisense (A1–A3) strand. (D) Schematic map of CpG 30-mer.
Protein interactions with CpG 30-mer
Protein interactions with CpG 30-mer were evaluated by EMSA. Unmethylated CpG 30-mer produces a single-shift complex with nuclear extract from Hep3B and a doublet with extracts from HeLa (Figure 7A; lane 3; arrow). Complex formation is blocked by a 50-fold excess of unmethylated self-competitor (Figure 7A; lane 4), but not by competitors containing an SP-1 site, the erythropoietin enhancer steroid receptor element direct repeat (DR2), or a fragment of the erythropoietin promoter (P56) (Figure 7B; Table). The HIF1 element from the erythropoietin enhancer partially blocks formation of the lower complex in HeLa extracts (Figure 7B; lane 5). These experiments confirm that nuclear proteins from Hep3B and HeLa bind an element in CpG 30-mer in a sequence-specific manner.
CpG 30-mer shares sequence homology with the aryl hydrocarbon nuclear translocator (ARNT)-binding element, suggesting that ARNT is involved in the DNA/protein complex. We tested this hypothesis by incubating HeLa and Hep3B nuclear extracts, unmethylated CpG 30-mer, and 2B10 (Affinity Bioreagents, Golden, CO), an antibody to ARNT. A small portion of the specific complex is further shifted, suggesting ARNT is a part of the complex (Figure 7A; lane 6; open circle). Control reactions—including an excess of self-competitor, omission of nuclear extract, and inclusion of an antibody to an unrelated antigen—confirm the specificity of the ARNT antibody (Figure 7A; lanes 7, 8, 9).
Effect of methylation of CpG 30-mer on protein binding
CpG 30-mer symmetrically methylated at all 3 CpGs does not bind protein from HeLa or Hep3B extracts (Figure 7A; lanes 1, 2), nor does it block the formation of specific complexes when used as an unlabeled competitor against unmethylated CpG 30-mer (Figure 7A; lane 5). Even hemimethylation of the 3 CpGs on the sense or antisense strand of CpG 30-mer blocks the formation of DNA/protein complexes (Figure 7A; lanes 10, 11). Because these CpGs are only partially methylated in cells that do not express erythropoietin (Figures 3, 6), we chose to test whether methylation of each individual CpG in CpG 30-mer could block the formation of sequence-specific protein complexes. Symmetric methylation of CpG2 prevents the formation of specific complexes with Hep3B extracts (Figure 7C; lane 3), and both symmetric methylation (Figure 7; lane 8) and hemimethylation (Figure 7; lanes 11, 14) of CpG2 render the DNA fragment ineffective as a competitor. Symmetric methylation of CpG1 or CpG3 has little effect on DNA/protein interactions (Figure 7C; lanes 2, 4 [probes] and lanes 7, 9 [competitors]). CpG 30-mer hemimethylated at CpG1 competes well for protein binding to unmethylated CpG 30-mer (Figure 7C; lanes 10, 13), but hemimethylation of CpG3 on the antisense strand impairs its ability to act as a competitor (Figure 7C; lane 15). Similar results were obtained with HeLa extracts (data not shown).
Methylation of the promoter or 5′-UTR versus transcriptional activity
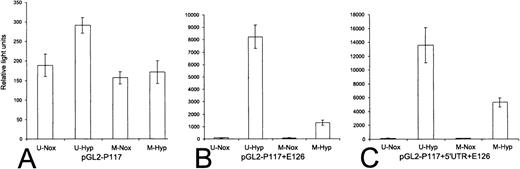
We examined whether methylation of the promoter or 5′-UTR could block the activity of the erythropoietin promoter in transient expression assays. The minimal promoter was selectively methylated at 11 CpGs in the context of the unmethylated pGL2 backbone ± the 126-bp erythropoietin enhancer (the enhancer was not methylated). In a separate experiment, the 5′-UTR was selectively methylated at 31 CpGs in the context of the unmethylated pGL2-P117+E126 backbone.
Without an enhancer, methylation of the CpGs in the erythropoietin promoter has a minor repressive effect on normoxic expression of luciferase. Under hypoxic conditions, however, the expression of luciferase falls by 40% (Figure 8A). In the context of the erythropoietin enhancer (unmethylated), methylation of erythropoietin promoter produces a substantial decrease in luciferase expression—the basal activity of the promoter (normoxia) falls by 35%, and the expression of the minigene under hypoxic conditions falls by 85% (Figure 8B). Methylation of the erythropoietin 5′-UTR in the context of an unmethylated erythropoietin promoter and enhancer results in a fall in luciferase activity of 30% for basal expression and 60% for inducible expression (Figure 8C).
The effect of methylation on the transcriptional activity of the erythropoietin promoter and 5′-UTR.
The minimal inducible erythropoietin promoter (P117) was methylated or mock methylated, then ligated into unmethylated pGL2 (pGL2-P117) or pGL2 with the unmethylated erythropoietin enhancer (pGL2-P117+E126). The 5′-UTR was methylated or mock methylated and ligated in its natural position into unmethylated pGL2-P117+E126, creating pGL2-P117 + 5′-UTR+E126. Reporter vectors were introduced into Hep3B cells, and luciferase assays were performed. Relative light units are expressed as raw light units divided by mU/min of β-galactosidase activity. (error bars) ±1 SD. (A) pGL2-P117 (n = 4). (B) pGL2-P117+E126 (n = 4). (C) pGL2-P117 + 5′-UTR+E126 (n = 5). U, unmethylated; M, methylated; Nox, normoxia; Hyp, hypoxia.
The effect of methylation on the transcriptional activity of the erythropoietin promoter and 5′-UTR.
The minimal inducible erythropoietin promoter (P117) was methylated or mock methylated, then ligated into unmethylated pGL2 (pGL2-P117) or pGL2 with the unmethylated erythropoietin enhancer (pGL2-P117+E126). The 5′-UTR was methylated or mock methylated and ligated in its natural position into unmethylated pGL2-P117+E126, creating pGL2-P117 + 5′-UTR+E126. Reporter vectors were introduced into Hep3B cells, and luciferase assays were performed. Relative light units are expressed as raw light units divided by mU/min of β-galactosidase activity. (error bars) ±1 SD. (A) pGL2-P117 (n = 4). (B) pGL2-P117+E126 (n = 4). (C) pGL2-P117 + 5′-UTR+E126 (n = 5). U, unmethylated; M, methylated; Nox, normoxia; Hyp, hypoxia.
Discussion
The minimal inducible promoter of the human erythropoietin gene consists of a 117-bp region upstream of the transcription start site (−240 bp proximal to the translation start site),54is highly G and C rich, and lacks a canonical TATA box. A GATA box located 25 to 30 bp upstream of the transcription start site may serve as the dock for TFIID/TBP and may bind a GATA family member.60 61 Despite the simplicity of the erythropoietin promoter and its similarity to housekeeping genes, erythropoietin expression is exquisitely tissue specific.
The cis-acting and trans-acting elements that control the hypoxic regulation of the erythropoietin gene have been carefully dissected. A hypoxia-inducible enhancer62 lies 3′ of the polyadenylation signal of the erythropoietin gene and cooperates with the minimal inducible erythropoietin promoter.54 Both the enhancer and the promoter interact with hypoxia-inducible factor-1 (HIF1),63 a heterodimer of HIF1-α, and ARNT. Other genes are activated by hypoxia through HIF1, including glycolytic enzymes,64,65 glucose transporters, 64-67 and vascular endothelial-derived growth factor.68 An intact hypoxia-signaling pathway is present in most cells69,70; hence, the silence of the erythropoietin gene in most tissues is perplexing. We previously demonstrated that sequence-specific binding proteins do not occupy the HIF1 element in the erythropoietin enhancer in HeLa cells,56 despite their expression of active HIF1 during hypoxia. The exclusion of HIF1 and other proteins from the enhancer suggests that the erythropoietin gene must assume a tissue-specific permissive state before activation by hypoxia. We believe the accessibility of the enhancer depends in part on the activity of the promoter, and we doubt that the permissive state of the promoter is determined by the ubiquitous factor HIF1. The presence of a CpG island in the promoter prompted us to study the methylation of the erythropoietin gene and to search for proteins that bind the gene in a methylation-dependent fashion.
The 5′-UTR of the erythropoietin gene has a region of high CpG density that is methylated in HeLa but is unmethylated in Hep3B cells. Although methylation of CpG islands in nonessential genes may be commonplace in cells lines and may not reflect the situation in primary tissues,71 we have shown that the methylation pattern of the erythropoietin 5′-UTR in normal peripheral blood mononuclear cells and HeLa cells is similar. Proteins from HeLa and Hep3B nuclear extracts bind to the methylated 5′-UTR in a manner consistent with methyl-CpG–binding proteins. Interactions between proteins and methyl-CpGs in the erythropoietin 5′-UTR were confirmed by in vivo footprints that prove the erythropoietin gene has different architectures in expressing and nonexpressing cells. Ex vivo methylation of the CpGs in the 5-UTR impedes transcription from the erythropoietin promoter during transient expression in Hep3B cells, consistent with the known repressive effects of methyl-CpG–binding proteins.
In contrast to the 5′-UTR, methylation of CpGs in the proximal erythropoietin promoter interferes with the binding of a sequence-specific DNA-binding protein. The binding element contains 3 CpGs that are methylated only in nonexpressing cells, and symmetric methylation or hemimethylation of the middle CpGs blocks formation of the DNA/protein complexes. The proteins in the complexes have not been identified, but they may include ARNT based on antibody recognition. Ex vivo methylation of the CpGs in the minimal erythropoietin promoter subverts the function of the promoter and substantially blocks the functional cooperation between the promoter and the enhancer.54 Recently, methylation of the HIF1 site in the erythropoietin enhancer has been shown to block HIF1 binding and to decrease the transcriptional activation conferred by the enhancer.72
A CpG near the transcription start site is hemimethylated in normoxic, but not in hypoxic, Hep3B cells. We have not determined whether hemimethylation at this site affects the transcriptional activity of the promoter. The role of asymmetric methylation in eukaryotic gene regulation is largely undefined.73-75
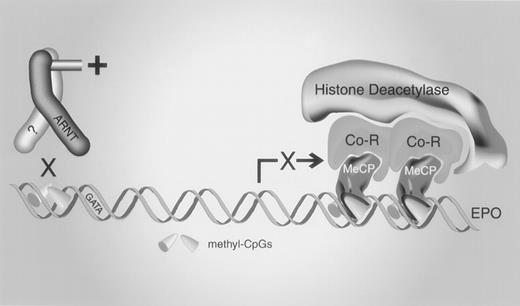
We have presented evidence that methylation of the promoter and 5′-UTR of the human erythropoietin gene contributes to tissue-specific regulation, and we propose the following model to explain the silencing of erythropoietin expression by methylation (Figure 9). Methylation provides 2 levels of control, both dependent on DNA/protein interactions. Methylation of 3 CpGs in the proximal promoter blocks the binding of essentialtrans-acting proteins, thereby indirectly repressing transcription. High-density CpG methylation in the 5′-UTR permits the binding of a methyl-CpG binding protein that either directly represses transcription or recruits corepressors, histone deacetylases, or both. Additional studies of primary human tissues will be necessary to verify that methylation is essential for the tissue-specific regulation of the erythropoietin gene and to identify the factors and control elements necessary for initiation and maintenance of the methylated state.
A model for silencing the erythropoietin gene by methylation.
Methylation of CpGs in a cis element upstream of the GATA box prevents the binding of a trans-acting protein, impairing transcriptional activation. The sequence-specific binding activity may contain aryl hydrocarbon nuclear translocator (ARNT). Methylation of high-density CpGs in the erythropoietin 5′-UTR recruits a methyl-CpG–binding protein (MeCP) that represses transcription by engaging corepressors (Co-R), histone deacetylases, or both.
A model for silencing the erythropoietin gene by methylation.
Methylation of CpGs in a cis element upstream of the GATA box prevents the binding of a trans-acting protein, impairing transcriptional activation. The sequence-specific binding activity may contain aryl hydrocarbon nuclear translocator (ARNT). Methylation of high-density CpGs in the erythropoietin 5′-UTR recruits a methyl-CpG–binding protein (MeCP) that represses transcription by engaging corepressors (Co-R), histone deacetylases, or both.
Acknowledgment
The authors thank Jeanne Housman for her emotional and financial support.
Supported by Public Health Service grant DK-46967 from the National Institute of Diabetes and Digestive and Kidney Diseases (KLB).
Reprints:Kerry L. Blanchard, Feist–Weiller Cancer Center, Louisiana State University Medical Center, 1501 Kings Highway, Shreveport, LA 71103; e-mail: kblanc@lsumc.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Fig. 4. Erythropoietin 5′-UTR recruits a methyl-CpG–binding protein. / Electrophoretic mobility shift assays were performed with methylated and unmethylated CG11 and EPO236 as probes and competitors. [32P]-end-labeled probes were incubated with nuclear extracts and, where indicated, with excess unlabeled competitors. (A) CG11 probe. (B) EPO236 probe. U, unmethylated; M, methylated probe or competitor. Probe identity and methylation status are noted below the panels, and competitors and sources of nuclear extracts are noted above the panels. (arrow) Methyl-CpG–binding complex. (asterisks) Complexes formed on unmethylated probes blocked by methylation. (arrowheads) Protein complexes formed on unmethylated and methylated probes, whose formation is blocked by methylated and unmethylated competitors.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/1/10.1182_blood.v95.1.111/5/m_bloo00120004w.jpeg?Expires=1769296368&Signature=2h7bg~MMoaqvTbX0OzqsRdF8jOQIVZ4avIn6HC6ubdY6OJVU9ZBn1gjx7huuvKKYB9SuLDvJzowc0uvpafdsm64hlUsR~AFHtNs9PJ5SUXIZNxlRZ-dQ21502tZXCLFIEsttCNGCtUKh8rAu2SFOAD1JKtgwRW5Iusr5gmOndGOavOKGHWv0GFS8KpBXrBNaPRCPQR4xgI3oKiW8qXl6udIHSqJ0YBpWI~Tcp0gGGRrruKjBBajyoDl1srv6eNDxjXbywPtqdJy0wzvuY6O06ujPnXnJkuSveKAvBTAc12qWEHGatiaOJ71p6TjFlkRadb4~-e7Vk41dZEyAFo~Hyg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal