Abstract
We have investigated the blood cells from a woman with a low degree of chronic nonspherocytic hemolytic anemia and frequent bacterial infections accompanied by icterus and anemia. The activity of glucose 6-phosphate dehydrogenase (G6PD) in her red blood cells (RBCs) was below detection level, and in her leukocytes less than 3% of normal. In cultured skin fibroblasts, G6PD activity was approximately 15% of normal, with 4- to 5-fold increased Michaelis constant (Km) for NADP and for glucose 6-phosphate. Activated neutrophils showed a decreased respiratory burst. Family studies showed normal G6PD activity in the RBCs from all family members, including both parents and the 2 daughters of the patient. Sequencing of polymerase chain reaction (PCR)-amplified genomic DNA showed a novel, heterozygous 514C→T mutation, predicting a Pro172→Ser replacement. Analysis of G6PD RNA from the patient’s leukocytes and fibroblasts showed only transcripts with the 514C→T mutation. This was explained by the pattern of X-chromosome inactivation, studied by means of the human androgen receptor (HUMARA) assay, which proved to be skewed in the patient, her mother, and one of the patient’s daughters. Thus, the patient has inherited a de novo mutation in G6PD from her father and an X-chromosome inactivation determinant from her mother, causing exclusive expression of the mutated G6PD allele. Purified mutant protein from an Escherichia coli expression system showed strongly decreased specific activity, increased Km for NADP and for glucose 6-phosphate, and increased heat lability, which indicates that the defective phenotype is due to 2 synergistic molecular dysfunctions: decreased catalytic efficiency and protein instability.
GLUCOSE 6-PHOSPHATE dehydrogenase (G6PD) catalyzes the conversion of glucose 6-phosphate into 6-phosphogluconolactone and the concomitant reduction of NADP into NADPH. This is the first step in the hexose monophosphate (HMP) pathway and, together with the second step in this pathway, the only source of NADPH in cells lacking mitochondria. Therefore, G6PD has an essential function in protecting red blood cells (RBCs) against oxidative stress.
Deficiency of G6PD is one of the most common genetic defects in humans, probably due to the protection of G6PD-deficient RBCs against proliferation of malaria parasites.1 The clinical expression of G6PD deficiency varies from mild hemolytic anemia induced by infections or drugs (World Health Organization [WHO] class 3) to chronic nonspherocytic hemolytic anemia with attacks of severe anemia induced by infections and drugs (class 1). This last type of G6PD deficiency is often first manifested by neonatal jaundice, because the liver function may be impaired as well.1 Because the G6PD gene is located on the X chromosome, the clinical symptoms of the disease are usually confined to hemizygous men, although female carriers with a marked expression of the aberrant allele may also suffer.
Occasionally, patients with G6PD deficiency have been described with decreased leukocyte functions.2-6 In general, the G6PD activity in the leukocytes needs to be below 5% of normal for such impairment to occur.3 In particular, phagocytic leukocytes use NADPH for the reduction of molecular oxygen to superoxide (O2−), a prerequisite for effective killing of micro-organisms by these cells. As a result, severe G6PD deficiency manifested in leukocytes may present as a form of chronic granulomatous disease, which is usually caused by intrinsic defects in the superoxide-generating enzyme.7
We now describe a female patient with severe G6PD deficiency, chronic hemolytic anemia, increased susceptibility to infections, and impaired superoxide generation. We have characterized the enzymatic properties of the mutant G6PD and determined the mutation responsible for these changes. The results indicate that replacement of proline-172 by serine causes drastic changes in the characteristics of the enzyme. We propose to call this variant enzyme G6PD Volendam, after the place of origin of the patient. Some of our findings were reported previously in abstract form.8 9
MATERIALS AND METHODS
Case History
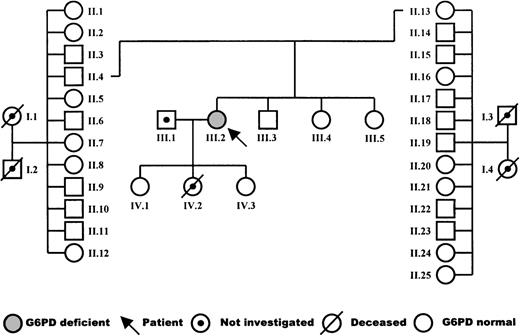
The patient is a woman born in 1960 from a nonconsanguineous marriage, with no hematologic abnormalities in the maternal or paternal family. The pedigree is shown in Fig 1. After birth, she presented with severe icterus. Thereafter, she experienced several episodes of hemolysis and icterus after the use of aspirin and during infections. In particular, she suffered from pneumonia due to pneumococcus infection, from cystitis, and from several noncharacterized infections of the airways. These infections were treated with oral antibiotics, but recovery was slow. During infection-free periods, a mild chronic hemolysis was apparent from the low-normal level of hemoglobin (11.1 to 11.6 g/dL; normal, 11.8 to 15.3), decreased levels of haptoglobin (<30 mg Hb bound per 100 mL serum; normal, 30 to 240), and increased fraction of reticulocytes (3.6% to 4.0%; normal, 0.2% to 2.0%). The mean RBC indices were as follows: mean corpuscular hemoglobin concentration (MCHC), 20.5 mmol/L (normal, 19.3 to 22.3); mean corpuscular hemoglobin (MCH), 2,330 attomol (normal, 1,720 to 2,170); and mean corpuscular volume (MCV), 114 fL (normal, 85 to 104). Blood transfusions were given to the patient after episodes of hemolysis and each time after she had given birth. The first child, a daughter, was born in 1988 after an uneventful, full-term pregnancy. The second child, a daughter, was born in 1990 after a 30-week pregnancy but died 12 hours later of intra-abdominal bleeding. The last child, a daughter, was born in 1992 after a 30-week pregnancy. She and her eldest sister are healthy.
Methods
Purification and culture of cells.
RBCs were obtained as described10 from citrated blood by centrifugation and aspiration of plasma and buffy coat, and were washed 3 times with physiologic saline before analysis. Leukocytes were prepared from heparinized blood as described11 by centrifugation, aspiration of plasma, and lysis of RBCs with isotonic ammonium chloride. The leukocytes were washed 3 times with saline before analysis. Neutrophils and lymphocytes were purified from leukocytes by centrifugation over isotonic Percoll of 1.077 g/mL.10 Skin fibroblasts were cultured until confluency in Ham’s F10 medium (Life Technologies, Breda, The Netherlands) supplemented with 20% fetal calf serum, and were harvested by trypsinization according to standard methods.
Enzyme determinations.
G6PD activity in RBCs was determined as described previously,10 with 1.3 mmol/L glucose 6-phosphate and 0.1 mmol/L NADP. Quantitative determination of G6PD activity in single erythrocytes was performed with a cytochemical staining method based on the reduction of tetra-nitroblue tetrazolium (tetra NBT).12Activities of glutathione reductase in RBCs and 6-phosphogluconate dehydrogenase in purified neutrophils and lymphocytes were determined as described previously.10 13
Partial purification of G6PD from cultured fibroblasts was performed by the first 2 steps of the procedure described by Yoshida.14Molecular characterization of partially purified G6PD was performed according to the recommendations of the WHO Report on the Study of G6PD,15 except for the electrophoresis on Cellogel (MALTA, Milano, Italy) instead of starch.
Neutrophil function tests.
Purified neutrophils were suspended in a medium that contained 138 mmol/L NaCl, 2.7 mmol/L KCl, 8.1 mmol/L Na2HPO4, 1.5 mmol/L KH2PO4, 0.6 mmol/L CaCl2, 1.0 mmol/L MgCl2, 5.5 mmol/L glucose, and 0.5% (wt/vol) human albumin (pH 7.4). Oxygen consumption was measured with an oxygen electrode11 and hydrogen peroxide production by oxidation of homovanillic acid.16 Semiquantitative assessment of superoxide generation in individual cells was performed with an NBT slide test.17 Activity of the hexose monophosphate pathway was measured by 14CO2formation from 1-14C-glucose.13 Phagocytosis, killing, and degradation of Escherichia coli by neutrophils was determined with the test described by Hamers et al.18
Molecular genetic studies.
Genomic DNA was isolated from circulating leukocytes.19Southern blot analysis of genomic DNA was performed by digestion with the restriction enzyme EcoRI and hybridization with the full-length G6PD cDNA probe pGD-T-5B. Fragments of the G6PD gene were amplified from genomic DNA by polymerase chain reaction (PCR) with the primers and under conditions specified by Beutler et al.20For nucleotide sequencing, the direct sequencing method was used.21 To confirm the mutation found in the patient, mismatch PCR was applied with the primers listed in Table1, followed by restriction fragment analysis with BanII. In addition, a PCR-amplified fragment of genomic DNA that contained nt C514 was also sequenced with an Amplitaq-21M13 Dye Primer Cycle Sequencing kit (Perkin-Elmer, Norwalk, CT) and analyzed with a model 373 DNA sequencer (Applied Biosystems, Foster City, CA). Primers and conditions are given in Table 1.
Primers Used for PCR Amplification
| Amplified Fragment . | Primer Sequence . |
|---|---|
| Mismatch primers | |
| bp 493-643 (150 bp) | S 5′-AACCGCATCATCGTGGAGGAG AS 5′-TCAGCACCATGAGGTTCTGC |
| Sequence primers for genomic DNA | |
| intron 5-bp 743 (270 bp) | S 5′-TGTAAAACGACGGCCAGTAGGAGGTTCTGGCCTCTACT AS 5′-TCAGCACCATGAGGTTCTGC |
| Sequence primers for complementary DNA | |
| bp 448-703 (255 bp) | S 5′-TGTAAAACGACGGCCAGTCACCAAGAACATTCACGAGTCC AS 5′-GGATAACGCAGGCGATGTTGTCCC |
| Amplified Fragment . | Primer Sequence . |
|---|---|
| Mismatch primers | |
| bp 493-643 (150 bp) | S 5′-AACCGCATCATCGTGGAGGAG AS 5′-TCAGCACCATGAGGTTCTGC |
| Sequence primers for genomic DNA | |
| intron 5-bp 743 (270 bp) | S 5′-TGTAAAACGACGGCCAGTAGGAGGTTCTGGCCTCTACT AS 5′-TCAGCACCATGAGGTTCTGC |
| Sequence primers for complementary DNA | |
| bp 448-703 (255 bp) | S 5′-TGTAAAACGACGGCCAGTCACCAAGAACATTCACGAGTCC AS 5′-GGATAACGCAGGCGATGTTGTCCC |
Bold print indicates mismatch nucleotide to permit BanII cutting of wild-type product into 3 fragments of 105 bp + 22 bp + 23 bp, but mutated 514T product into 2 fragments of 105 bp + 45 bp. Italics indicate binding site for dye primers. Amplification conditions for genomic DNA in Air Thermo-cycler 1605 (Idaho Technology, Idaho Falls, ID): 50 rounds of 5 seconds at 95°C (denaturation), 10 seconds at 50°C (annealing), and 30 seconds at 72°C (extension), at full speed. Amplification conditions for cDNA in Air Thermocycler: 50 rounds of 5 seconds at 95°C, 30 seconds at 60°C, and 15 seconds at 72°C, at full speed.
RNA was isolated from circulating mononuclear leukocytes or cultured fibroblasts and converted into cDNA.21 A fragment of G6PD cDNA from nt 408 to nt 703 was amplified by PCR with the primers and under conditions specified in Table 1. The product of this reaction was sequenced by cycle sequencing as described earlier.
Expression of G6PD in E coli.
The pMPM24 series of arabinose-inducible expression vectors was a kind gift of Dr M.P. Mayer (Institute for Biochemistry and Molecular Biology, Albert Ludwigs University, Freiburg, Germany). To facilitate cloning into the pMPM series of vectors, G6PD cDNA was prepared as a partial NcoI/XhoI fragment. This was cloned into NcoI/XhoI-cleaved pMPM-A4Ω. In this vector, there is an EcoRI site immediately 5′ of theNcoI site. Subsequently, the EcoRI/XhoI fragment containing G6PD cDNA was cloned into pBluescript (Stratagene, La Jolla, CA) for manipulation. Site-directed mutagenesis was performed with the Transformer mutagenesis kit obtained from Clontech (Palo Alto, CA) and used according to the manufacturer’s recommendations. The template for mutagenesis was a plasmid that consisted of the NcoI/XhoI fragment of G6PD cDNA cloned into pBluescript. By means of the oligonucleotide 5′CGTGGAGAAGAGCTTCGGGAGGGA3′, the sequence 5′GCCCTT3′ was changed into 5′GAGCTT3′. This changed the residue 172 from Pro to Ser and introduced an MboII restriction site 5′GAAGA3′, which was subsequently used to identify clones carrying the mutation. After mutagenesis, the cDNA was cloned into pMPM-A4Ω by means of EcoRI and XhoI sites. Recombinant (r) human G6PD B and G6PD Volendam were expressed in DR612 E coli lacking endogenous G6PD activity25 by induction with 2% arabinose.24 Purification of rG6PDs was performed by affinity chromatography as previously reported.26,27 Elution of rG6PD Volendam in the presence of 20 μmol/L glucose 6-phosphate was achieved with 100 μmol/L NADP instead of the usual 50 μmol/L NADP due to the increased binding of G6PD Volendam to 2′,5′-ADP-Sepharose 4B. The electrophoretic mobility of the recombinant enzymes was determined by means of electrophoresis in native polyacrylamide in Tris buffer (pH 8.8) at 4°C,28 followed by G6PD activity staining with phenazine methosulfate (PMS) and methylthiotetrazole (MTT).15 Thermostability assays were performed according to the WHO protocol.15 Briefly, samples with purified recombinant enzyme were incubated for 60 minutes at 51°C; aliquots were drawn every 20 minutes and assayed for G6PD activity.
RESULTS
Table 2 shows that the G6PD activity in the patient’s RBCs was below the detection limit of our assay. In contrast, the glutathione reductase activity was increased, as was the number of reticulocytes in the patient’s blood. None of these abnormalities were found in the patient’s relatives: her parents, 11 paternal uncles and aunts, 12 maternal uncles and aunts, 1 brother, 2 sisters, and 2 daughters (Fig 1). Nevertheless, paternity investigations showed a 99.4% probability of the patient being the child of her legal parents.
Enzyme Activities in Purified Blood Cells
| Cell Type . | G6PD . | 6PGD . | GSR . |
|---|---|---|---|
| RBCs (n = 4), U/g hemoglobin | 0.1 (4.8-7.2) | ND | 6.2 (2.4-4.8) |
| Leukocytes (n = 1), U/g protein | 3.6 (776) | ND | ND |
| Neutrophils (n = 2), U/1010 cells | 5.2 (97-200) | 138 (35-107) | 27 (17-37) |
| Cell Type . | G6PD . | 6PGD . | GSR . |
|---|---|---|---|
| RBCs (n = 4), U/g hemoglobin | 0.1 (4.8-7.2) | ND | 6.2 (2.4-4.8) |
| Leukocytes (n = 1), U/g protein | 3.6 (776) | ND | ND |
| Neutrophils (n = 2), U/1010 cells | 5.2 (97-200) | 138 (35-107) | 27 (17-37) |
Values are derived from (n) measurements with patient cells (range obtained with 10 healthy donors is also given).
Abbreviations: 6PGD, 6-phosphogluconate dehydrogenase; GSR, glutathione reductase; ND, not determined.
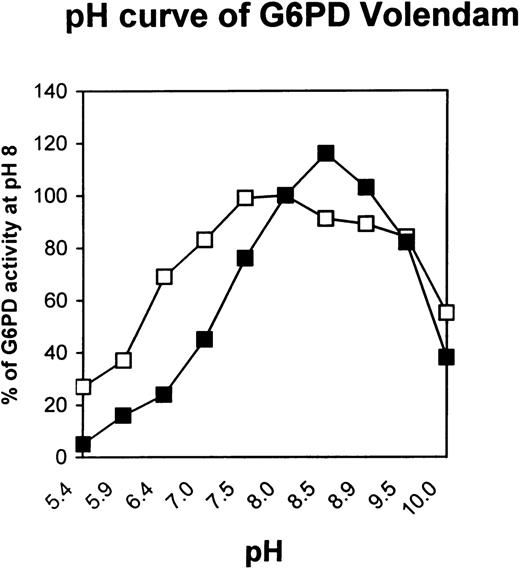
The cultured fibroblasts from the patient contained approximately 15% of normal G6PD activity (Table 3). On electrophoresis the enzyme formed 2 anodal bands that migrated approximately 75% and 95% of the distance of the normal G6PD B enzyme on Cellogel in 75 mmol/L Tris-EDTA-citric acid (TEC) buffer at pH 7.5.29 The Michaelis constants (Km) for NADP and glucose 6-phosphate were increased by a factor of 4 and 5, respectively, whereas the inhibitor constant (Ki) for NADPH was normal (Table 3). The utilization of the substrate analog deamino-NADP was one third of normal (Table 3). Furthermore, the pH optimum was found to be a narrow peak at pH 8.5, instead of a broad range at pH 7.0 to 8.5 for G6PD B (Fig 2).
Molecular Characteristics of Partially Purified G6PD From Skin Fibroblasts
| . | Patient . | Normal . |
|---|---|---|
| Specific activity (U/g protein) | 49 | 369 |
| Km glucose 6-P 37°C (μmol/L) | 328 | 60 |
| Km NADP 37°C (μmol/L) | 51 | 14 |
| Ki NADPH 25°C (μmol/L) | 33 | 40 |
| Substrate analog utilization | ||
| 2-deoxy-glucose 6-P (%) | 4.4 | <4 |
| Galactose 6-P (%) | 1.9 | 2.4 |
| Deamino-NADP (%) | 20.8 | 60 |
| NAD (%) | 0 | 0 |
| Heat stability at 50°C | Slightly decreased | Normal |
| . | Patient . | Normal . |
|---|---|---|
| Specific activity (U/g protein) | 49 | 369 |
| Km glucose 6-P 37°C (μmol/L) | 328 | 60 |
| Km NADP 37°C (μmol/L) | 51 | 14 |
| Ki NADPH 25°C (μmol/L) | 33 | 40 |
| Substrate analog utilization | ||
| 2-deoxy-glucose 6-P (%) | 4.4 | <4 |
| Galactose 6-P (%) | 1.9 | 2.4 |
| Deamino-NADP (%) | 20.8 | 60 |
| NAD (%) | 0 | 0 |
| Heat stability at 50°C | Slightly decreased | Normal |
Mean values of 2 measurements performed in duplicate on 2 different days. The mean values of the 2 duplicates did not differ by >5%.
pH curve of G6PD Volendam. The enzymatic activity of G6PD partially purified from cultured fibroblasts of a healthy individual (□) or from the patient (▪) was measured at various pH values. Results are percentage of normal G6PD-B activity at pH 8. Each data point is the mean of 2 independent measurements performed in duplicate; the mean values of the 2 duplicates did not differ by more than 5%.
pH curve of G6PD Volendam. The enzymatic activity of G6PD partially purified from cultured fibroblasts of a healthy individual (□) or from the patient (▪) was measured at various pH values. Results are percentage of normal G6PD-B activity at pH 8. Each data point is the mean of 2 independent measurements performed in duplicate; the mean values of the 2 duplicates did not differ by more than 5%.
Because the patient suffered from undue susceptibility to bacterial infections of the airways, granulocyte functions and enzyme content were tested. Table 2 shows that G6PD deficiency was also manifested in the total leukocytes and in the purified neutrophils of the patient. Moreover, as shown in Table 4, her neutrophils displayed a profoundly diminished oxidative capacity after activation with opsonized zymosan particles (STZ) or phorbol-myristate acetate (PMA). Phagocytosis, killing, and degradation of E coliin vitro were normal (data not shown).
Oxidative Metabolism of Neutrophils
| Parameter . | Patient . | Controls . |
|---|---|---|
| O2consumption | ||
| STZ (nmol/106 cells · min) | 2.1 ± 0.9 (3) | 8.9 ± 1.4 (20) |
| PMA (nmol/106 cells · min) | 1.7, 1.4 (2) | 7.7 ± 2.8 (8) |
| H2O2 production | ||
| STZ (nmol/106 cells · min) | 0.7 | 4.3 ± 1.1 (10) |
| HMP pathway activity | ||
| STZ (nmol/106 cells · 30 min) | 2.9 | 8.3 ± 3.0 (10) |
| Parameter . | Patient . | Controls . |
|---|---|---|
| O2consumption | ||
| STZ (nmol/106 cells · min) | 2.1 ± 0.9 (3) | 8.9 ± 1.4 (20) |
| PMA (nmol/106 cells · min) | 1.7, 1.4 (2) | 7.7 ± 2.8 (8) |
| H2O2 production | ||
| STZ (nmol/106 cells · min) | 0.7 | 4.3 ± 1.1 (10) |
| HMP pathway activity | ||
| STZ (nmol/106 cells · 30 min) | 2.9 | 8.3 ± 3.0 (10) |
Values are given as mean ± SD of (n) determinations.
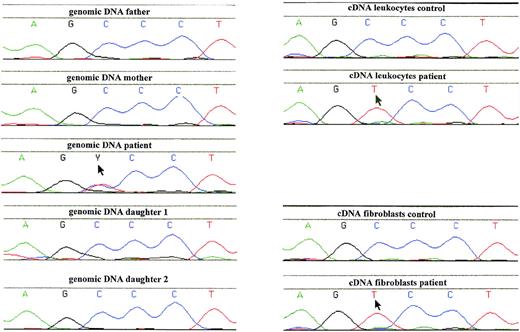
To identify the mutation in the G6PD gene responsible for the enzyme deficiency, genomic DNA from leukocytes was PCR-amplified and sequenced. This showed a heterozygous 514C→T mutation, predicting a Pro172→Ser substitution (Fig3). Restriction enzyme analysis of PCR-amplified genomic DNA from hair roots also indicated the patient to be heterozygous for the 514C→T mutation (not shown). No other mutations in the coding region of the G6PD gene were found. We also examined the G6PD gene for a possible deletion by restriction fragment length polymorphism (RFLP) analysis of genomic DNA after incubation with EcoRI, probing with full-length G6PD cDNA, but found only normal fragments in the patient, her parents, sisters, brother, and eldest daughter. These fragments were stained with normal intensity (in the women with an intensity twice as big as in the men). In leukocyte DNA from these family members, the 514C→T mutation was not found (Fig 3).
Analysis of genomic and complementary DNA from the patient with G6PD Volendam and her family. A PCR product, containing the region around 514C of the G6PD gene, was generated from genomic or cDNA obtained from blood leukocytes or cultured fibroblasts, and was analyzed by Dye Primer Cycle sequencing. Starting material: left, genomic DNA from leukocytes of the father and the mother of the patient, the patient, and her 2 daughters; right, cDNA from leukocytes of a healthy control and the patient; and cDNA from cultured fibroblasts of a healthy control and the patient. Arrows indicate mutant sequence.
Analysis of genomic and complementary DNA from the patient with G6PD Volendam and her family. A PCR product, containing the region around 514C of the G6PD gene, was generated from genomic or cDNA obtained from blood leukocytes or cultured fibroblasts, and was analyzed by Dye Primer Cycle sequencing. Starting material: left, genomic DNA from leukocytes of the father and the mother of the patient, the patient, and her 2 daughters; right, cDNA from leukocytes of a healthy control and the patient; and cDNA from cultured fibroblasts of a healthy control and the patient. Arrows indicate mutant sequence.
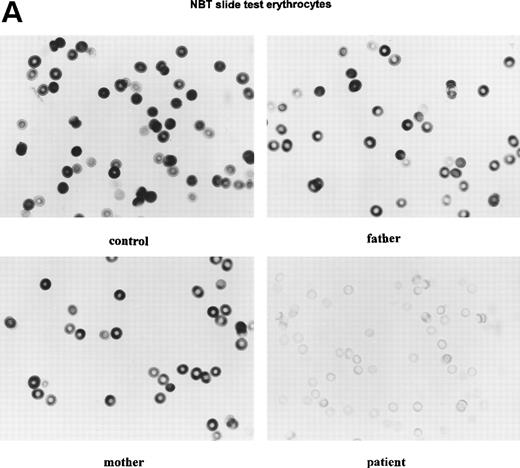
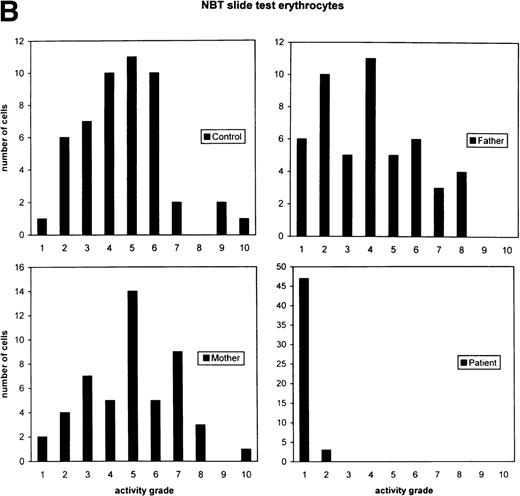
Because the G6PD activity in the patient’s cells was so low, we suspected that the normal allele was hardly transcribed. Indeed, when we tested the RBCs with a cytochemical assay that scores the G6PD activity quantitatively in each separate cell, we found 94% of the patient’s cells without any activity and 6% with less than 5% of the normal mean activity (Fig 4). Thus, there was no indication for a population of RBCs with normal G6PD activity. Moreover, in the patient’s purified neutrophils, a similar cytochemical test for the distribution of the NADPH oxidase activity (which generates superoxide) showed all cells to contain a low activity. Apparently, the G6PD deficiency was manifested in all cells, in neutrophils as well as in erythrocytes. Analysis of PCR-amplified cDNA from the patient’s leukocytes and cultured fibroblasts revealed only the 514T sequence in the G6PD cDNA (Fig 3), indicating the preferential transcription of the mutated allele in these cells.
Cytochemical localization of G6PD activity in erythrocytes from the patient with G6PD Volendam and her family. The test was performed as described.12 G6PD activity in individual erythrocytes was analyzed as production of formazan by reduction of tetra NBT and quatitated by means of cytophotometry through a microscope. The mean integrated absorbance per cell was measured in 50 randomly chosen cells, corrected for aspecific light loss in cells incubated in the absence of glucose 6-phosphate and expressed in activity grades from 1 to 10 (grade 1 = 0 to 10 arbitrary units, grade 2 = 10 to 20 arbitrary units, etc, up to grade 10 = 90 to 100 arbitrary units). (A) Histochemical localization of G6PD activity in erythrocytes from a healthy control, the father and mother of the patient, and the patient herself. Original magnification x 400. (B) Quantitative scores of these reactions are shown for the control, the father, the mother, and the patient.
Cytochemical localization of G6PD activity in erythrocytes from the patient with G6PD Volendam and her family. The test was performed as described.12 G6PD activity in individual erythrocytes was analyzed as production of formazan by reduction of tetra NBT and quatitated by means of cytophotometry through a microscope. The mean integrated absorbance per cell was measured in 50 randomly chosen cells, corrected for aspecific light loss in cells incubated in the absence of glucose 6-phosphate and expressed in activity grades from 1 to 10 (grade 1 = 0 to 10 arbitrary units, grade 2 = 10 to 20 arbitrary units, etc, up to grade 10 = 90 to 100 arbitrary units). (A) Histochemical localization of G6PD activity in erythrocytes from a healthy control, the father and mother of the patient, and the patient herself. Original magnification x 400. (B) Quantitative scores of these reactions are shown for the control, the father, the mother, and the patient.
Thus, the patient, a heterozygous female, only appears to be expressing the mutated G6PD gene. This situation may arise by nonrandom X-chromosome inactivation. We investigated X-chromosome inactivation patterns in her leukocytes and in those from some of her relatives by means of the HUMARA assay. The results (Fig5) show that she had a completely skewed X-chromosome inactivation pattern, with only 1 chromosome active in all her cells. Her mother and 1 of the patient’s daughters also had completely skewed patterns, while her 2 sisters showed a random pattern. Data from the other daughter were uninformative.
Segregation of a skewed pattern of X-chromosome inactivation. The autoradiograph shows the results of amplification of the HUMARA polymorphism either without (−) or with (+) prior digestion with HpaII. Each allele appears as a doublet. With reference to the family tree in Fig 1, the subjects analyzed are as follows: 1 =II:13 (mother of the patient); 2 = II:4 (father of the patient); 3 = III:2 (patient); 4 = III:4 (sister of the patient); 5 = III:5 (sister of the patient); 6 = IV:1 (daughter of the patient); 7 = IV:3 (daughter of the patient). Notice that 1, 3, and 7 show a skewed pattern of X-chromosome inactivation, 4 and 5 are random, and 6 is uninformative.
Segregation of a skewed pattern of X-chromosome inactivation. The autoradiograph shows the results of amplification of the HUMARA polymorphism either without (−) or with (+) prior digestion with HpaII. Each allele appears as a doublet. With reference to the family tree in Fig 1, the subjects analyzed are as follows: 1 =II:13 (mother of the patient); 2 = II:4 (father of the patient); 3 = III:2 (patient); 4 = III:4 (sister of the patient); 5 = III:5 (sister of the patient); 6 = IV:1 (daughter of the patient); 7 = IV:3 (daughter of the patient). Notice that 1, 3, and 7 show a skewed pattern of X-chromosome inactivation, 4 and 5 are random, and 6 is uninformative.
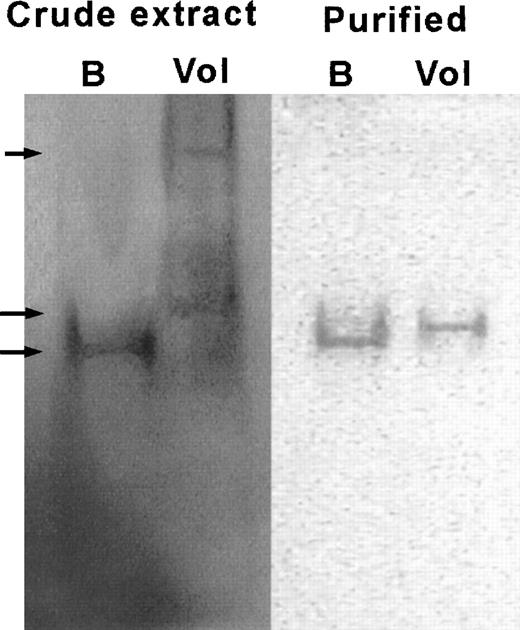
Finally, we also analyzed rG6PD Volendam514T purified fromE coli. The results (Table5) were in full agreement with those obtained with the partially purified enzyme from the patient’s fibroblasts (Table 3). The specific activity of the rG6PD Volendam was only 17% of that observed with rG6PD B, and the Km for NADP and glucose 6-phosphate was increased by a factor of 2.5 and 3, respectively. The Ki for NADPH was normal. Utilization of 2-deoxy-glucose 6-phosphate, galactose 6-phosphate, and deamino-NADP by rG6PD Volendam was decreased. The electrophoretic mobility of purified rG6PD Volendam was near normal, with a band migrating at 95% of the distance traveled by rG6PD B. However, electrophoresis of crude bacterial lysate containing rG6PD Volendam resulted in a G6PD-specific smear between 70% and 95% of the distance migrated by G6PD B in a crude lysate (Fig 6). The thermostability of purified rG6PD Volendam at 51°C was strongly decreased (90% loss of activity after 20 minutes v 20% loss in rG6PD B). These results indicate that the Pro172→Ser substitution in G6PD induces both altered enzymatic properties and protein instability.
Molecular Characteristics of Purified Recombinant G6PD
| . | G6PD Volendam . | G6PD B . |
|---|---|---|
| Specific activity (IU/mg protein) | 36 ± 3 | 210 ± 15 |
| Km glucose 6-P 25°C (μmol/L) | 211 ± 16 | 65 ± 4 |
| Km NADP 25°C (μmol/L) | 30 ± 3 | 12 ± 2 |
| Ki NADPH 25°C (μmol/L) | 20 | 14 ± 3 |
| Substrate analog utilization | ||
| 2-deoxy-glucose 6-P (%) | 1.5 | 4.0 |
| Galactose 6-P (%) | 1.1 | 8.0 |
| Deamino-NADP (%) | 14 | 59 |
| Electrophoretic mobility (%) | 96 | 100 |
| Heat stability at 51°C | Strongly decreased | Normal |
| . | G6PD Volendam . | G6PD B . |
|---|---|---|
| Specific activity (IU/mg protein) | 36 ± 3 | 210 ± 15 |
| Km glucose 6-P 25°C (μmol/L) | 211 ± 16 | 65 ± 4 |
| Km NADP 25°C (μmol/L) | 30 ± 3 | 12 ± 2 |
| Ki NADPH 25°C (μmol/L) | 20 | 14 ± 3 |
| Substrate analog utilization | ||
| 2-deoxy-glucose 6-P (%) | 1.5 | 4.0 |
| Galactose 6-P (%) | 1.1 | 8.0 |
| Deamino-NADP (%) | 14 | 59 |
| Electrophoretic mobility (%) | 96 | 100 |
| Heat stability at 51°C | Strongly decreased | Normal |
Values are means ± SD of 3 independent duplicate measurements.
Electrophoretic mobility of rG6PD B and rG6PD Volendam. rG6PD B and rG6PD Volendam were electrophoresed in native 5% polyacrylamide gel. The gels were stained for G6PD activity by PMS and MTT, as described.15 Left: Bacterial crude extract of E coli DR612 containing pMPM-A4 encoding human G6PD B (B) or G6PD Volendam (Vol). From top to bottom, arrows indicate the 70%, 95%, and 100% mobilities. Right: rG6PD B and rG6PD Volendam purified by affinity chromatography.
Electrophoretic mobility of rG6PD B and rG6PD Volendam. rG6PD B and rG6PD Volendam were electrophoresed in native 5% polyacrylamide gel. The gels were stained for G6PD activity by PMS and MTT, as described.15 Left: Bacterial crude extract of E coli DR612 containing pMPM-A4 encoding human G6PD B (B) or G6PD Volendam (Vol). From top to bottom, arrows indicate the 70%, 95%, and 100% mobilities. Right: rG6PD B and rG6PD Volendam purified by affinity chromatography.
DISCUSSION
The 514C→T mutation found in the G6PD gene of the patient was not detected in the DNA from her father or her mother. However, the mutation was present in the patient’s leukocytes, as well as in her hair-root cells, and the enzyme deficiency was found in her blood cells as well as in her fibroblasts. Therefore, we conclude that the mutation must have arisen in the germ-line cells of one of her parents. In addition, the heterozygous mutation was expressed in an apparently homozygous way, which can only be explained by inactivation of the X chromosome that carries the normal G6PD allele in almost all of her cells. Detection of only the mutated G6PD RNA sequence in the leukocytes and in the fibroblasts of the patient confirms this idea.
Our study of X-chromosome inactivation patterns in the family showed that the patient had completely skewed X-chromosome inactivation with only the X chromosome containing the mutated G6PD allele active in all of her cells. Her mother and 1 of her daughters also showed complete skewing. Thus, skewed X-chromosome inactivation appears to be segregating in this family as a Mendelian X-linked trait. In other X-linked traits, such as Wiskott-Aldrich syndrome and Duchenne muscular dystrophy, female carriers who manifest the disease have been observed, sometimes in families in which skewed X-chromosome inactivation is inherited (reviewed by Belmont30 and Puck and Willard31). One mechanism to explain this phenomenon is a deleterious genetic lesion on the X chromosome that is always inactivated. An alternative mechanism is a genetic defect in the X-chromosome inactivation process itself.31 In the patient and her mother, this skewed X-chromosome inactivation trait is linked to a different HUMARA allele than in the patient’s daughter, which indicates that a recombination event has taken place between the inactivation determinant and the HUMARA allele at Xq11.2. Because the patient inherited the X chromosome carrying the inactivation determinant from her mother, the de novo mutation in the G6PD gene must have occurred on the paternal X chromosome.
G6PD molecules form catalytically active dimers by binding NADP. The proline-to-serine substitution does not alter the charge of the protein, but it did diminish the affinity for the negatively charged NADP (Table 3), which may have led to an apparent increase in positive charge of the protein. Thus, the 95%-mobility band may have contained mutant dimers with bound NADP, and the 75%-mobility band may have consisted of monomers or dimers without NADP. Indeed, the fully NADP-saturated, purified rG6PD Volendam showed a mobility that was 95% of normal, whereas the same mutant enzyme in crude E colilysate (not NADP-saturated) displayed a smear with a mobility of 70% to 95% of normal.
Although there is a cluster of class 1 mutations in exons 10 and 11 (residues 351 to 455), some other substitutions producing chronic nonspherocytic hemolytic anemia have been identified elsewhere (as far away as exons 3 and 4). Pro172 of the human enzyme corresponds to Pro149 in G6PD from the bacterium Leuconostoc mesenteroides,32 the only G6PD from which the three-dimensional structure has been solved up to date.33 This residue is neither in the subunit interface nor in the NADP-binding site. Instead, this proline is in a completely conserved protein coil between the secondary structural elements beta-sheet-E and alpha-helix-e, important for structural stability. Probably, this coil is also involved in allowing the 2 domains of each G6PD subunit to adjust their positions when NADP and glucose 6-phosphate are bound.32 With secondary structural content prediction (SSCP,34,35 the 7 amino acids of this coil (EKPFGRD) can be predicted to have a 100% coil content, whereas in the mutant configuration (EKSFGRD) it shows 80% coil content, 10% alpha helix content, and 10% beta sheet content. Moreover, Pro149 in theL mesenteroides G6PD structure refined at 2 Å resolution presents a different conformation (cis and trans) in each monomer of the G6PD dimer and is directly involved in positioning of the substrate pocket to bind the phosphate group of glucose 6-phosphate.33 Extra flexibility induced by substitutions at this position may be expected to both decrease the affinity for glucose 6-phosphate and, by opening up the structure at a point that should be rigid, render the enzyme more susceptible to denaturation. Indeed, the Km value of G6PD Volendam is among the highest described to date for any G6PD mutant,36 and the recombinant mutant enzyme was found to be highly unstable. Moreover, the total lack of enzymatic activity in the RBCs of the patient (mean age in the circulation, ≈30 days), the 3% to 4% of normal activity in her neutrophils (mean age in the circulation, ≈14 days) and the 15% of activity in her fibroblasts (doubling time in culture, 4 days) suggests that G6PD Volendam may be unstable in vivo as well. Probably, both synergistic effects of the mutation are needed to result in class 1 hemolytic anemia, because G6PD Orissa, caused by another mutation in the surroundings of the catalytic pocket, shows a high Km value for glucose 6-phosphate, but is a class 3 variant that leads to anemia only in the presence of stress.37
Increased susceptibility to bacterial or fungal infections has been described in a few G6PD-deficient patients.2-6 Only when the G6PD activity in the neutrophils is less than 5% of normal, is the substrate supply for the NADPH oxidase in these cells insufficient for the production of normal amounts of superoxide, needed for efficient killing of micro-organisms.3 Indeed, we found markedly diminished activity of the HMP pathway, oxygen consumption, and hydrogen peroxide formation in the activated neutrophils from our patient, albeit not as low as found in neutrophils from patients with chronic granulomatous disease.7 The capacity of the neutrophils from our patient to kill E coli in vitro was undisturbed, also in contrast to the findings in chronic granulomatous disease,18 and the clinical symptoms were less severe. However, the in vivo capacity of the G6PD-deficient neutrophils may be insufficient to locally kill large numbers of bacteria and clear an infected area quickly.
Address reprint requrests to Dirk Roos, PhD, Central Laboratory of the Netherlands Red Cross Blood Transfusion Service, Plesmanlaan 125, 1066 CX Amsterdam, The Netherlands; e-mail:d_roos@clb.nl.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Deceased.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal