Abstract
The gene PIGA encodes one of the protein subunits of the 1-6-N acetylglucosaminyltransferase complex, which catalyses an early step in the biosynthesis of glycosyl phosphatidylinositol (GPI) anchors. PIGA is somatically mutated in blood cells from patients with paroxysmal nocturnal hemoglobinuria (PNH), leading to deficiency of GPI-linked proteins on the cell surface. To investigate in detail how inactivating mutations of the PIGA gene affect hematopoiesis, we generated a mouse line, in whichloxP-mediated excision of part of exon 2 occurs on the expression of Cre. After crossbreeding with EIIa-cre transgenic mice, recombination occurs early in embryonic life. Mice that are mosaics for the recombined Piga gene are viable and lack GPI-linked proteins on a proportion of circulating blood cells. This resembles the coexistence of normal cells and PNH cells in patients with an established PNH clone. PIGA(−) blood cells in mosaic mice have biologic features characteristic of those classically seen in patients with PNH, including an increased sensitivity toward complement mediated lysis and a decreased life span in circulation. However, during the 12-month follow-up, the PIGA(−) cell population did not increase, clearly showing that a Piga gene mutation is not sufficient to cause the human disease, PNH.
IN PATIENTS WITH paroxysmal nocturnal hemoglobinuria (PNH), an acquired hemolytic anemia, the X-linkedPIGA gene is mutated in a proportion of blood cells, leading to partial or complete absence of glycosyl phosphatidylinositol (GPI)-linked proteins on the cell surface of the affected cells.1-3 The PIGA gene encodes a protein subunit of the α1-6-N acetylglucosaminyltransferase complex, an enzyme essential in the biosynthesis of GPI anchor molecules.4 As a result, PNH red cells have an increased sensitivity to complement mediated lysis,5 because 2 of the complement inhibitors, normally GPI-linked, are missing on their cell surface.6,7As multiple blood lineages may be affected, the somatic mutation in thePIGA gene must occur in a multipotent hematopoietic progenitor cell.8,9 Although PIGA mutations account for the cellular phenotype, it is not clear why and how cells deficient in GPI-linked proteins can expand and contribute substantially to hematopoiesis in patients. PNH is often associated with aplastic anemia.10 This observation has led to the hypothesis that hematopoietic progenitor cells deficient in GPI-anchored proteins might have a selective growth advantage in a bone marrow environment in which normal hematopoiesis is impaired.11 12
Complete absence of GPI-linked proteins is not compatible with life.13,14 This limits the possibility of studying the biologic consequences of GPI-anchor deficiency in hematopoiesis by conventional knockout technology. Here we describe the generation of a mouse suitable for spatially and temporally controlled inactivation of the Piga gene by flanking the largest coding exon ofPiga with 2 loxP sites (in the following “PIGA” refers to the human gene, “Piga” to the murine gene, and “PIGA” to either gene product15). Cre mediated inactivation of Piga was obtained by crossbreeding these mice with mice transgenic for thecre recombinase under the control of the adenoviral EIIa promoter (EIIa-cre16). Because the EIIa promoter is only active early in embryonic development we are able to obtain viable offspring that are mosaic for a nonfunctional Piga gene. The ease of crossbreeding provides us with an unlimited number of animals and thus circumvents the limitations associated with chimeras obtained by blastocyst injection of PIGA(−) murine embryonic stem (ES) cells.13,14 In mosaic animals, we observe a proportion of blood cells lacking GPI-linked proteins [also referred to as PIGA(−) or PNH phenotype]. The coexistence of normal blood cells and blood cells that lack GPI-linked proteins mimics the findings in patients with PNH. In mosaic mice, no further Piga inactivation occurs after birth.17 This enabled us to study the fate of blood cells deficient in GPI-linked proteins during the lifetime of a mouse and compare it with hematopoiesis in PNH patients.
MATERIALS AND METHODS
Targeting construct lox-Piga-LTNL-lacZ.
In a 9.7-kb genomic XhoI/NotI subclone of Piga, the EcoRI site in intron 1 was destroyed. Then a 7.2-kbSalI/BamHI Piga fragment was subcloned into Bluescript (Stratagene, La Jolla, CA). The following oligonucleotide containing the loxP sequence was inserted into the uniqueSmaI site of exon 2: 5′ CATAACTTCGTATAATGTATGCTATACGAAGTTATC 3′. This destroyed theSmaI site. To lengthen the homologous sequences 5′ to thelox-Piga-LTNL-lacZ targeting vector, a 2.5-kbXhoI (filled)/BamHI genomic Piga fragment was cloned into the targeting construct cut with BamHI/XbaI (filled).
For the LTNL lacZ fusion, first the ApaI/ClaI and ClaI/BamHI region of pnLacF (obtained from Andreas Kottman, Columbia University, New York, NY) was subcloned into the KS Bluescript vector cut with ApaI/BamHI. Then an Asp718 (filled)/XbaI (filled) fragment was ligated to the LTNL selectable marker cassette (loxP, thymidine kinase gene, neomycin resistance gene, 3′loxP site)18 cut with PacI and recessed. Finally, the XbaI (filled) fragment containing the LTNL lacZ fusion was cloned into the unique EcoRI (filled) site of the lox-Piga-LTNL-lacZ targeting vector.
Homologous recombination and Cre recombination in ES cells.
The 129 SV CJ7 ES cell line19 was transfected with theSalI linearized targeting constructlox-Piga-LTNL-lacZ as described.14 In total, 239 ES cell clones were picked and analyzed for homologous recombination by Southern blot hybridization.20 DNA Probe 6, located in exon 6, was used as an external probe14 and DNA probe 1 and probe 2 were used as 2 internal probes. Probe 1 was generated by subcloning a HindIII/SacII fragment located upstream of exon 1 into the Bluescript vector. Probe 2 was generated by subcloning a 400-bp polymerase chain reaction (PCR) product of exon 2 with the forward primer 5′ GTACATATTTGTTCGGGA 3′ and the reverse primer 5′ CTTTTCTGTAAACAAGTCTG 3′ into the pGem vector. Southern blot analysis was done by digesting genomic DNA with EcoRI to screen for the loss of theEcoRI site in intron 1. Then the ES cells were screened for the presence of the 5′ loxP sequence at the SmaI site by PCR: forward primer (SacII) 5′ GCCGCGGACCACCTCAGCATGGCCAA 3′, reverse primer (I2) 5′ AAAGCCACCATACAGAATGA 3′. The PCR products were digested withSmaI. A 700-bp restriction fragment identified the wild-typePiga gene, whereas the lox-Piga-lacZ gene was identified by an 836-bp fragment. In 39 ES clones, (16%) homologous recombination at the Piga locus had occurred. However, 17 (44%) of the recombined ES clones did not contain the 5′loxP site. Clone 1G2 was electroporated as a single-cell suspension of 5 × 106 cells at 240 mV, 500 μF with 30 μg supercoiled pMC-Cre plasmid.21 After 72 hours, the cells were selected with 1 μmol/L Gancyclovir (Cytovene, Syntex, Palo Alto, CA) for 10 days. Clones were analyzed for excision of the LTNL selectable marker cassette by Southern blot and by PCR analysis with the above described primer pair. In 2 of 751 ES clones, 1G2/87 and 1D4, the LTNL marker cassette was excised. In 17 clones, both exon 2 and the LTNL marker cassette had been excised, and 732 ES clones were of wild-type origin. Targeted ES cells with Cre recombination that preserves only the LTNL cassette were not detected because of selection against the presence of the Herpes simplex virus (HSV) thymidine kinase.
Generation of the lox-Piga-lacZ mice.
Clones 1G2/87 and 1D4 showed a normal karyotype of 40 XY22and were injected into C57BL/6J blastocysts. Of 10 chimeric animals (3 females and 7 males), 4 male chimeras, all generated with clone 1G2/87, transmitted the targeted Piga gene to their progeny (lox-Piga-lacZ mice). Heterozygous lox-Piga-lacZfemales were bred to C57BL/6 males to produce hemizygouslox-Piga-lacZ males in the N2 generation.
FVB/NJ mice homozygous for the EIIa-cre transgene (EIIa-cre [+/+] mice) were generously provided by Heiner Westphal (NIH, Bethesda, MD).16Lox-Piga-lacZ (L) mice were crossbred with EIIa-cre (+/+) (E) mice as indicated. Thelox-Piga-lacZ/EIIa-cre offspring analyzed were of mixed genetic background. (In this article, the different breedings are abbreviated by 2 letters, EL and LE. The first letter represents the origin of the mother and the second the origin of the father.)
Flow cytometric analysis (FCA).
FCA on fetal and adult peripheral blood cells was performed as described previously.14 The following fluorochrome conjugated antibodies toward GPI-linked surface antigens were used: CD24 (Heat Stable Antigen, HSA) for red blood cells (RBCs); Gr-1 for granulocytes; and CD48 for lymphocytes. Lineage specificity was determined by using fluorescein isothiocyanate or phycoerythrin (PE)-conjugated anti-B220 antibodies for B lymphocytes, anti-TCRαβ for T lymphocytes, and anti-CD11b for granulocytes. All the antibodies were purchased from Pharmingen (San Diego, CA). One to 2 μL of blood was used for the immunostaining of RBCs and 25 μL was used for the staining of white blood cells.
Blood cell counts.
Blood samples were obtained by puncture of the retroorbital plexus of anesthetized mice by using heparinized capillary tubes. RBC count, white blood cell count, hemoglobin, mean RBC volume, hematocrit, and platelet count were determined with an automated blood cell counter, Hemavet 850 (CDC Technology, Oxford, CT). Differential white blood cell counts were performed on May-Grünwald-Giemsa–stained blood films.23
Reticulocyte counts.
The fluorescence dye thiazole orange (Retic-COUNT, Becton Dickinson, San Jose, CA) was used to detect reticulocytes in conjunction with PE-conjugated CD24 to distinguish PIGA(+) from PIGA(−) RBCs.24 The half-life of PIGA(−) red cells was calculated for individual mice in steady state (t1/2PIGA(+) = 15 days25):
Sensitivity toward complement mediated lysis.
We studied the sensitivity of PIGA(+) and PIGA(−) RBCs toward complement mediated lysis. Many inbred laboratory mouse strains are deficient in C5.26 According to information obtained from the Jackson Laboratory (Bar Harbor, ME), C57BL/6 and 129SV have a normal level of C5, whereas FVB/N are C5 deficient. Lox-Piga-LacZ mice were a mixture of 129SV and C57BL/6, whereas the EIIa-cre animals had the FVB/N genetic background. Mosaic offspring are therefore heterozygous for C5 deficiency and are expected to have a C5 level of 50% from normal. C5 is not a limiting factor in complement activation. In vivo whole complement activity in our mosaic mice and most biologic tests for complement activity in vitro are expected to be indistinguishable from mice with 100% of C5 levels. Mouse serum is known to have a low lytic activity in vitro.27 We used, as have previously others, rat or human serum as a source of complement.27 28 Aliquots were stored in liquid nitrogen and thawed only once before use. Possible natural antibodies were removed by RBC absorption. Absorption was performed on ice for 15 minutes with 100 μL of packed washed RBCs in 1,500 μL of serum. Absorption was repeated 4 times. Half of the absorbed serum was heat inactivated for 60 minutes at 56°C (heat-inactivated serum [HS]). To activate complement serum was acidified to a pH of 6.9 by adding HCl to a final concentration of 20 mmol/L. Washed blood red cells (10 μL) from mosaic mice or an age matched control mouse were incubated with dilutions of fresh or heat inactivated serum in 154 mmol/L NaCl in a final volume of 122 μL. MgCl2 was added to a final concentration of 4.18 mmol/L. Lysis of PIGA(−) RBCs was measured by flow cytometry after 60 and 120 minutes of incubation. The hemoglobin content was determined by measuring absorbance at 412 nm. Lysis (100%) corresponded to the release of hemoglobin after hypotonic lysis of an equal volume of RBCs.
DNA preparation and amplification.
ES cells were lysed overnight at 55oC in 100 mmol/L Tris-HCl pH 8.5, 5 mmol/L ethylenediamine tetraacetate, 0.2% sodium dodecyl sulfate, 200 mmol/L NaCl, 100 μg/mL proteinase K. DNA was precipitated in equal volume isopropanol and analyzed by Southern blot and PCR analysis. Genomic DNA from mouse tails was obtained as described by Laird et al.29
Genotype analysis was done by PCR analysis from tail DNA: Wild-typePiga and lox-Piga-lacZ were amplified with the forward primer “−13 + 8” 5′ GGACCACCTCAGCATGGCCAA 3′ and the reverse primer “Piga 600 rev” 5′ TATTTCAGGATTCAGTGCTGC 3′. The SmaI restriction site in exon 2 was destroyed by the introduction of the 5′ loxPsite. Digestion of the amplification product with SmaI, therefore, yielded a 551-kb fragment for the wild-type (wt) gene and a 655-kb fragment for lox-Piga-lacZ.Lox-ΔPiga-lacZ was amplified with the forward primer “−13 + 8” and the reverse primer “lacZ 500 rev” 5′ CGACAGTATCGGCCTCAGGAAGA 3′, yielding a 382-bp fragment.
Reverse transcription (RT)-PCR analysis.
Total RNA was isolated with RNAzol B (Biotecx Laboratories, Houston, TX) and treated with RNase free DNase. RNA (1 μg) was subjected to cDNA synthesis by using the Superscript II system (GIBCO-BRL, Gaithersburg, MD) followed by PCR amplification. The primer pairs for the Cre expression cre1 (forward): 5′ TAAAATGTCCAATTTACTGACCGTACACAA 3′ and cre2 (reverse): 5′ CTGGCAATTTCGGCTATACGTAACAGGGTG 3′ amplifies a band of 520 bp. In simultaneous PCR experiments, the primer pair SacII/293 (SacII: see above; Primer 293: 5′ GGGTGACAGTTATGACCTTGTG 3′), which amplifies a 268-bp fragment of the Piga cDNA, was used to control the integrity of the isolated RNA followed by cDNA synthesis.
RESULTS
Generation of the lox-Piga-lacZ mouse line.
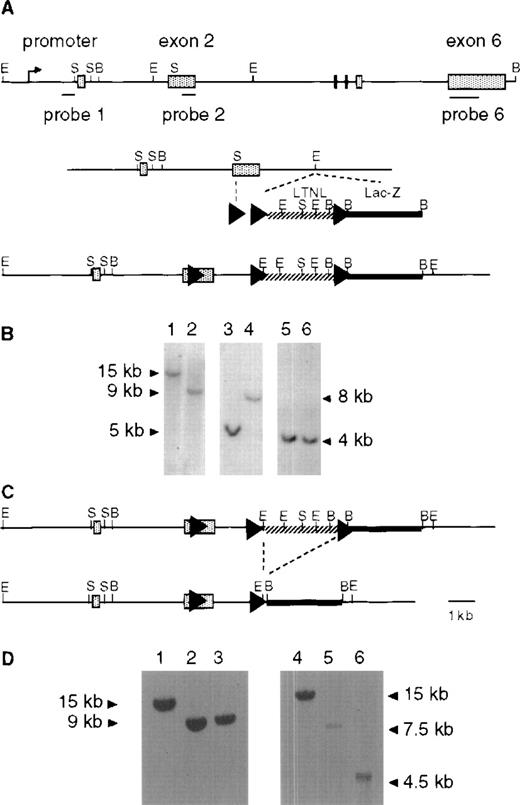
To enable Cre-mediated recombination to take place at the Pigalocus, we introduced 3 loxP sites (Fig 1A). In the replacement vectorlox-Piga-LTNL-lacZ, a 5′ loxP sequence was inserted into exon 2, downstream of the first AUG. This insertion extends the Piga open reading frame by 12 amino acids and does not interfere with PIGA function in vivo (see below). Two additional loxP sequences were introduced, flanking a positive/negative LTNL selection cassette. Downstream of the selection cassette we placed the coding region of the lacZ gene with a nuclear localization signal but without the translation initiating ATG. Cre mediated recombination of the 5′ and the 3′loxP sites will generate a fusion gene between Piga andlacZ. This recombination will excise a large portion ofPiga exon 2 resulting in complete loss of PIGA function.14
Generation of lox-Piga-lacZ ES cells. (A) Top panel shows the genomic structure of the mouse Piga gene. The internal probes used for Southern blot analysis map to exon 1 (probe 1) and to exon 2 (probe 2), and the external probe maps to exon 6 (probe 6). Middle panel displays the targeting construct (lox-Piga-LTNL-lacZ.). Bottom panel shows thePiga gene after homologous recombination (lox-LTNL-Piga-lacZ). Shaded boxes represent exon 1-6, arrowheads represent loxP sites, hatched line represents the selectable marker cassette and the bold black line represents the lacZ gene. LTNL = loxPthymidine kinase, neomycin resistance geneloxP. Restriction enzyme sites S=SmaI, B=BamHI, E=EcoRI. (B) Southern blot of homologous recombined ES clone 1G2. Lanes 1, 3, and 5 represent DNA from wild-type ES cells and lanes 2, 4, and 6 represent DNA from recombined ES clone 1G2. Lanes 1 and 2 were digested with BamHI and hybridized with probe 6. DNA in lanes 3 and 4 was digested with EcoRI and hybridized with probe 1. DNA in lanes 5 and 6 was digested withBamHI and hybridized with probe 1. Because Piga maps to the X chromosome and the ES cells are of male origin, no other allele remains. (C) Top panel shows thelox-LTNL-Piga-lacZ gene. Bottom panel shows thelox-Piga-lacZ gene after Cre-mediated excision of the selectable marker cassette LTNL. Dashed line represents the desired Cre-mediated excision. (D) Southern blot of the desired Cre-mediated recombination. Lanes 1 and 4 represent DNA from wild-type ES cells, lanes 2 and 5 represent DNA from recombined ES clone 1G2, and lanes 3 and 6 represent DNA from Cre recombined ES clone 1G2/87. DNA in lanes 1-3 was hybridized with probe 6; DNA in lanes 4-6 was hybridized with probe 2; DNA in lanes 1-6 was digested with BamHI.
Generation of lox-Piga-lacZ ES cells. (A) Top panel shows the genomic structure of the mouse Piga gene. The internal probes used for Southern blot analysis map to exon 1 (probe 1) and to exon 2 (probe 2), and the external probe maps to exon 6 (probe 6). Middle panel displays the targeting construct (lox-Piga-LTNL-lacZ.). Bottom panel shows thePiga gene after homologous recombination (lox-LTNL-Piga-lacZ). Shaded boxes represent exon 1-6, arrowheads represent loxP sites, hatched line represents the selectable marker cassette and the bold black line represents the lacZ gene. LTNL = loxPthymidine kinase, neomycin resistance geneloxP. Restriction enzyme sites S=SmaI, B=BamHI, E=EcoRI. (B) Southern blot of homologous recombined ES clone 1G2. Lanes 1, 3, and 5 represent DNA from wild-type ES cells and lanes 2, 4, and 6 represent DNA from recombined ES clone 1G2. Lanes 1 and 2 were digested with BamHI and hybridized with probe 6. DNA in lanes 3 and 4 was digested with EcoRI and hybridized with probe 1. DNA in lanes 5 and 6 was digested withBamHI and hybridized with probe 1. Because Piga maps to the X chromosome and the ES cells are of male origin, no other allele remains. (C) Top panel shows thelox-LTNL-Piga-lacZ gene. Bottom panel shows thelox-Piga-lacZ gene after Cre-mediated excision of the selectable marker cassette LTNL. Dashed line represents the desired Cre-mediated excision. (D) Southern blot of the desired Cre-mediated recombination. Lanes 1 and 4 represent DNA from wild-type ES cells, lanes 2 and 5 represent DNA from recombined ES clone 1G2, and lanes 3 and 6 represent DNA from Cre recombined ES clone 1G2/87. DNA in lanes 1-3 was hybridized with probe 6; DNA in lanes 4-6 was hybridized with probe 2; DNA in lanes 1-6 was digested with BamHI.
We next proceeded to generate lox-Piga-lacZ ES cells according to the 2-step technique described by Gu et al30in 1994. In a first step, the replacement vectorlox-Piga-LTNL-lacZ was introduced into the Pigalocus by homologous recombination (Fig 1A and B). In a second step, the selection cassette was removed by transient expression of the Cre recombinase in targeted ES cells (Fig 1C and D).
For the generation of chimeras, 2 ES cell clones were injected into blastocysts. Germ line transmission was obtained from clone 1G2/87. In mice as in humans Piga maps to the X chromosome,31 32 therefore lox-Piga-lacZ shows an X-linked transmission pattern. No hematological or developmental abnormalities were seen in homozygous females and in hemizygous males (data not shown), confirming that the introduction of the loxPsites and of the lacZ gene does not interfere with the normal function of the Piga gene in vivo.
Generation of mosaic mice.
Cre mediated recombination was obtained by crossbreeding thelox-Piga-lacZ mice with transgenic mice that express Cre under the adenoviral EIIa promoter. Figure 2summarizes the factors that influence the extent of mosaicism in the offspring of the 2 possible breeding pair. In the absence of its natural transactivating coactivator E1A, the promoter of the adenovirus EIIa gene is thought to be active only in oocytes and in preimplantation embryos.17 In the offspring of the EIIa-cre x lox-Piga-lacZ intercross, we, therefore, expect that recombination of the lox-Piga-lacZ gene will only take place early in embryogenesis, just after fertilization, and will cease after implantation around day E4.5. The possibility that Cre is expressed in bone marrow cells seems unlikely, because we failed to detect Cre transcripts by RT-PCR analysis of bone marrow RNA from EIIa-cre (+/+) transgenic mice (see Fig 3).
Choice of the parent husbandry and sex of the offspring influence the extent of blood mosaicism [proportion of PIGA(−) cells] in lox-Piga-lacZ x EIIa-cre offspring: Thelox-Piga-lacZ gene follows an X-linked inheritance pattern. Male offspring of the EL breeding therefore do not inherit alox-Piga-lacZ gene and therefore do not undergo Pigagene recombination. The expression of Cre determines the time span during which Piga gene recombination may occur. If maternally derived, EIIa promoter activity starts already in the oocyte and ceases around day E4.5. In contrast, if paternally derived, Cre expression starts at the time when male gene expression is initiated which is around E2.5. The contribution of PIGA(−) cells to hematopoiesis depends on the contribution of PIGA(−) cells to the stem cell pool. Male PIGA(−) cells are subject to early negative selection.43 In female cells however, the PIGA(−) phenotype is only expressed after the wild-type Piga gene has been inactivated by X chromosome inactivation. Selection against PIGA(−) cells starts therefore later in ontogenesis. This explains the higher contribution of PIGA(−) hematopoietic cells found in female mice compared to male mice from the same breeding and compared to the PIGA(−) cell contribution previously reported in chimeric mice obtained after injection of PIGA(−) XY ES cells.13 14 For the LE breeding only the offspring carrying a lox-Piga-lacZgene are illustrated. The genotype is shown in italics, pindicates paternally derived, m maternally derived genes.
Choice of the parent husbandry and sex of the offspring influence the extent of blood mosaicism [proportion of PIGA(−) cells] in lox-Piga-lacZ x EIIa-cre offspring: Thelox-Piga-lacZ gene follows an X-linked inheritance pattern. Male offspring of the EL breeding therefore do not inherit alox-Piga-lacZ gene and therefore do not undergo Pigagene recombination. The expression of Cre determines the time span during which Piga gene recombination may occur. If maternally derived, EIIa promoter activity starts already in the oocyte and ceases around day E4.5. In contrast, if paternally derived, Cre expression starts at the time when male gene expression is initiated which is around E2.5. The contribution of PIGA(−) cells to hematopoiesis depends on the contribution of PIGA(−) cells to the stem cell pool. Male PIGA(−) cells are subject to early negative selection.43 In female cells however, the PIGA(−) phenotype is only expressed after the wild-type Piga gene has been inactivated by X chromosome inactivation. Selection against PIGA(−) cells starts therefore later in ontogenesis. This explains the higher contribution of PIGA(−) hematopoietic cells found in female mice compared to male mice from the same breeding and compared to the PIGA(−) cell contribution previously reported in chimeric mice obtained after injection of PIGA(−) XY ES cells.13 14 For the LE breeding only the offspring carrying a lox-Piga-lacZgene are illustrated. The genotype is shown in italics, pindicates paternally derived, m maternally derived genes.
Expression of Cre in EIIa-cre transgenic mice by RT-PCR. Lanes 1-6, cDNA amplified with control primer pairSacII/293. Lanes 7-12, cDNA amplified with primer pair cre1/cre2. Lanes 2, 4, 6, 8, 10, 12 amplified in the presence of RT. Lanes 1, 3, 5, 7, 9, 11 controls amplified without RT. Lanes 1, 2, 7, 8 RNA isolated from leg muscle. Lanes 3, 4, 9, 10 RNA isolated from ovaries. Lanes 5, 6, 11, 12 RNA isolated from bone marrow. Lane 13 molecular marker. Note that primer pair cre1/cre2 amplifies a band of 520 bp and control primer pair SacII/293 amplifies a band of 268 bp.
Expression of Cre in EIIa-cre transgenic mice by RT-PCR. Lanes 1-6, cDNA amplified with control primer pairSacII/293. Lanes 7-12, cDNA amplified with primer pair cre1/cre2. Lanes 2, 4, 6, 8, 10, 12 amplified in the presence of RT. Lanes 1, 3, 5, 7, 9, 11 controls amplified without RT. Lanes 1, 2, 7, 8 RNA isolated from leg muscle. Lanes 3, 4, 9, 10 RNA isolated from ovaries. Lanes 5, 6, 11, 12 RNA isolated from bone marrow. Lane 13 molecular marker. Note that primer pair cre1/cre2 amplifies a band of 520 bp and control primer pair SacII/293 amplifies a band of 268 bp.
Male mice hemizygous for lox-Piga-lacZ(+), or heterozygouslox-Piga-lacZ(+/−) females, were crossbred with homozygous EIIa-cre(+/+) mice. Of 185 newborn mice, 67 carried a lox-Piga-lacZ and an EIIa-cre gene. Fifty-five animals had more than 2% CD24(−) erythrocytes. The number of PIGA(−) blood cells in newborn mice was dependent on the breeding and on the sex of the offspring (see Fig 2). Female offspring of the LE breeding (LE; see Fig 2) had a higher contribution of PIGA(−) RBCs at birth (median 8.5%) than male offspring from the same breeding (median 3.5). The highest contribution of PIGA(−) cells (up to 53%) was observed in offspring with a paternally derivedlox-Piga-lacZ gene and a maternally derived EIIa-cregene (offspring of the EL breeding, genotype;lox-Piga-lacZ(−/+p); EIIa-cre(+m/−); Fig 4 and Table1). Offspring of the EL breeding had a higher perinatal lethality (13.4% v 3.7%) and a clearly biased sex distribution in favor of male newborns not carrying a lox-Piga-lacZ allele (male/female = 1.8). This suggests that very high levels ofPiga gene recombination may occur more frequently in EL offspring but are associated with an increased intrauterine and perinatal lethality.
Proportion of CD24-deficient RBCs in male and female newborn mice obtained from the 2 different breedings. Fifty-five animals that carry both the lox-Piga-lacZ and the EIIa-cregene are shown. (The analysis of the first 12 animals [11 EL females and 1 LE female] were excluded because of initial problems caused by clumping of CD24 positive cells during the staining procedure.) Four male mice that did not inherit thelox-Piga-lacZ gene served as negative controls. Each box plot displays the 10th, 25th, 50th, 75th, and 90th percentile. The median is indicated. Values above the 90th or below the 10th percentile are plotted.
Proportion of CD24-deficient RBCs in male and female newborn mice obtained from the 2 different breedings. Fifty-five animals that carry both the lox-Piga-lacZ and the EIIa-cregene are shown. (The analysis of the first 12 animals [11 EL females and 1 LE female] were excluded because of initial problems caused by clumping of CD24 positive cells during the staining procedure.) Four male mice that did not inherit thelox-Piga-lacZ gene served as negative controls. Each box plot displays the 10th, 25th, 50th, 75th, and 90th percentile. The median is indicated. Values above the 90th or below the 10th percentile are plotted.
Genotype and Blood Phenotype of Offspring From Lox-Piga-lacZ X EIIa-cre Breeding
| Breeding Parents . | Mother Father . | EL . | LE . |
|---|---|---|---|
| EIIa-cre(+/+) Lox-Piga-LacZ (+) . | Lox-Piga-lacZ(+/−) Ella-cre(+/+) . | ||
| No. of litters | 16 | 8 | |
| No. of pups | 156 | 54 | |
| No. of pups alive after 48 h | 132 | 53 | |
| Male:female ratio | 1.8 | 0.86 | |
| Males: | Total | 85 (64%) | 25 (47%) |
| Lox-Piga-lacZ(+)EIIa-cre(−/+) genotype | 0 | 10 (19%) | |
| >2% PIGA(−) RBCs* | NA | 10 (19%) | |
| >5% PIGA(−) white cells† | NA | 3 (6%) | |
| Females: | Total | 47 (36%) | 28 (53%) |
| Lox-Piga-lacZ(+/−)EIIa-cre(−/+) genotype | 47 (36%) | 10 (19%) | |
| >2% PIGA(−) RBCs* | 35 (27%) | 10 (19%) | |
| >5% PIGA(−) white cells† | 28 (21%) | 8 (15%) | |
| Breeding Parents . | Mother Father . | EL . | LE . |
|---|---|---|---|
| EIIa-cre(+/+) Lox-Piga-LacZ (+) . | Lox-Piga-lacZ(+/−) Ella-cre(+/+) . | ||
| No. of litters | 16 | 8 | |
| No. of pups | 156 | 54 | |
| No. of pups alive after 48 h | 132 | 53 | |
| Male:female ratio | 1.8 | 0.86 | |
| Males: | Total | 85 (64%) | 25 (47%) |
| Lox-Piga-lacZ(+)EIIa-cre(−/+) genotype | 0 | 10 (19%) | |
| >2% PIGA(−) RBCs* | NA | 10 (19%) | |
| >5% PIGA(−) white cells† | NA | 3 (6%) | |
| Females: | Total | 47 (36%) | 28 (53%) |
| Lox-Piga-lacZ(+/−)EIIa-cre(−/+) genotype | 47 (36%) | 10 (19%) | |
| >2% PIGA(−) RBCs* | 35 (27%) | 10 (19%) | |
| >5% PIGA(−) white cells† | 28 (21%) | 8 (15%) | |
Numbers in parentheses refer to the percentage of pups alive after 48 hours.
Abbreviation: NA, not applicable.
At age of 24-48 h.
At the age of 4 weeks.
PIGA(−) blood cells in adult mice mosaic for a nonfunctional Piga gene.
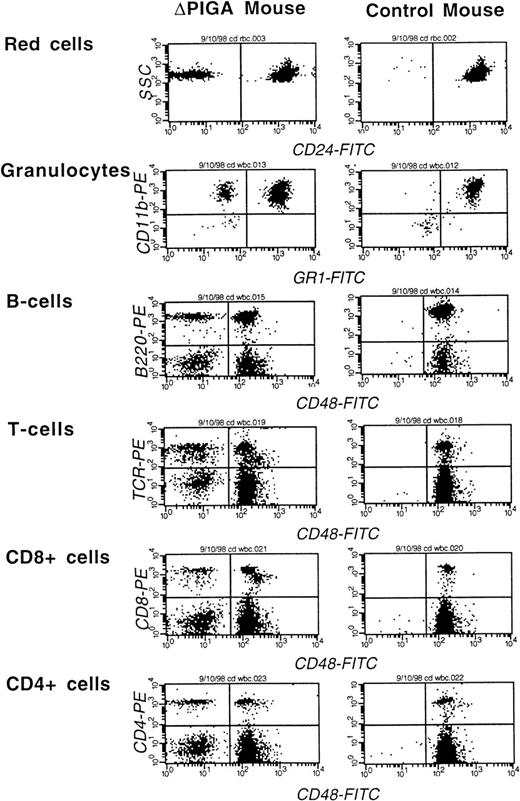
Thirty-nine animals with a RBC contribution of PIGA(−) cells greater than 2% and a white blood cell contribution greater than 5% at the age of 4 weeks were followed for an observation period of 8 to 12 months. During the observation period, 3 animals died. In 2 animals the cause of death was most likely related to the anesthesia during the bleeding procedure. One mouse developed a metastatic tumor of the uterus at the age of 1 year (data not shown). The contribution of PIGA(−) cells to the different blood cell lineages in peripheral blood was variable. A representative example of a FCA is shown in Fig 5. Animals with a high proportion of PIGA(−) cells in one blood cell lineage had usually also a high contribution of PIGA(−) cells in all other blood cell lineages. The magnitude of differences in the proportion of PIGA(−) cells was dependent on the cell type (Fig 6A and B).
FCA of peripheral blood cells of a mouse mosaic forlox-▵Piga-lacZ (▵PIGA) and a normal age matched control mouse. Monoclonal antibodies against CD24, Gr−1, and CD48 were used to determine the proportion of PIGA(−) cells in individual blood cell lineages (see Materials and Methods). The left upper quadrant displays the proportion of PIGA(−) cells in the individual cell lineage. Proportion of PIGA(−) cells in the ▵PIGA mouse and values of the control mouse in parenthesis: RBCs: 10% {0%}, granulocytes: 22% {1%}, B cells 15% {0%}, T cells: 35% {0%}, CD8+: 31% {0%}, CD4+: 33% {0%}.
FCA of peripheral blood cells of a mouse mosaic forlox-▵Piga-lacZ (▵PIGA) and a normal age matched control mouse. Monoclonal antibodies against CD24, Gr−1, and CD48 were used to determine the proportion of PIGA(−) cells in individual blood cell lineages (see Materials and Methods). The left upper quadrant displays the proportion of PIGA(−) cells in the individual cell lineage. Proportion of PIGA(−) cells in the ▵PIGA mouse and values of the control mouse in parenthesis: RBCs: 10% {0%}, granulocytes: 22% {1%}, B cells 15% {0%}, T cells: 35% {0%}, CD8+: 31% {0%}, CD4+: 33% {0%}.
Proportion of PIGA(−) cells in peripheral blood from mosaic mice. (A) Proportion of PIGA(−) RBCs during the observation period of 12 months. (B) Proportion of PIGA(−) granulocytes (PMN), T cells, B cells, CD8+, and CD4+ cells during the observation period of 12 months. Each box plot displays the 10th, 25th, 50th, 75th, and 90th percentile. Values above the 90th or below the 10th percentile are plotted [Table 2 shows the values for median, the number of animals analyzed, and the level of significance of the difference in the percentage of PIGA(−) cells between 2 successive time points]. (C) Proportion of PIGA(−) reticulocytes in relation to the proportion of PIGA(−) RBCs, granulocytes (PMN), and lymphocytes (Lyc) in 11 animals more than 16 weeks of age. The values for the median are indicated and shown by a horizontal line.
Proportion of PIGA(−) cells in peripheral blood from mosaic mice. (A) Proportion of PIGA(−) RBCs during the observation period of 12 months. (B) Proportion of PIGA(−) granulocytes (PMN), T cells, B cells, CD8+, and CD4+ cells during the observation period of 12 months. Each box plot displays the 10th, 25th, 50th, 75th, and 90th percentile. Values above the 90th or below the 10th percentile are plotted [Table 2 shows the values for median, the number of animals analyzed, and the level of significance of the difference in the percentage of PIGA(−) cells between 2 successive time points]. (C) Proportion of PIGA(−) reticulocytes in relation to the proportion of PIGA(−) RBCs, granulocytes (PMN), and lymphocytes (Lyc) in 11 animals more than 16 weeks of age. The values for the median are indicated and shown by a horizontal line.
The proportion of PIGA(−) RBCs was highest after birth reaching 53%. Within the first 4 weeks of life, the proportion of PIGA(−) RBCs dropped drastically and continued to drop slowly thereafter, reaching a plateau at the age of 16 weeks of around 4% (Fig 6A and Table 2). After 16 weeks, the proportion of PIGA(−) RBCs did not change significantly.
Proportion of PIGA(−) Blood Cells in Peripheral Blood in PIGA(−) Mosaic Mice
| Age (wk) . | PIGA(−) RBC % (n) . | PIGA(−) PMN % (n) . | PIGA(−) T Cells % (n) . | PIGA(−) B Cells % (n) . | PIGA(−) CD4 Cells % (n) . | PIGA(−) CD8 Cells % (n) . |
|---|---|---|---|---|---|---|
| Birth | 17.8 (32†) | ND | ND | ND | ND | ND |
| 4 | 8.4 (19†) | 12.2 (37) | 16.3 (7‡) | 6 (7‡) | ND | ND |
| P < .05 | ||||||
| 16 | 4 (39) | 11.2 (39) | 18 (39) | 8.6 (39) | 16.8 (37) | 16.9 (39) |
| P = NS | P = NS | P = NS | P < .05 | |||
| 32 | 4.1 (38*) | 10.5 (37) | 18.6 (37) | 11 (37) | 13.3 (37) | 20.1 (37) |
| P = NS | P = NS | P = NS | P < .05 | P < .05 | P < .05 | |
| 52 | 3 (37*) | 10.5 (37) | 18.8 (37) | 7.6 (35) | 13.5 (37) | 24.5 (36) |
| P = NS | P = NS | P = NS | P < .05 | P < .05 | P < .05 |
| Age (wk) . | PIGA(−) RBC % (n) . | PIGA(−) PMN % (n) . | PIGA(−) T Cells % (n) . | PIGA(−) B Cells % (n) . | PIGA(−) CD4 Cells % (n) . | PIGA(−) CD8 Cells % (n) . |
|---|---|---|---|---|---|---|
| Birth | 17.8 (32†) | ND | ND | ND | ND | ND |
| 4 | 8.4 (19†) | 12.2 (37) | 16.3 (7‡) | 6 (7‡) | ND | ND |
| P < .05 | ||||||
| 16 | 4 (39) | 11.2 (39) | 18 (39) | 8.6 (39) | 16.8 (37) | 16.9 (39) |
| P = NS | P = NS | P = NS | P < .05 | |||
| 32 | 4.1 (38*) | 10.5 (37) | 18.6 (37) | 11 (37) | 13.3 (37) | 20.1 (37) |
| P = NS | P = NS | P = NS | P < .05 | P < .05 | P < .05 | |
| 52 | 3 (37*) | 10.5 (37) | 18.8 (37) | 7.6 (35) | 13.5 (37) | 24.5 (36) |
| P = NS | P = NS | P = NS | P < .05 | P < .05 | P < .05 |
Numbers represent the median of the proportion of PNH cells within the subpopulation of cells at the indicated age.
Abbreviations: n, number of mice analyzed; NS, not significant; ND, not done.
P < .05; in comparison to the previous analysis values are statistically significant different (paired Ttest).
Mouse died under anesthesia.
Because of RBC clumping, initial RBC analysis was discarded.
Lymphocyte subset analysis for the majority of mice was started at the age of 16 weeks (B cells and T cells).
Amongst white blood cells, the proportion of PIGA(−) cells was highest for the T cells reaching a maximum of 40% and lowest for the B cells with a maximum of 21%. The highest proportion of PIGA(−) granulocytes observed was 29% (Fig 6B and Table 2). In contrast to the RBCs within the granulocytes and T cells, the proportion of PIGA(−) cells was stable and did not change significantly during the observation period. Within the B cells an initial small increase of the proportion of PIGA(−) B cells was observed up to 8 months, followed by a decrease at the age of 1 year. Within the T-cell subpopulation (CD4+ and CD8+ lymphocytes), the proportion of PIGA(−) CD4+ cells decreased slowly, whereas the proportion of PIGA(−) CD8+ increased during the observation period of 1 year. It remains uncertain if the changes observed within the B cells and the T-cell subpopulations are of biologic significance.
Peripheral blood cell counts in mice mosaic for PIGA(−) cells.
Peripheral blood cell counts including reticulocyte counts and white blood cell differentials were obtained from mosaic mice and age-matched controls. At all times, blood cell counts from mosaic mice did not differ from blood cell counts of age-matched control mice. In particular, there was no anemia, cytopenia, or reticulocytosis (Table 3). Red and white blood cell morphologies were normal, as determined by visual examination of May-Grünwald-Giemsa–stained blood films. Hemoglobinuria was not observed. There was one salient exception: mosaic mouse 406.2 showed an unexplained transient leukocytosis of 23.3 ×103/ μL at the age of 12 months (see Table 3). There were no signs of infection or other physical findings that would have explained the leukocytosis in this animal. The proportion of PNH cells in the individual blood cell lineages did not change.
Hemoglobin, Neutrophils, Platelet, and Reticulocyte Counts of Lox-▵Piga-lacZ and Wild-Type Mice
| Age . | Lox-ΔPiga-LacZ . | Wild Type . | ||||||
|---|---|---|---|---|---|---|---|---|
| Hb . | PMN . | Plt . | Retic . | Hb . | PMN . | Plt . | Retic . | |
| wk | g/dL | 103/μL | 103/μL | % | g/dL | 103/μL | 103/μL | % |
| 4 | 14.3 ± 0.5 | ND | ND | ND | 14.5 ± 0.5 | ND | ND | ND |
| 16 | 15.2 ± 1.2 | 1.2 ± 0.3 | 1,291 ± 188 | 5.7 ± 1.4 | 15 ± 1.7 | 1.4 ± 4 | 1,218 ± 202 | 4.7 ± 1.4 |
| 32 | 14.9 ± 1 | 1.2 ± 0.5 | 1,303 ± 300 | 6.6 ± 1.9 | 14.4 ± 3 | 1.5 ± 0.9 | 1,342 ± 272 | 6.2 ± 1.3 |
| 52 | 15 ± 1.4 | 1.8 ± 1.13-150 | 1,524 ± 269 | 6.2 ± 1.7 | 1.8 ± 2.2 | 1.2 ± 0.3 | 1,562 ± 246 | 7.6 ± 1.5 |
| Age . | Lox-ΔPiga-LacZ . | Wild Type . | ||||||
|---|---|---|---|---|---|---|---|---|
| Hb . | PMN . | Plt . | Retic . | Hb . | PMN . | Plt . | Retic . | |
| wk | g/dL | 103/μL | 103/μL | % | g/dL | 103/μL | 103/μL | % |
| 4 | 14.3 ± 0.5 | ND | ND | ND | 14.5 ± 0.5 | ND | ND | ND |
| 16 | 15.2 ± 1.2 | 1.2 ± 0.3 | 1,291 ± 188 | 5.7 ± 1.4 | 15 ± 1.7 | 1.4 ± 4 | 1,218 ± 202 | 4.7 ± 1.4 |
| 32 | 14.9 ± 1 | 1.2 ± 0.5 | 1,303 ± 300 | 6.6 ± 1.9 | 14.4 ± 3 | 1.5 ± 0.9 | 1,342 ± 272 | 6.2 ± 1.3 |
| 52 | 15 ± 1.4 | 1.8 ± 1.13-150 | 1,524 ± 269 | 6.2 ± 1.7 | 1.8 ± 2.2 | 1.2 ± 0.3 | 1,562 ± 246 | 7.6 ± 1.5 |
Each value represents mean ± SD obtained from mosaic (n = see Table 2) and control mice (n = 12) at the indicated age.
Abbreviations: Hb, hemoglobin; PMN, neutrophils; Plt, platelets; Retic, reticulocytes; ND, not done; n, number of mice.
Mosaic mouse 406.2 showed a transient leukocytosis of 23.3 × 103/μL (PMN 6.1; lymphocytes 14.9 × 103/μL) and Plts of 945 × 103/μL.
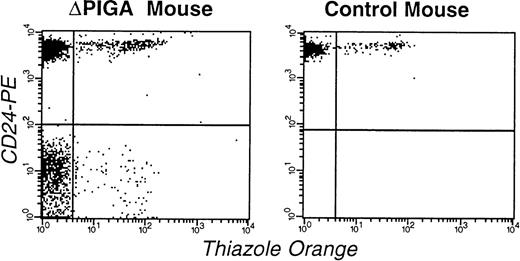
Half-life of PIGA(−) RBCs.
To further investigate the decrease of PIGA(−) red blood cells after birth we analyzed in 11 mice the proportion of circulating PIGA(−) reticulocytes (Fig 7) and compared it with the proportion of PIGA(−) RBCs, granulocytes, and lymphocytes within the same animal. In all 11 animals, the proportion of PIGA(−) reticulocytes was significantly higher (median 18.2%, range 7.9 to 27.6, P < .05) than the proportion of PIGA(−) RBCs (median 9.5%, range 5.5 to 21.3) but not significantly different from the number of PIGA(−) granulocytes (median 18.3%, range 12.4 to 26.7, P = 0.6) from the same animal (Fig 6C and Table 4). The mean calculated half-life of PIGA(−) RBCs was 7.3 days and ranged from 5.3 to 11.9 (Table 4), which is significantly lower than the half-life of normal mouse RBCs of 15 days.25
FCA of RBCs and reticulocytes in a mouse mosaic forlox-▵Piga-lacZ (▵PIGA, mouse I.D. 3.1; Table 4) and a normal age matched control mouse. RBCs were stained with thiazole orange and PE-conjugated anti-CD24 antibodies. The left upper quadrant shows the proportion of mature normal RBCs (83%), the left lower quadrant corresponds to the mature PIGA(−) cells (11%), right upper quadrant normal reticulocytes (4%), and the right lower quadrant displays PIGA(−) reticulocytes (1%).
FCA of RBCs and reticulocytes in a mouse mosaic forlox-▵Piga-lacZ (▵PIGA, mouse I.D. 3.1; Table 4) and a normal age matched control mouse. RBCs were stained with thiazole orange and PE-conjugated anti-CD24 antibodies. The left upper quadrant shows the proportion of mature normal RBCs (83%), the left lower quadrant corresponds to the mature PIGA(−) cells (11%), right upper quadrant normal reticulocytes (4%), and the right lower quadrant displays PIGA(−) reticulocytes (1%).
Percentage of Reticulocytes Within PIGA(+) and PIGA(−) RBCs in Lox-▵Piga-lacZ Mice and the Calculated Half-Life of PIGA(−) RBCs
| Mouse ID . | PIGA(−) RBC % . | % Retic Within PIGA(+) RBCs . | % Retic Within PIGA(−) RBCs . | t1/2 of PIGA(−) RBCs (d) . |
|---|---|---|---|---|
| 1.3 | 12.2 | 4.8 | 8.8 | 8.2 |
| 12.21 | 9.5 | 3.9 | 8.9 | 6.6 |
| 15.1 | 9.5 | 4.6 | 13.1 | 5.3 |
| 406.5 | 13.1 | 4.5 | 7.9 | 8.5 |
| 406.12 | 10.1 | 6.1 | 15.1 | 6.1 |
| 20.4 | 8.6 | 3.9 | 10 | 5.9 |
| 28.5 | 7.1 | 3.4 | 4.6 | 11.1 |
| 6.1 | 21.3 | 6.1 | 7.7 | 11.9 |
| 12.2 | 5.5 | 6 | 14.6 | 6.2 |
| 11.2 | 6.4 | 3 | 7.6 | 5.9 |
| 12.11 | 7.6 | 4.6 | 13 | 5.3 |
| Mean | 10.1 | 4.6 | 10.1 | 7.3 |
| Controls | t1/2 of PIGA(+) RBC | |||
| C 1.45 | 0.1 | 5.1 | 0 | 154-150 |
| C 15 | 0 | 3.9 | 0 | 154-150 |
| Mouse ID . | PIGA(−) RBC % . | % Retic Within PIGA(+) RBCs . | % Retic Within PIGA(−) RBCs . | t1/2 of PIGA(−) RBCs (d) . |
|---|---|---|---|---|
| 1.3 | 12.2 | 4.8 | 8.8 | 8.2 |
| 12.21 | 9.5 | 3.9 | 8.9 | 6.6 |
| 15.1 | 9.5 | 4.6 | 13.1 | 5.3 |
| 406.5 | 13.1 | 4.5 | 7.9 | 8.5 |
| 406.12 | 10.1 | 6.1 | 15.1 | 6.1 |
| 20.4 | 8.6 | 3.9 | 10 | 5.9 |
| 28.5 | 7.1 | 3.4 | 4.6 | 11.1 |
| 6.1 | 21.3 | 6.1 | 7.7 | 11.9 |
| 12.2 | 5.5 | 6 | 14.6 | 6.2 |
| 11.2 | 6.4 | 3 | 7.6 | 5.9 |
| 12.11 | 7.6 | 4.6 | 13 | 5.3 |
| Mean | 10.1 | 4.6 | 10.1 | 7.3 |
| Controls | t1/2 of PIGA(+) RBC | |||
| C 1.45 | 0.1 | 5.1 | 0 | 154-150 |
| C 15 | 0 | 3.9 | 0 | 154-150 |
Abbreviations: PMN, granulocytes; retic, reticulocytes.
t1/2 of normal murine RBCs = 15 days.25
Complement sensitivity of PIGA(−) RBCs from mice mosaic for lox-ΔPiga-LacZ.
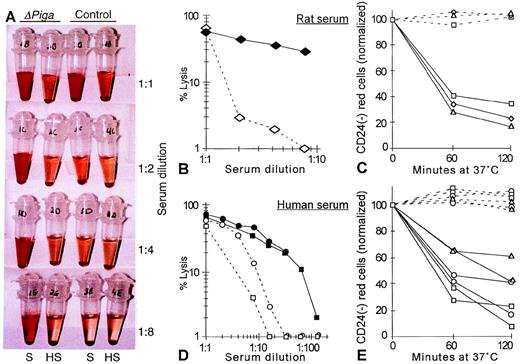
To determine whether the decreased life span of PIGA(−) RBCs observed in our mosaic mice could be caused by complement mediated intravascular lysis we investigated if the PIGA(−) RBCs have an increased sensitivity toward complement (Fig 8A). As mouse serum has previously been shown to have a poor lytic activity in vitro27 and our mosaic mice have a mixed genetic background, we chose rat and human serum as the source of complement.27 28 Serial dilutions of the serum was used to titrate complement activity. Exposed to acidified serum at 37oC, RBCs from lox-ΔPiga-lacZ mosaic mice showed a substantially increased susceptibility toward the lytic action of activated complement when compared with RBCs obtained from a normal control animal (Fig 8A, B, and D). Lysis was complement dependent as it only occurred in samples with fresh serum but not in samples with heat inactivated serum. Complement dependent lysis of PIGA(−) RBCs in mosaic animals was assessed by flow cytometry. After 1 hour of incubation in either rat or human serum, a significant decrease of the proportion of PIGA(−) RBCs was noted, indicating that PIGA(−) RBCs are preferentially lysed after complement activation (see Fig 8C and 8E).
Complement sensitivity of PIGA(−) RBCs. (A) Sensitivity toward complement mediated lysis. Lysis of RBCs from mosaic mice (▵PIGA) and from normal control mice were exposed to different dilutions of acidified absorbed rat serum (S) or acidified absorbed rat serum after heat inactivation (HS; see Materials and Methods) for 60 minutes at 37oC. The supernatant shows the degree of hemolysis. (B, D) Lysis of RBCs from mosaic mice (filled symbols) and from normal control mice (empty symbols) at different serum dilutions from rat and (B) human (D). (C, E) Preferential lysis of PIGA(−) RBCs by rat and (C) human serum (E) at serum dilutions that lyse only blood cells from mosaic mice but not from normal control mice (full lines). Dotted lines show the percentage of PIGA(−) RBCs exposed to heat inactivated serum. Serum dilutions shown in C: triangle 1:2, rectangle: 1:4, squares: 1:8; in E: rectangle: 1:4, squares: 1:8, circles: 1:16, and triangles: 1:32.
Complement sensitivity of PIGA(−) RBCs. (A) Sensitivity toward complement mediated lysis. Lysis of RBCs from mosaic mice (▵PIGA) and from normal control mice were exposed to different dilutions of acidified absorbed rat serum (S) or acidified absorbed rat serum after heat inactivation (HS; see Materials and Methods) for 60 minutes at 37oC. The supernatant shows the degree of hemolysis. (B, D) Lysis of RBCs from mosaic mice (filled symbols) and from normal control mice (empty symbols) at different serum dilutions from rat and (B) human (D). (C, E) Preferential lysis of PIGA(−) RBCs by rat and (C) human serum (E) at serum dilutions that lyse only blood cells from mosaic mice but not from normal control mice (full lines). Dotted lines show the percentage of PIGA(−) RBCs exposed to heat inactivated serum. Serum dilutions shown in C: triangle 1:2, rectangle: 1:4, squares: 1:8; in E: rectangle: 1:4, squares: 1:8, circles: 1:16, and triangles: 1:32.
DISCUSSION
The molecular basis of the human disease PNH has been elucidated,33-35 but we still need to explain how one or few PNH clones virtually take over hematopoiesis in PNH patients. For this purpose a mouse model would be extremely useful. Because Pigainactivation is embryonic lethal, previous attempts had to depend on the laborious task of producing chimeras.13,14 The mouse model we have reported here has 4 distinct advantages: (1) By matinglox-Piga-lacZ with EIIa-cre mice we can produce any desired number of animals that undergo Piga inactivation at the somatic cell level (as occurs in PNH patients). (2) Cre expression under the EIIa promoter is temporally restricted to early embryonic life and no Piga gene recombination occurs thereafter (see also Fig 3). This allows us to study growth and differentiation of hematopoietic stem cells that lack GPI-linked proteins. (3) In the previously reported chimeric mice, the PIGA(−) ES-cells were from strain 129, whereas the PIGA(+) host blastocysts were from strain C57BL/6.13,14 Therefore, in those animals, PIGA(+) and PIGA (−) hematopoiesis were genetically different.36,37 By contrast, in our loxP-ΔPiga-lacZ mosaic animals, all cells are genetically identical except for the Piga mutation. (4) In comparison to the chimeric mice13 14 the contribution of PIGA(−) cells to hematopoiesis is significantly higher (up to 30-fold).
Piga gene inactivation in lox-ΔPiga-lacZ mice.
Concurring with the “Ohno’s law”31 the mousePiga gene, like its human homologue, maps to the X chromosome at position F3-F4, which is syntenic to human Xp22.1.32 The presence of GPI-deficient cells in female mice heterozygous for the mutated Piga gene indicates that the murine Piga gene, like the human gene, is subject to X-chromosome inactivation. The fact that Piga is X-linked has been a distinct advantage in our work, because knocking out a single allele produces complete loss of gene product and in female offspring the PIGA(−) phenotype is only expressed after X-chromosome inactivation of the chromosome carrying the wild type Piga gene. The latter also accounts for the high contribution of PIGA(−) cells in female offspring (see also Figs 2 and 4 and Table 1).
Mice that are mosaic for a nonfunctional Piga gene have blood cells with the PNH phenotype.
In lox-ΔPiga-lacZ offspring some but not all of RBCs, granulocytes, T lymphocytes, and B lymphocytes are deficient in GPI-linked proteins. The extent of PIGA(−) cell contribution is dependent on the breeding and on the sex of the offspring (see above and Figs 2 and 4 and Table 1). The coexistence of normal cells and PIGA(−) blood cells mirrors the phenotype seen in patients with PNH and confirms that in mice, like in humans, PIGA(−) stem cells are able to contribute to hematopoiesis and to differentiate into mature blood cells of all lineages. The contribution of PIGA(−) cells to the peripheral blood was dependent on the blood cell lineage (see Fig 6A and B). No relative increase in PIGA(−) blood cells was observed in our mosaic mice over the observation period of 12 months. Because in our animals PIGA(+) and PIGA(−) cells are otherwise isogenic, the fact that PIGA(−) cells do not have a growth advantage proves conclusively that Piga inactivation is not sufficient to bring about any progressive expansion of PNH cells.
PIGA(−) RBCs have a reduced half-life and an increased sensitivity toward complement mediated lysis.
One of the classical clinical characteristics of patients with PNH is hemoglobinuria (as implied by the name of the disease). In humans, hemoglobinuria is caused by complement mediated intravascular hemolysis of PNH RBCs because of their lack of GPI-linked complement regulatory molecules (DAF and CD59). The half- life of PNH RBCs in circulation is reduced, which is reflected by a usually higher proportion of PNH reticulocytes compared with the proportion of mature PNH RBCs.38,39 The presence of RBCs with an increased sensitivity to lysis by activated complement was used for the diagnosis of PNH for many years. The various diagnostic tests mainly differ in their means of complement activation. The Ham test was the standard diagnostic test for PNH over half a century.5 It measures indirectly the functional consequences of the lack of complement regulatory proteins. In the Ham test PNH red blood cells, but not phenotypically normal RBCs, are lysed by the action of complement activated through the alternative pathway by lowering the serum pH5. Although in our mice hemoglobinuria was not observed and there were no obvious signs of hemolysis (Table 3), we found a reduced half-life of PIGA(−) RBCs (Fig 6C) and the in vitro sensitivity of PIGA(−) RBCs toward complement mediated lysis was increased (Fig 8). The defect in regulation of complement on the cell surface offers an elegant explanation of the decreased life span of PIGA(−) RBCs in vivo, suggesting that our mice may experience some degree of intravascular hemolysis, which might account for the comparably low contribution of PIGA(−) RBCs in peripheral blood and, at least in part, for the rapid decrease of PIGA(−) RBCs in newborn mosaic animals. The fact that laboratory mice live in an artificial clean environment, that the proportion of PIGA(−) RBCs in our mice is rather small, and that murine RBCs, in contrast to human RBCs, express, in addition to the GPI-linked CD55 and CD59 molecules, also a transmembrane molecule Crry/p65, which regulates complement activation,40 may explain why obvious signs of hemolysis such as hemoglobinuria, reticulocytosis, and anemia were not observed in mice with PIGA(−) RBCs. Although the increased sensitivity to complement is an attractive explanation for the reduced life span of PIGA(−) RBCs in vivo, we cannot exclude the possibility that other mechanisms, different from hemolysis, may be responsible for or contribute to the decreased half-life of PIGA(−) RBCs. An example could be the removal of RBCs lacking GPI-linked proteins by macrophages in the reticuloendothelial system.
Mice mosaic for a nonfunctional Piga gene; a mouse model of PNH.
Hematopoiesis of our mosaic mice resembles, in many ways, hematopoiesis seen in patients with a small established PNH clone. However, there are distinct interesting differences; for example in the contribution of PIGA(−) cells to the lymphoid lineage. Recent studies suggest that most patients with PNH have circulating PNH lymphocytes.41 However, the proportion of lymphocytes affected is always much smaller than the proportion of PNH granulocytes or RBCs. In contrast in our mosaic mice, the proportion of PIGA(−) cells was highest within the lymphocytes (sum of CD48-cells) and significantly lower within the granulocytes or RBCs. Moreover, in humans, the proportion of PNH B cells and T cells is variable,42 whereas in our mice the proportion of PIGA(−) B cells was always lower (about 50%) compared with the proportion of PIGA(−) T cells (Fig 6B). Of course we cannot exclude that some of these differences in PIGA(−) hematopoiesis between mice and humans are due to the overall contribution of PIGA(−) cells in our mosaic mice. However, studies on the contribution of PIGA(−) cells in various tissues of mosaic mice revealed a strong selection against PIGA(−) cells during embryogenesis in all tissues with the salient exception of hematopoietic cells.43 Nonetheless, transplantation experiments in a congenic background will be necessary to finally confirm that our findings are intrinsic to PIGA(−) hematopoiesis. The most important difference, however, is that, in our mice, because of the nature of how Piga gene recombination was obtained, hematopoiesis is mosaic and most likely derived from several PIGA(−) hematopoietic stem cells. But, in humans PNH hematopoiesis is clonal, caused by the expansion of one9 or a few12 early hematopoietic progenitor cells. The ability of the mutated progenitor cell to contribute equally to all blood cell lineages might be limited. Clonal expansion of PIGA(−) cells was not observed in our mice, suggesting that in humans in addition to thePIGA mutation a second factor is responsible for the PNH clone to expand and cause the disease of PNH.
The study of hematopoiesis after Cre mediated recombination in ourlox-ΔPiga-lacZ animals is a promising tool for defining these additional factors, which are crucial in the pathogenesis of PNH.
ACKNOWLEDGMENT
We thank Lucio Luzzatto for his generous support and helpful discussions. We are grateful to John P. Atkinson for his expertise in complement activation. We thank Dan Link, Rosario Notaro, Tassos Karadimitris, Doudja Nafa, David Araten for stimulating discussions, Philip J. Mason for reading the manuscript, Hubert Amrein and Andreas Kottman for providing the lacZ and Cre vectors, and Peter Mombaerts for the LTNL vector. CJ7 ES cells were obtained from Tom Gridley. EIIa-cre transgenic mice were kindly provided by Heiner Westphal.
Work at the Memorial Sloan-Kettering Cancer Center was supported by a National Institutes of Health (NIH) Grant No. RO1-HL-56678-01 to L. Luzzatto. Work at Washington University is supported by the McDonnell Foundation, Howard Hughes Medical Institute, Barnes Jewish Hospital Foundation, and Edward Mallinckrodt Foundation. P.P.P. is a Scholar of the Leukemia Society of America. P.K. is funded by the Herzog-Egli Stiftung. M.B. receives the Junior Faculty Award of the American Society of Hematology.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to M. Bessler, MD, PhD, Division of Hematology, Department of Medicine, Washington University School of Medicine, 660 S Euclid Ave, Campus Box 8125, St Louis, MO 63110; e-mail: mbessler@im.wustl.edu.

![Fig. 2. Choice of the parent husbandry and sex of the offspring influence the extent of blood mosaicism [proportion of PIGA(−) cells] in lox-Piga-lacZ x EIIa-cre offspring: Thelox-Piga-lacZ gene follows an X-linked inheritance pattern. Male offspring of the EL breeding therefore do not inherit alox-Piga-lacZ gene and therefore do not undergo Pigagene recombination. The expression of Cre determines the time span during which Piga gene recombination may occur. If maternally derived, EIIa promoter activity starts already in the oocyte and ceases around day E4.5. In contrast, if paternally derived, Cre expression starts at the time when male gene expression is initiated which is around E2.5. The contribution of PIGA(−) cells to hematopoiesis depends on the contribution of PIGA(−) cells to the stem cell pool. Male PIGA(−) cells are subject to early negative selection.43 In female cells however, the PIGA(−) phenotype is only expressed after the wild-type Piga gene has been inactivated by X chromosome inactivation. Selection against PIGA(−) cells starts therefore later in ontogenesis. This explains the higher contribution of PIGA(−) hematopoietic cells found in female mice compared to male mice from the same breeding and compared to the PIGA(−) cell contribution previously reported in chimeric mice obtained after injection of PIGA(−) XY ES cells.1314 For the LE breeding only the offspring carrying a lox-Piga-lacZgene are illustrated. The genotype is shown in italics, pindicates paternally derived, m maternally derived genes.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/9/10.1182_blood.v94.9.2945/4/m_blod42135002x.jpeg?Expires=1767912539&Signature=BnzsbGFHUMt5WiXt~nhNxfhuLqryuX54zgNTA6OWvYmlhSqDNGN80FAGx8ipk4YvBvjLuIFXd~EnxFotkmc~7WQeXwMy3moW1HNNAaut0EakzSRUs-j9qV8P879mL-SpUayayW~09yzBjrrAZoYQg-4USD1bcPQwm8c-fh8twYJ4U4MQhGAebaKSnndXDsoOApul-lavZx6-~PYl7wcLjGXWtSZ8EhT0I~KoqOspU4OHK~viMG1LdpQ~dKCqo2RONS0xY9r9i5E14~pksLKNQb9IinQCu7sib0~FbGZ9lPw2jlBYaBlmmiGLmUtHC0ZLxlw-0OUlSBGGd85YTcRRfQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 4. Proportion of CD24-deficient RBCs in male and female newborn mice obtained from the 2 different breedings. Fifty-five animals that carry both the lox-Piga-lacZ and the EIIa-cregene are shown. (The analysis of the first 12 animals [11 EL females and 1 LE female] were excluded because of initial problems caused by clumping of CD24 positive cells during the staining procedure.) Four male mice that did not inherit thelox-Piga-lacZ gene served as negative controls. Each box plot displays the 10th, 25th, 50th, 75th, and 90th percentile. The median is indicated. Values above the 90th or below the 10th percentile are plotted.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/9/10.1182_blood.v94.9.2945/4/m_blod42135004x.jpeg?Expires=1767912539&Signature=idWyWOs-HQ7rPinaARCLzLdHPpT4yzKpk0EWZyzMtK2QrIZWIshMjK20884toclgCIG0j3PuMtsztVVjtFhlicT1bXPW0IZhvOCvJ5XaBg2J88KKvBb9SAu2S7NVXI9TVyeK5QQQrsHcPBjpdU83J~yQj8-944AplBUZDVpRyDA8T8UUq77h13VtcOmvyjPKqkJI~7eBWZFFL3Vp4pzQ2oqnnuE1McIla5JjhSgIuex3DB9GaJzeD9fPLqxpbuaeo-6AVIpRZDvkKnBJfm-MhUVa6t-uBVfK0rQwq-IV8irm6NzSSDf6Q0RJdZF8FnE-TMZ2S4gGn4KtUjIkYBA0FQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 6. Proportion of PIGA(−) cells in peripheral blood from mosaic mice. (A) Proportion of PIGA(−) RBCs during the observation period of 12 months. (B) Proportion of PIGA(−) granulocytes (PMN), T cells, B cells, CD8+, and CD4+ cells during the observation period of 12 months. Each box plot displays the 10th, 25th, 50th, 75th, and 90th percentile. Values above the 90th or below the 10th percentile are plotted [Table 2 shows the values for median, the number of animals analyzed, and the level of significance of the difference in the percentage of PIGA(−) cells between 2 successive time points]. (C) Proportion of PIGA(−) reticulocytes in relation to the proportion of PIGA(−) RBCs, granulocytes (PMN), and lymphocytes (Lyc) in 11 animals more than 16 weeks of age. The values for the median are indicated and shown by a horizontal line.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/9/10.1182_blood.v94.9.2945/4/m_blod42135006x.jpeg?Expires=1767912539&Signature=f6SXOdhBp5qDFizZrflYUlO0~Rmj6PEMuxFAgR69y7ChuxzmlOjmmrE8XxLC2hbxkgj3wnh2hFCJnX5HiwGe~M0Rvc-nK6tOfRLxCXIeXAJr7e6yH~Xro65WgDpNByN9NV4lpX~plLeKBKEY7ABPmrzQOc1dEa8JCQOCqccMA7TsS9BTTUd-On830zxsgLW9trZ6JIKTRmpERKjtvexSqwwxEMRNU~sAnVj1lR-CMBPkgHdd19lTqqoCLksKfeBYXU0EUSY~2u7R4gCOtFMWJOx2Zq3zf8qvoNzBPxPsK0HB9sCV~JQ4waJThbiahSRL4B3mqXWHkPXJWlF1as-Mcw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal