Chronic granulomatous disease (CGD) is a rare inherited disorder of phagocytes in which defective production of microbicidal oxidants leads to severe recurrent infections. CGD is caused by mutations in any of 4 genes encoding components of nicotinamide adenine dinucleotide phosphate (reduced form; NADPH) oxidase, the multisubunit enzyme that produces the precursor of these oxidants, superoxide. Approximately 5% of CGD patients have an autosomal recessive form of disease caused by a severe deficiency of p67-phox, a 526-amino acid subunit of the oxidase that appears to regulate electron transport within the enzyme. Here we report the biochemical and molecular characterization of 6 unrelated kindreds with p67-phox deficiency. These studies show that, as in gp91-phox and p22-phox deficiencies, the p67-phox CGD patients show a high degree of heterogeneity in the genetic defects that underlie their disease. Five different mutant alleles were identified: (1) a nonsense mutation in exon 4 (C304 → T); (2) a 5-nucleotide (nt) deletion in exon 13 (nts 1169-1173); (3) a splice mutation in the first nucleotide of intron 4 (G → A); (4) a deletion of 1 nt in exon 9 (A728); and (5) a 9-nt in-frame deletion in exon 2 (nts 55-63). The splice mutation was seen in 3 unrelated kindreds, while the 5-nt deletion was seen in 2 apparently unrelated families (both of Palestinian origin). Homozygosity was present in 4 of the kindreds, 2 of which had consanguineous parentage. In the isolated neutrophils of each of the affected patients in the 6 kindreds, there was no measurable respiratory burst activity and no p67-phox protein detected by immunoblot analysis. The level of 67-phox mRNA was less than 10% of normal in the mononuclear leukocytes from 3 of the 4 patients analyzed by Northern blot studies. Thus, this heterogeneous group of mutations in p67-phox all lead to marked instability of mRNA or protein (or both) that results in the complete loss of NADPH oxidase activity.
THE PHAGOCYTIC CELLS play a fundamental role in the innate host defense. This function is achieved through a coordinated interaction between reactive oxygen metabolites and antimicrobial proteins released to the phagolysosome during phagocytosis.1 When phagocytes are stimulated, oxygen is consumed at a dramatically increased rate and is converted to superoxide anion (O2−) by a multisubunit flavocytochrome oxidase termed nicotinamide adenine dinucleotide phosphate (reduced form; NADPH) oxidase.2,3 Superoxide is a reducing agent with little direct bactericidal action; however, it is used to produce other reactive oxygen species that include hydrogen peroxide, hypohalous acids, and hydroxyl radical, all of which have potent microbicidal activity and a high capacity to produce tissue damage.1 The importance of NADPH oxidase-dependent microbicidal mechanisms is made evident by an uncommon inherited syndrome called chronic granulomatous disease (CGD), a disorder in which there is a profound failure in phagocytes to produce O2−.4-6 This disorder is caused by mutations in any 1 of the 4 components of the NADPH oxidase complex and is characterized by recurrent severe infections (especially with catalase-positive microorganisms) that affect subcutaneous tissues, bones, lungs, liver, and lymph nodes.4 5
The central redox component of the phagocyte oxidase is an unusual flavocytochrome b that has a very low redox potential (Em7.0 = −245 mV).7 This cytochrome, termed cytochrome b-245 or cytochrome b558 for its α band absorbance, is located in both the plasma membrane and the membranes of specific granules. It is composed of 2 tightly bound nonidentical subunits, p22-phox (phagocytic oxidase) and the heavily glycosylated gp91-phox.8,9 The normal transfer of electrons from NADPH through the flavin adenine dinucleotide (FAD) and heme moieties of cytochrome b558 after phagocyte activation is dependent on several cytosolic proteins that translocate from the cytosol to the membrane to be in close contact with cytochrome b558.10Three cytosolic factors of NADPH oxidase have been characterized: a 47-kD phosphoprotein (p47-phox), a protein of 67 kD (p67-phox), and a 40-kD protein (p40-phox); these proteins share an important homology, which is related to the presence of several src homology (SH3) domains.11-14 Moreover, NADPH oxidase requires the participation of rac 2 or rac 1, members of the p21 ras family of low-molecular-weight guanosine 5′-triphosphatases (GTPases).15 16
Mutations in the gp91-phox gene, located on the short arm of the X chromosome at p21.1, are responsible for 60% to 65% of cases of CGD.4 The remaining 35% of patients have an autosomal recessive mode of inheritance due to mutations in the p47-phoxgene (located on chromosome 7q11.23; 25% of all CGD cases), the p22-phox gene (chromosome 16q24; 5% of cases), or p67-phox (chromosome 1q25; 5% of cases).4,6 There is a high degree of heterogeneity in the molecular alterations that produce gp91-phox and p22-phox deficiencies that explains, at least in part, the wide range of clinical variability of CGD in these groups. In contrast, the vast majority of p47-deficient patients have only one type of mutant allele: a GT deletion at the beginning of exon 24,6 that appears to have arisen through recombination events with a highly homologous p47-phoxpseudogene that contains this GT deletion.17,18 Relatively little is known about p67-deficient CGD, as just 6 patients have been reported in whom the molecular genetics have been described.6 19-23
Here we report the molecular characterization of 6 unrelated kindreds with p67-phox deficiency. The CGD diagnosis was confirmed by biochemical analysis and p67-phox deficiency was determined by Western blot analysis or cytosol complementation studies in the cell-free oxidase activation system. These studies show that, as in gp91-phox and p22-phox deficiencies, the p67-phox CGD patients show a high degree of heterogeneity in the genetic defects that underlie their disease.
MATERIALS AND METHODS
CGD patients and families.
Blood samples were obtained from CGD patients and family members referred to the investigators or were sent to the CGD reference laboratories at The Scripps Research Institute or Stanford University Medical School. Procedures and consent forms were approved by the Committees on the Protection of Human Subjects in Research of The Scripps Research Institute and of the Stanford University Medical School.
Case 1.
LC is the 8-year-old daughter of unrelated Hispanic parents. The diagnosis of CGD was made at age 8 months when she presented with a right upper lobe pneumonia caused by Serratia marcescens. Her neutrophils had absent nitroblue tetrazolium (NBT) reduction, undetectable O2− generation in response to phorbol myristate acetate (PMA), and no activity in the dichlorofluorescein assay for H2O2. The absence of p67-phox was established by Western blot.
Case 2.
MD is the 10-year-old daughter of first cousins native of Jordan. During the first year of life, she had recurrent abscesses caused by gram negative bacteria. At the age of 5 years, she developed inflammatory bowel disease. The diagnosis of CGD was made on the basis of absent NBT reduction and O2−production by her PMA-stimulated neutrophils. A deficiency of p67-phox was demonstrated by Western blot. Her sister, AD, is 2 years old and also affected with CGD; however, she has yet to develop serious illnesses or require hospitalization.
Case 3.
AA is the 2-year-old son of Palestinian parents who are first cousins. He presented with failure to thrive at age 2 months and peritonitis caused by Candida lusitania. CGD was suspected from a family history showing a sister who died at 40 days of age as a consequence of septicemia and multiorgan failure. Radiographic studies on AA showed a mesenteric mass, pulmonary nodules, and mediastinal adenopathy. The CGD diagnosis was confirmed by the absence of NBT reduction by his stimulated neutrophils. No detectable p67-phox protein was seen in Western blots of his neutrophils.
Case 4.
Patient IP was a 4-year-old girl at the time of her death. Her parents were nonconsanguineous and natives of a small town in Mexico. The diagnosis of CGD was suspected when she presented with skin abscesses containing Enterobacter spp and Klebsiella spp and confirmed by a negative NBT test and absent O2− production. In her last year of life, she presented with an Aspergillus fumigatus pneumonia, which was successfully treated with amphotericin B and 5-fluorocytosine. However, a few months later, she presented with a severe respiratory infection characterized by necrotizing granulomas and acid-fast bacteria that was treated with antituberculosis therapy. She died a few weeks later. The p67-phox defect was identified using the cell-free activation complementation assay11 and subsequently by Western blot analysis of Epstein-Barr virus (EBV)-transformed B cells from the patient. The molecular genetic studies were performed with DNA and RNA also obtained from the EBV-transformed B cells. Her siblings are not affected with CGD.
Case 5.
In this kindred, 2 sisters were affected. SG is 15 years old and CG is 12 years old; their unrelated parents are Native American (mother) and African American (father). The diagnosis of CGD was made by a negative NBT test, no superoxide production by PMA-stimulated neutrophils and undetectable p67-phox by Western blot analysis. At age 3 years, SG presented with a heel abscess associated with popliteal and inguinal lymphadenitis positive for Serratia marcescens. She has suffered from recurrent impetigo, pneumonia, bilateral axillary, and cervical lymphadenopathy with Pseudomonas spp, aphthous stomatitis, and a combined bacterial and fungal pneumonia. CG developed facial and periorbital cellulitis at age 1 month. At this time, the diagnosis of CGD was made in both sisters. In her first year of life, CG also presented with bilateral axillary lymphadenitis with S marcescens, as well as osteomyelitis at multiple sites associated with pneumonia and septic emboli caused by Staphylococcus aureus. Since that time, CG has had recurrent cervical lymphadenitis and 1 episode of Aspergillus pneumonia.
Case 6.
NS is the 16-year-old daughter of unrelated parents who are natives of Mexico. She presented with a S marcescens cellulitis at age 6 months and had several episodes of otitis media in the first year of life. The diagnosis of CGD was made based on a negative NBT test. Her neutrophils had no detectable p67-phox by Western blot. She has suffered from urinary tract infections, pharyngitis, a hepatic abscess, oral candidiasis, and a fungal pneumonia. One brother died previously at age 20 months secondary to a severe infection; he was not tested for CGD. Five brothers are not affected.
Preparation of cells.
Neutrophils were prepared from whole blood anticoagulated with acid-citrate-dextrose and isolated by dextran sedimentation, hypotonic lysis of erythrocytes, and Ficoll-Hypaque (Pharmacia, Piscataway, NJ) density gradient centrifugation as previously described.24Mononuclear cells were isolated from buffy coats from heparinized whole blood by Ficoll-Hypaque centrifugation.25 EBV-B–cell lines from patients were established following a previously described method.26
Biochemical analysis of the respiratory burst.
The generation of superoxide (O2−) by intact neutrophils was determined by superoxide dismutase-inhibitable ferricytochrome c reduction as previously described.27 The presence of p67-phox, p47-phox, and p40-phox in patients and controls was determined by immunoblot analysis of granulocytes and EBV-B–cell lines that were treated with diisopropylfluorophosphate (Sigma, St Louis, MO), lysed by sonication, and centrifuged as previously described.28 The p67-phoxcomponent was identified using 2 different antibodies: a rabbit polyclonal antibody produced against peptide 447-460 of human p67-phox (DEPKESEKADANNQ) that was purified by affinity chromatography (using the cognate peptide) and a second antibody obtained from a rabbit immunized with human recombinant p67-phox protein. A protein G-purified rabbit antihuman recombinant p47-phox antibody was used to detect the p47-phox protein. An antipeptide antibody raised in a rabbit against residues 134-148 (YQSPYDSEQVPQAIR) of the p40-phoxcomponent of NADPH oxidase was used as the primary antibody in immunoblots for p40-phox.
Preparation of genomic DNA, total RNA, mRNA, and cDNA.
Most of the DNA samples were extracted from the buffy coat from peripheral blood using an automated system (DNA Extractor; Applied Biosystems, Foster City, CA). The remaining DNA samples were prepared using the QIAamp Blood Kit (Qiagen Inc, Chatsworth, CA) from either blood or EBV-B cells. Total RNA and mRNA were isolated from peripheral blood mononuclear leukocytes or EBV-transformed B cells using the Stratagene Total RNA and Poly(A) Quick mRNA Isolation Kits (Stratagene, La Jolla, CA). For cDNA production, the StrataScript reverse transcriptase-polymerase chain reaction (RT-PCR) kit (Stratagene) was used.
Northern blots.
Total RNA obtained from EBV-transformed B cells was analyzed by Northern blot according to standard procedures.29 The p67-phox and gp91-phox transcripts were hybridized with full-length cDNA human probes. Procedures for sequential cycles of prehybridization, hybridization, washes, and filter stripping were performed as previously described.30 Equal loading of RNA in each lane was demonstrated by examination of gels after ethidium bromide staining. Quantitative data on the Northern blots was obtained by direct measurement of 32P decay on a phosphoimager analyzer.
PCR and DNA sequencing.
Oligonucleotide primers were designed using the Oligo software from National Biosciences, Inc (Plymouth, MN).31 Primers for amplification and sequencing of genomic DNA and cDNA are shown in Table 1. The numbering of nucleotides (nts) and amino acids used in this report was according to genomic DNA sequence deposited in GenBank based on Kenney (GenBank AccessionU00776)32 and cDNA sequence described by Leto et al13 (GenBank Accession M32011). PCR was performed as previously described33 using a GeneAmp 9600 thermal cycler (Perkin Elmer Corp, Norwalk, CT). Cycle sequencing of PCR amplified genomic or cDNA was performed using a commercial kit (fmol DNA Sequencing System; Promega, Madison, WI).34
Oligonucleotide Primers Used for Analysis of p67-phox
| Primers for Genomic DNA Analysis . | Primers for cDNA Analysis . |
|---|---|
| p67-5′L ACCTCTGGCTGGTGACAC p67-Ex1R CCTCCCTGGTGATAATGACA p67-Ex2L GGCCCAGAAAGTGAACAC p67-Ex2R ATTGGGGTTGAGAATCATAAT p67-Ex3L CTCACACCTCCTCACTGC p67-Ex3R ACTGCTCAAACACCAAGC p67-Ex4L GCTGCATTTATTTCTCCA p67-Ex4R TGACTGTCAGCCATCCAT p67-Ex5L TTTTATGTTTGCGGTCTGT p67-Ex5R ATGGCATGTCCTCTGAGA p67-Ex6L CGTCACCCCCATTTTCAC p67-Ex6R TCCCACCTTGCTCCACAT p67-Ex7L AGAAGAATGGAAACAGTGC p67-Ex7R AGGAGCCCTTACAATCAG p67-Ex8L TTCTGGAAGAATGCTCAAAT p67-Ex8R CCCCACCTTCATCTTCTT p67-Ex9L CTGGCTCCAAGTTCAGTG p67-Ex9R CAAAGAAGGCAGCAGATACT p67-Ex10L TGTGGGTACTGATGAGCA p67-Ex10R TCCTGACAACACCTCTTTT p67-Ex11L GTGTTTCCCCACATCCAC 67-Ex11R CAGGGAGAGGAACTCAGGA pP67-Ex12L CCCTATTTGAAGAGGTTT p67-Ex12R ATGTCTGTGGTTGATAGCC p67-Ex13L TGATCCAGGATGTTGAGAGAA p67-Ex13R TCCTCTTCCCTCCTCCAG p67-Ex14L ATGGTTTTTGTGATGATGTT p67-Ex14R ACAGAAGGTGCTTGATAAAT p67-Ex15L ATTTCGCTGTTTCCTTTA p67-Ex15R TTGACCTTGTTTCTGCTA p67-Ex16L AGCCAGACAGGGTAATCT p67-Ex16R GCAGAAGGGTGCTAAATC | p67-44LcDNA TGGCCTCCTAGTTTCTACC p67-94LcDNA CTGGAATGAAGGGGTGCT p67-254LcDNA AGCATTAACCGAGACAAGC p67-460LcDNA TGCCAAGAAGGAGGAATG p67-644LcDNA CTGGCCAAGAAGGATTACC p67-835LcDNA AGAAGAGCTCCAGGTCATG p67-1085LcDNA GAGCCTAAGGAAGTGAAGC p67-1404LcDNA ATGCTAATAACCAGACAACAG p67-455RcDNA CCTCCTTCTTGGCATACAT p67-825RcDNA GGAGCTCTTCTTTTGTCTCAG pP67-1102RcDNA GGCATGGGAACACTGAGC p67-1320RcDNA CACAGAGTCAGGCAGTAGTTTT p67-1476RcDNA AGGTCCTCTGGTTGGGTAG p67-1658RcDNA TTTTCTTCAGCTTTGTAGTTTG |
| Primers for Genomic DNA Analysis . | Primers for cDNA Analysis . |
|---|---|
| p67-5′L ACCTCTGGCTGGTGACAC p67-Ex1R CCTCCCTGGTGATAATGACA p67-Ex2L GGCCCAGAAAGTGAACAC p67-Ex2R ATTGGGGTTGAGAATCATAAT p67-Ex3L CTCACACCTCCTCACTGC p67-Ex3R ACTGCTCAAACACCAAGC p67-Ex4L GCTGCATTTATTTCTCCA p67-Ex4R TGACTGTCAGCCATCCAT p67-Ex5L TTTTATGTTTGCGGTCTGT p67-Ex5R ATGGCATGTCCTCTGAGA p67-Ex6L CGTCACCCCCATTTTCAC p67-Ex6R TCCCACCTTGCTCCACAT p67-Ex7L AGAAGAATGGAAACAGTGC p67-Ex7R AGGAGCCCTTACAATCAG p67-Ex8L TTCTGGAAGAATGCTCAAAT p67-Ex8R CCCCACCTTCATCTTCTT p67-Ex9L CTGGCTCCAAGTTCAGTG p67-Ex9R CAAAGAAGGCAGCAGATACT p67-Ex10L TGTGGGTACTGATGAGCA p67-Ex10R TCCTGACAACACCTCTTTT p67-Ex11L GTGTTTCCCCACATCCAC 67-Ex11R CAGGGAGAGGAACTCAGGA pP67-Ex12L CCCTATTTGAAGAGGTTT p67-Ex12R ATGTCTGTGGTTGATAGCC p67-Ex13L TGATCCAGGATGTTGAGAGAA p67-Ex13R TCCTCTTCCCTCCTCCAG p67-Ex14L ATGGTTTTTGTGATGATGTT p67-Ex14R ACAGAAGGTGCTTGATAAAT p67-Ex15L ATTTCGCTGTTTCCTTTA p67-Ex15R TTGACCTTGTTTCTGCTA p67-Ex16L AGCCAGACAGGGTAATCT p67-Ex16R GCAGAAGGGTGCTAAATC | p67-44LcDNA TGGCCTCCTAGTTTCTACC p67-94LcDNA CTGGAATGAAGGGGTGCT p67-254LcDNA AGCATTAACCGAGACAAGC p67-460LcDNA TGCCAAGAAGGAGGAATG p67-644LcDNA CTGGCCAAGAAGGATTACC p67-835LcDNA AGAAGAGCTCCAGGTCATG p67-1085LcDNA GAGCCTAAGGAAGTGAAGC p67-1404LcDNA ATGCTAATAACCAGACAACAG p67-455RcDNA CCTCCTTCTTGGCATACAT p67-825RcDNA GGAGCTCTTCTTTTGTCTCAG pP67-1102RcDNA GGCATGGGAACACTGAGC p67-1320RcDNA CACAGAGTCAGGCAGTAGTTTT p67-1476RcDNA AGGTCCTCTGGTTGGGTAG p67-1658RcDNA TTTTCTTCAGCTTTGTAGTTTG |
cDNA primers were numbered from the first nucleotide of sequence obtained from GenBank.
Abbreviations: L, sense; R, antisense.
RESULTS
DNA polymorphisms.
A total of 6 differences were identified between the published p67-phox nt sequences13,32 and the results obtained in this study with genomic DNA from normal subjects. The homozygous substitutions T895 → C and A983→ G were found in all normals sequenced both at the genomic and cDNA levels. The former predicts a silent amino acid change (Leu299 → Leu), whereas the latter predicts a conservative modification (Lys328 → Arg). In 50% of the subjects analyzed (both normals and patients), an additional apparent polymorphism was identified, A542 → G, that predicts a conservative Lys181 → Arg substitution. All 3 of these nt substitutions have been previously considered as polymorphisms of the p67-phox gene.21Furthermore, in the genomic DNA of 2 normal subjects used as controls, 2 other nt differences were observed. One of these individuals was heterozygous for A235 → G in exon 3, a change that predicts a Met79 → Val substitution. The other normal control showed a C1167 → A transversion in exon 13 that predicts the replacement of His389 with Gln. These last 2 changes might represent rare polymorphisms in the p67-phox gene or could indicate a carrier state for p67-phox–deficient CGD. To address these possibilities, site-directed mutagenesis of p67-phox is currently in progress. Finally, an unexpected finding was a highly frequent polymorphism at position −21 of intron 10 in which A is shifted to G. Some normal individuals were heterozygous for this substitution, while others were homozygous.
Three other apparent polymorphisms were identified in the 5′ regulatory region. These came to light in the study of the fifth kindred (SG/CG). This family had a cluster of 3 point mutations upstream of exon 1 of the p67-phox gene: G−185 → A, G−181→ A, and C−24 → T (both patients, their mother, and their half-sister were heterozygous for all 3 mutations). None of these changes reside in known transcriptional regulatory motifs. They likely represent polymorphisms, as all 3 were seen in several normal controls as follows (of a total of 18 individuals encompassing 36 alleles): G−185→ A (2 of 36 alleles), G−181 → A (6 of 36 alleles, 2 individuals homozygous), and C−24→ T (2 of 36 alleles).
Identification of mutations in CGD patients with p67-phoxdeficiency.
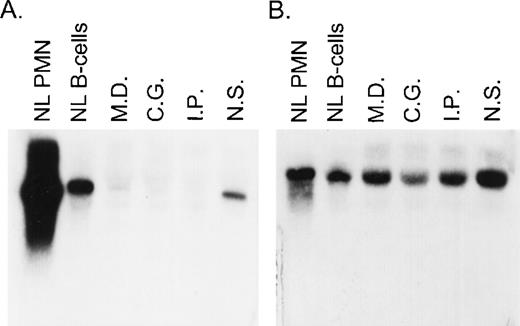
A total of 8 CGD patients (7 females and 1 male) from 6 different kindreds were analyzed in this study. In each of these families, the proband had absent respiratory burst activity and undetectable levels of the p67-cytosolic component of NADPH oxidase by immunochemical analysis (data not shown). The levels of p47-phox, gp91-phox, p22-phox, and cytochrome b558were normal in each patient, thus establishing the diagnosis of p67-phox–deficient CGD. Northern blot analysis in the 4 kindreds studied showed a variable level of expression of p67-phox RNA that appeared, in each case, to be approximately normal in size. The levels of expression were as follows (each as a percent of control): MD (kindred 2) 9.3%, IP (kindred 4) 5%, CG (kindred 5) 7%, and NS (kindred 6) 39% (Fig 1). In all of these patients, the expression of gp91-phox mRNA was similar to the control.
Northern blot analysis of p67-phox and gp91-phox mRNA transcripts. Each lane contained 10 μg of total cellular RNA from normal neutrophils (PMN) or EBV-B cells from a normal control or from the indicated patients. The figure shows a blot probed with 32P-labeled full-length p67-phox cDNA (A) and then stripped and reprobed with gp91-phox cDNA (B).
Northern blot analysis of p67-phox and gp91-phox mRNA transcripts. Each lane contained 10 μg of total cellular RNA from normal neutrophils (PMN) or EBV-B cells from a normal control or from the indicated patients. The figure shows a blot probed with 32P-labeled full-length p67-phox cDNA (A) and then stripped and reprobed with gp91-phox cDNA (B).
To identify the underlying p67-phox mutation in these cases, all 16 exons of the gene were sequenced in each patient using genomic DNA first, and in 6 of the 8 patients, cDNA as well. In addition, approximately 40 nts on either side of each exon were sequenced in the gDNA samples to identify potential splice mutations. Furthermore, to identify possible regulatory mutations, approximately 400 nts immediately upstream (5′) of exon 1 were also sequenced.
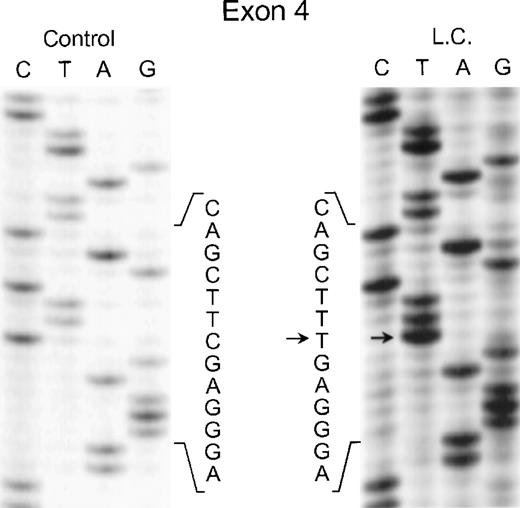
In the genomic DNA from case 1 (LC), a single mutation was identified that was seen in both strands of the p67-phox gene: a C304 → T transition in exon 4. As shown in Fig 2, the patient was homozygous for this mutation, while each of her parents was heterozygous for the defect (data not shown) despite being apparently unrelated. This mutation predicts the replacement of Arg102 with a TGA stop codon.
Sequence analysis of PCR-amplified exon 4 of the p67-phox gene from genomic DNA in patient LC. Sequences of patient and control are shown. The arrows indicate the C to T transition. The 5′ end of the DNA is at the top of each sequencing panel.
Sequence analysis of PCR-amplified exon 4 of the p67-phox gene from genomic DNA in patient LC. Sequences of patient and control are shown. The arrows indicate the C to T transition. The 5′ end of the DNA is at the top of each sequencing panel.
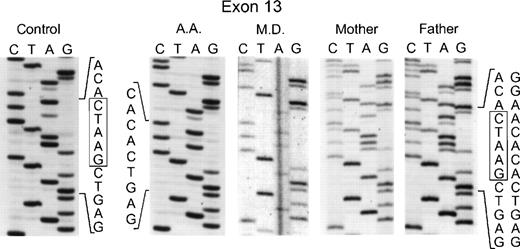
In the genomic DNA of the 2 affected sisters in the second kindred (MD and AD), a single mutation was again seen: a deletion of 5 nts at the 3′ end of exon 13 involving nts 1169 through 1173 (Fig 3). In this case, the homozygosity in the proband was due to consanguinity of her parents who were each heterozygous for this mutation, as evidenced by the presence of 2 superimposed sequences after position 1169: 1 from the normal allele and the other from the allele with the deletion (Fig 3). This mutation in the genomic DNA was confirmed by sequencing the cDNA from the 2 affected sisters and their parents (data not shown). Interestingly, the same deletion in the 3′ end of exon 13 was found in an unrelated patient (AA from kindred 3). As in the case of the second kindred, the proband was homozygous for the deletion, the parents were first cousins and each heterozygous for the same mutation, and the family was of Palestinian descent (Fig 3). Of the 4 surviving siblings, 2 were heterozygous carriers of this p67-phox mutation, while the other 2 were homozygous for the normal allele (DNA was not available for sequencing from a female sibling who had died previously at age 40 days from septicemia). The 5-nt deletion found in these 2 unrelated families produces a translational frameshift beginning in codon 391 and predicts a premature stop codon (TGA) at amino acid position 401 in exon 14. If translated, the mutant transcript would generate a protein only 76% of the full-length of p67-phox that would lack the C terminal 126 amino acids. There was no evidence, however, that this truncated protein was expressed in either neutrophils or EBV-transformed B lymphocytes based on immunoblot studies using a polyclonal anti-p67 antibody raised to the full-length recombinant p67-phox.
Sequence analysis of PCR-amplified exon 13 of the p67-phox gene from genomic DNA. In the two left panels are the sequences of a normal control and patient AA. The right three panels show patient MD and her parents. Boxes indicate the 5 nts in the normal sequence that are deleted in the mutated alleles. The 5′ end of the DNA is at the top of each sequencing panel.
Sequence analysis of PCR-amplified exon 13 of the p67-phox gene from genomic DNA. In the two left panels are the sequences of a normal control and patient AA. The right three panels show patient MD and her parents. Boxes indicate the 5 nts in the normal sequence that are deleted in the mutated alleles. The 5′ end of the DNA is at the top of each sequencing panel.
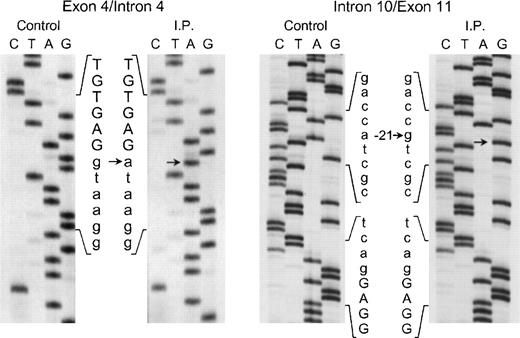
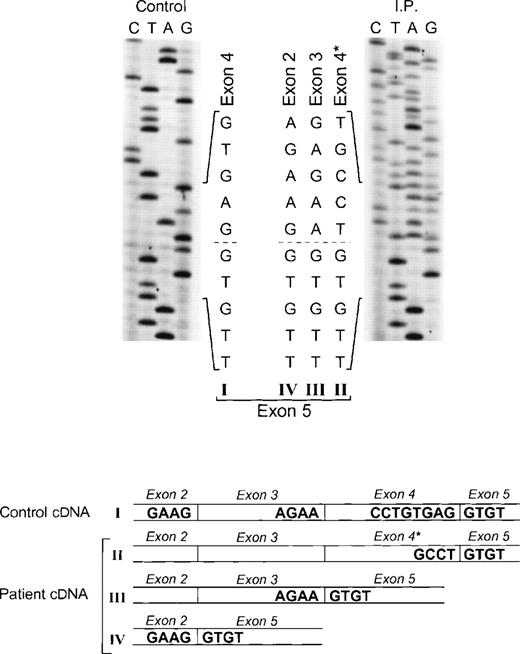
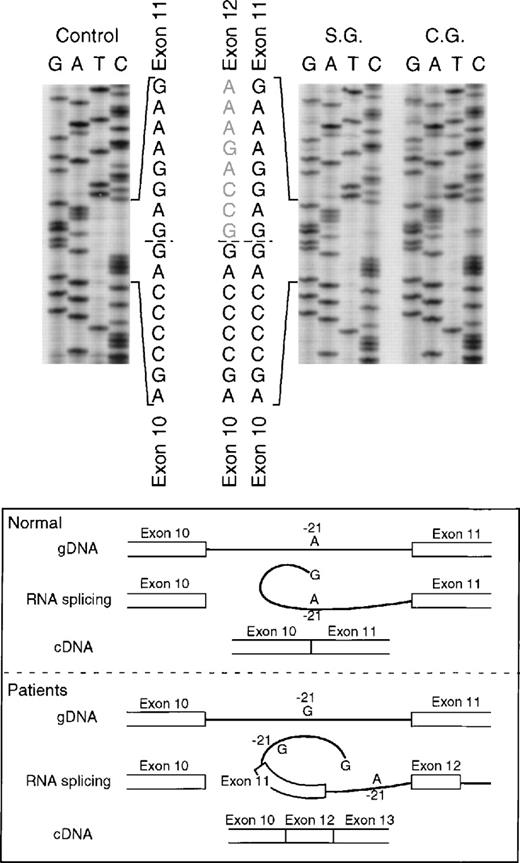
The proband in the fourth kindred (IP) showed a homozygous mutation even though her parents were apparently unrelated. The mutation was first identified in the patient’s genomic DNA: a G → A transition in the first nt in the consensus splice site of intron 4 (Fig 4). This mutation at position +1 of intron 4 predicts abnormal splicing during the pre-mRNA processing. To confirm this, the PCR product of the 5′ end of p67-phoxcDNA of this patient was sequenced. The results showed 3 different species of cDNA: 1 that lacks exons 3 and 4, a second with a deletion of exon 4, and a third that lacked only the last 5 nts of exon 4 (Fig 5). These 3 different sequences are the consequence of alternative splicing that results from the mutation in the donor splice sequence of intron 4. This mutation can cause the skipping of exons 3 and 4 together, exon 4 alone, or just a portion of exon 4 through the use of a cryptic splice site in the 3′ terminus of exon 4 (GTGAG). The deletion of exons 3 and 4 generates a mRNA species that is 186 nts shorter than control and that predicts an in-frame deletion of 62 amino acids (Fig 5, splice version IV). The other 2 mRNA species contain reading frame shifts that predict premature stop codons at positions 145 (TAG) and 141 (TGA), respectively. The exon 4 deletion predicts an N-terminal truncated protein that corresponds to the first 107 amino acids of p67-phox (Fig 5, splice version III), while the mRNA with the cryptic splice site predicts a truncated protein of 138 amino acids (Fig 5, splice version II). Interestingly, this patient was also homozygous for the A → G transition 21 nts before the beginning of the exon 11 (Fig 4). As noted above, this nt change appears to be a polymorphism in the p67-phox gene. Because this sequence belongs to the branch point sequence necessary for pre-mRNA splicing, we analyzed mRNA to detect whether any defective processing of exon 11 occurred. The cDNA amplification of this region did, in fact, show an abnormal transcript that contained an out-of-frame deletion of exon 11 (see below). Despite the fact that this mutation was homozygous, a normal transcript containing exon 11 was also detected by PCR, indicating that this intron 10-branch point sequence is important, but not essential, for normal pre-mRNA processing. As noted above and in Fig 1, the Northern blot in this patient showed only a normal-sized p67-phox transcript expressed at a level 5% of control. This most likely represents the mRNA molecules with the normal splicing of exon 11 and the deletion of the last 5 nts of exon 4 (splice version II described above). The other possible species of p67-phox mRNA were not seen, presumably due to their marked instability, and therefore detectable only with the highly sensitive RT-PCR method.
Sequence analysis of the exon 4/intron 4 and intron 10/exon 11 regions of the p67-phox gene amplified by PCR using genomic DNA from patient IP and a normal control. The exonic sequences are represented by capital letters, while the intronic sequences are indicated by lower case letters. Arrows indicate transitions of g → a at position +1 of intron 4 and of a → g at position −21 of the branch point sequence of intron 10. The 5′ end of the DNA is at the top of each sequencing panel.
Sequence analysis of the exon 4/intron 4 and intron 10/exon 11 regions of the p67-phox gene amplified by PCR using genomic DNA from patient IP and a normal control. The exonic sequences are represented by capital letters, while the intronic sequences are indicated by lower case letters. Arrows indicate transitions of g → a at position +1 of intron 4 and of a → g at position −21 of the branch point sequence of intron 10. The 5′ end of the DNA is at the top of each sequencing panel.
Sequence analysis of p67-phox cDNA in patient IP. The region between exons 2 and 5 was amplified by PCR using primers p67-94LcDNA and p67-455RcDNA. The upper panel shows the sequences of the patient and control with the 5′ end of the cDNA sequence at the top. The dashed line indicates the beginning of exon 5, the point at which the cDNA sequence returns to normal after the 3 abnormal types of splicing (splice versions labeled II, III, and IV). The lower panel shows a diagram of the different cDNA molecules derived from the alternative splicing. The GT dinucleotide in the 3′ end of exon 4 in the normal cDNA species (version I) is the cryptic splice site used for generating the abnormally spliced version II (exon 4*).
Sequence analysis of p67-phox cDNA in patient IP. The region between exons 2 and 5 was amplified by PCR using primers p67-94LcDNA and p67-455RcDNA. The upper panel shows the sequences of the patient and control with the 5′ end of the cDNA sequence at the top. The dashed line indicates the beginning of exon 5, the point at which the cDNA sequence returns to normal after the 3 abnormal types of splicing (splice versions labeled II, III, and IV). The lower panel shows a diagram of the different cDNA molecules derived from the alternative splicing. The GT dinucleotide in the 3′ end of exon 4 in the normal cDNA species (version I) is the cryptic splice site used for generating the abnormally spliced version II (exon 4*).
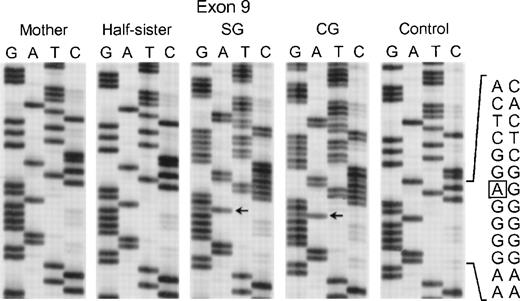
In the fifth kindred, both sisters affected with CGD (SG and CG) were heterozygous for the mutant allele identified in kindred 4 above: the G → A transition at the +1 position of intron 4 and the A → G substitution at position −21 in the 3′ end of intron 10. The cDNA sequencing studies showed multiple mRNA species consistent with this pair of genomic DNA mutations in a pattern similar to that seen in kindred 4: alternative splicing due to the intron 4 mutation (cf. patient IP in Fig 5) and deletion of exon 11 due to the branch point sequence defect in intron 10 (Fig 6) (however, the autoradiographic intensity of the abnormal sequence without exon 11 was weak). The mother and a half-sister (by the same mother) were heterozygous for these mutations. The defect in the paternal allele was identified as deletion of A728 in exon 9 (Fig7); this point mutation causes a translational frameshift beginning at codon 243 and predicts a premature stop codon (TGA) at amino acid position 270. Although the DNA of the father of the 2 affected patients was not available to confirm the origin of this mutation, the fact that the mother and half-sister did not carry the A728 deletion is consistent with this being the mutant paternal allele.
Sequence analysis of p67-phox cDNA in patients SG and CG. The region between exons 10 and 14 was amplified by PCR using primers p67-835LcDNA and p67-1102RcDNA. The upper panel of the figure shows the cDNA sequences of a normal control and patients SG and CG with the 3′ end at the top. The dashed line indicates the 3′ end of exon 10 and the beginning of 2 different sequences in each patient. The lower panel of the figure shows a schematic representation of the splice defect produced by the nt substitution in the branch point sequence of intron 10. The A → G transition at position −21 of intron 10, important for RNA lariat formation, produces a partial defect in splicing of exon 11 that can allow alternative splicing between exons 10 and 12.
Sequence analysis of p67-phox cDNA in patients SG and CG. The region between exons 10 and 14 was amplified by PCR using primers p67-835LcDNA and p67-1102RcDNA. The upper panel of the figure shows the cDNA sequences of a normal control and patients SG and CG with the 3′ end at the top. The dashed line indicates the 3′ end of exon 10 and the beginning of 2 different sequences in each patient. The lower panel of the figure shows a schematic representation of the splice defect produced by the nt substitution in the branch point sequence of intron 10. The A → G transition at position −21 of intron 10, important for RNA lariat formation, produces a partial defect in splicing of exon 11 that can allow alternative splicing between exons 10 and 12.
Sequence analysis of PCR-amplified exon 9 of the p67-phox gene from genomic DNA from patients SG and CG, their mother, half-sister (maternal), and a normal control. The box in the normal sequence indicates the adenine that is deleted in SG and CG, with only normal sequence seen in the mother and maternal half-sister. The 3′ end of the DNA is at the top of each sequencing panel.
Sequence analysis of PCR-amplified exon 9 of the p67-phox gene from genomic DNA from patients SG and CG, their mother, half-sister (maternal), and a normal control. The box in the normal sequence indicates the adenine that is deleted in SG and CG, with only normal sequence seen in the mother and maternal half-sister. The 3′ end of the DNA is at the top of each sequencing panel.
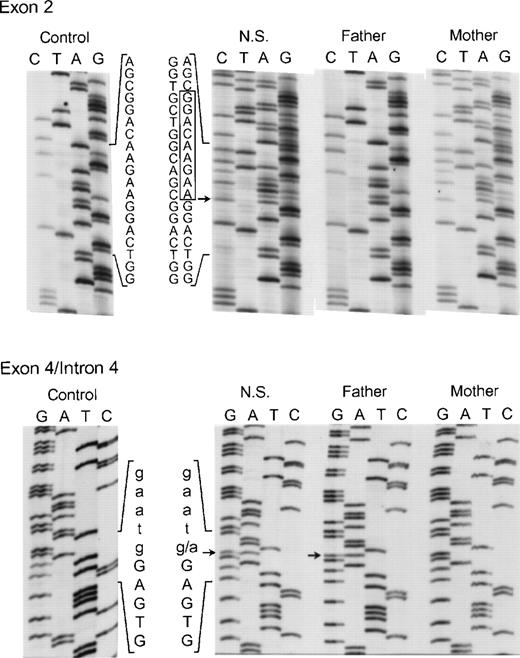
The proband in the sixth and final kindred (NS) was a compound heterozygote daughter of unrelated parents. On 1 allele an in-frame deletion of 9 nts (AAGAAGGAC) involving bp 55 through 63 in exon 2 was identified (Fig 8, top panel) that predicts the elimination of Lys19, Lys20, and Asp21 from the p67-phox protein. Her mother was heterozygous for the same mutation in exon 2. In addition, 2 other changes were found in the maternal allele of this patient. In exon 6 the above-mentioned polymorphism, A542 → G (Lys181 → Arg) was present, while in exon 14, a C1183 → T transition was identified that predicts a nonconservative substitution of Arg395 → Trp (not shown). As discussed below, the Lys19Lys20 Asp21 deletion and, to a lesser extent, the Arg395 → Trp substitution, lead to a loss of activity in p67-phox. The other allele in NS (from the father) appeared identical to the one identified in kindreds 4 and 5, as it contained both the intron 4 (position +1) mutation (Fig 8, bottom panel) and the intron 10 (position −21) polymorphism (not shown). In contrast to the cDNA results in kindred 4, however, the transcript from this defective allele was not detected in either NS or her father, even after RT-PCR. This suggests that the splicing defects have a negative effect on p67-phox mRNA stability and therefore allow a preferential amplification of the nearly full-length maternal allele. These findings agree with the Northern blot result in which a 39% of normal level of p67-RNA was detected, probably reflecting the transcript from the maternal allele.
Genomic DNA sequence analysis of exon 2 (upper panel) and the exon 4/intron 4 region (lower panel) of the p67-phox gene from patient NS and her parents. In the upper panel, the box indicates the 9 nts deleted from the normal exon 2 sequence seen in both NS and her mother. The arrow adjacent to the box indicates the site where the 9-nt deletion starts in exon 2. The 5′ end of the DNA is at the top. In the lower panel, the arrows indicate the g → a transition in the first nt in intron 4 seen in both NS and her father. The upper case letters represent exon 4 sequence, while lower case letters indicate the intronic sequence. The 3′ end of the DNA is at the top of this panel.
Genomic DNA sequence analysis of exon 2 (upper panel) and the exon 4/intron 4 region (lower panel) of the p67-phox gene from patient NS and her parents. In the upper panel, the box indicates the 9 nts deleted from the normal exon 2 sequence seen in both NS and her mother. The arrow adjacent to the box indicates the site where the 9-nt deletion starts in exon 2. The 5′ end of the DNA is at the top. In the lower panel, the arrows indicate the g → a transition in the first nt in intron 4 seen in both NS and her father. The upper case letters represent exon 4 sequence, while lower case letters indicate the intronic sequence. The 3′ end of the DNA is at the top of this panel.
DISCUSSION
CGD caused by mutations in the p67-phox gene is an uncommon inherited defect accounting for less than 5% of all cases of CGD. Until now, only a few p67-phox deficiency patients have been characterized at the molecular level. The patient studied by de Boer et al,19 a 12-year-old daughter of nonconsanguineous parents, was homozygous for a missense mutation (Gly78 → Glu) in exon 3 that appeared to cause instability of the p67-phox protein. Both of the parents, as well as a sister, proved to be heterozygous for this mutation. The second patient described by Tanugi-Cholley et al20 was a 19-year-old daughter of consanguineous parents who was homozygous for a T → C transition at the +2 position at the 5′ end of the intron 3 that led to a highly unstable mRNA species lacking exon 3. The third patient reported by Nunoi et al,21 a 19-year-old man, had a homozygous insertion of an AG dinucleotide at position 399 in exon 5 that resulted in a frame shift predicting a premature stop codon at nt 433. Both parents were heterozygous for this mutation. The fourth case of p67-phoxdeficiency was heterozygous for an in-frame deletion of a single GAA codon in exon 3 that predicted the deletion of Lys58.6 The other allele contained a large 11- to 13-kb deletion. The fifth case reported was a 24-year-old man whose parents were first cousins. He showed a homozygous G → A substitution at position +1 of intron 9 that produced the skipping of exons 8 and 9 from his mRNA.22 The most recent patient to be described showed 2 heterozygous nt substitutions in exon 5 resulting in a double nonconservative amino acid change (Lys160 → Glu and Asp161 → Val) in p67-phox.23
As summarized in Table 2, our patients presented with family-specific mutations as well, none of which have been previously reported. In the first patient (LC), a homozygous C → T transition in exon 4 was identified that produces a premature stop codon at amino acid 102. This type of nonsense mutation is frequently found in other genetic diseases, including CGD4-6 and is usually associated with a severe deficiency of the protein due, at least in part, to its instability secondary to the loss of part of its sequence. Furthermore, it has been demonstrated that premature stop codons often result in substantial decreases in the mRNA levels.35 RNA from this patient was not available to quantify the level of p67-phox transcript. The next 2 kindreds (MD/AD and AA in Table2) also presented with homozygous mutations. They showed a deletion of 5 nts in the 3′ end of exon 13 that predicts an early stop codon 24 nts downstream. This mutation appears to have a severe effect on the stable expression of both transcript and protein: the level of p67-phox RNA observed in the Northern blot was only 10% of normal, while the p67-phox protein was undetectable in the patients’ neutrophils by immunoblot. To the extent that it is synthesized, the truncated protein is probably not properly folded and therefore is easily degraded within the cells of the patients.
Summary of Molecular Findings in p67-phox–Deficient Patients
| Patients . | p67-phox Protein (Immunoblotting) . | p67-phox mRNA (Northern blot) . | Sequence Findings in Genomic DNA . | Predicted Defects . |
|---|---|---|---|---|
| LC | Not detected | ND | C304 → T (Homozygous) | Arg102 → Stop |
| MD and AD | Not detected | 9.3% | Del 1169-1173 (Homozygous) | Premature stop at codon 401 |
| AA | Not detected | ND | Del 1169-1173 (Homozygous) | Premature stop at codon 401 |
| LP | Not detected | 5% | G → A + 1 In 4 (Homozygous) | Skipping of Ex 3 and 4, Ex 4, or 5 last nts of Ex 4 |
| SG and CG | Not detected | 7% | G → A + 1 In 4 (Heterozygous) | Skipping of Ex 3 and 4, Ex 4, or 5 last nts of Ex 4 |
| Del A728 (Heterozygous) | Premature stop at codon 270 | |||
| G−185 → A | Possible polymorphisms | |||
| G−181 → A | ||||
| C−24 → T (Heterozygous) | ||||
| NS | Not detected | 38.7% | G → A + 1 In 4 (Heterozygous) | Skipping of Ex 3 and 4, Ex 4, or 5 last nts of Ex 4 |
| Del 55-63 (Heterozygous) | Del Lys19, Lys20, Asp21 | |||
| C1183 → T (Heterozygous) | Arg395 → Trp |
| Patients . | p67-phox Protein (Immunoblotting) . | p67-phox mRNA (Northern blot) . | Sequence Findings in Genomic DNA . | Predicted Defects . |
|---|---|---|---|---|
| LC | Not detected | ND | C304 → T (Homozygous) | Arg102 → Stop |
| MD and AD | Not detected | 9.3% | Del 1169-1173 (Homozygous) | Premature stop at codon 401 |
| AA | Not detected | ND | Del 1169-1173 (Homozygous) | Premature stop at codon 401 |
| LP | Not detected | 5% | G → A + 1 In 4 (Homozygous) | Skipping of Ex 3 and 4, Ex 4, or 5 last nts of Ex 4 |
| SG and CG | Not detected | 7% | G → A + 1 In 4 (Heterozygous) | Skipping of Ex 3 and 4, Ex 4, or 5 last nts of Ex 4 |
| Del A728 (Heterozygous) | Premature stop at codon 270 | |||
| G−185 → A | Possible polymorphisms | |||
| G−181 → A | ||||
| C−24 → T (Heterozygous) | ||||
| NS | Not detected | 38.7% | G → A + 1 In 4 (Heterozygous) | Skipping of Ex 3 and 4, Ex 4, or 5 last nts of Ex 4 |
| Del 55-63 (Heterozygous) | Del Lys19, Lys20, Asp21 | |||
| C1183 → T (Heterozygous) | Arg395 → Trp |
Abbreviation: ND, not done.
The last 3 kindreds summarized in Table 2 shared an allele with the same mutation (G → A at position +1 of intron 4). Patient IP was homozygous, whereas the 3 patients in the other 2 kindreds were heterozygous for this mutation. This nt is part of the consensus splice site at the exon 4-intron 4 boundary necessary for the processing of mRNA. This mutation causes an alteration of mRNA splicing, as well as a marked instability of the transcript. Three different cDNA species were identified: 1 with a deletion of exons 3 and 4, another with exon 4 missing, and the third with only 5 nts from exon 4 absent. This would indicate that the splicing machinery uses different signals during the course of mRNA processing: sometimes using the 5′ consensus donor sequence of intron 2, other times using that of the exon 3, and in some cases recognizing a cryptic splice site in the 3′ end of exon 4. The finding of this mutant allele in 3 apparently unrelated families is intriguing. It is possible that this allele might have a common ancestry in these families. Kindreds 4 and 6 are unrelated, but are both native to Mexico, while the mother in the kindred 5 (who is also carrying this defective allele) is Native American.
The A to G transition in the branch point sequence of intron 10 found in the mutant allele also shared by these 3 kindreds may explain the observed skipping of exon 11 in the mRNA from patients IP and SG/CG. During pre-mRNA processing, the 5′ splice donor site of the intron is cleaved generating an exon with a 3′-hydroxyl group. The phosphate group at the now free 5′ terminus of the intron is esterified with the 2′-hydroxyl group of a conserved adenosine within the branch point sequence (−18 to −37 nts from the 3′ end of the same intron) to form an RNA lariat. This interaction is necessary for the second catalytic step that takes place, the cleavage of the 3′ splice site of the intron that occurs in concert with the ligation of the 2 exons to produce a mature mRNA.36,37 It is possible that the substitution A → G found in the branch point sequence of these patients alters the pre-mRNA splicing, although not completely, as other similar sequences upstream from the intron can be used to form the lariat.36 It is important to note that this nt transition was found in several normal subjects, both in a heterozygous and homozygous state. It thus appears to be a normal DNA polymorphism. Current studies are in progress to analyze the mRNA splicing in these normal individuals and to determine whether there is any defect in p67-phox protein expression, NADPH oxidase activity, and susceptibility to bacterial and fungal infections.
The affected sisters in the fifth kindred (SG/CG) were compound heterozygotes: the maternal allele contained the splice mutations just described, while the paternal allele had a deletion of an adenine nt at position 728 at the 5′ end of exon 9 that modifies the open reading frame and leads to a premature stop codon at amino acid position 270 (Table 2). Both the mRNA transcript and the truncated p67-phox protein encoded by this mutant paternal allele appear quite unstable, as the p67-phox mRNA level was only 7% of normal and the p67-phox protein undetectable. An unexpected finding in this kindred was a cluster of 3 point substitutions (G−185 → A, G−181→ A, and C−24 → T) in the 5′ regulatory region of the p67-phox gene found in the maternal allele (Table 2). These probably represent a group of polymorphisms based on studies of 18 normal individuals (see Results). Although there are no known transcriptional regulatory motifs in the vicinity of these nt transitions, the 1 at position −24 is located sufficiently close to the start site of transcription that it could play an important regulatory role.
Patient NS in kindred 6 was also a compound heterozygote for mutations in the p67-phox gene (Table 2). The paternal allele contained the intron 4/intron 10 splice mutations discussed above, while the maternal allele contained an in-frame deletion of 9 nts in the middle of exon 2 predicting the absence of Lys19, Lys20, and Asp21. Although this in-frame deletion in exon 2 allowed the synthesis of a nearly complete p67-phox transcript (this was the only patient with a high level of expression of an apparently normal-sized p67-phoxRNA), the protein was not detected by immunoblot analysis. It is possible that the absence of the 3 amino acids produces a highly unstable protein that is degraded soon after its synthesis. To the extent that this mutant form of p67-phox is present, studies with recombinant p67-phox protein containing the KKD deletion indicate that it is devoid of activity in the cell-free NADPH oxidase activation system (Curnutte et al, unpublished results, September 1996). This suggests that the 3 deleted amino acids either play an important structural role or are directly involved in the function of p67-phox. The mechanism by which p67-phoxcontributes to NADPH oxidase activity is not known in detail, although it appears to play some role in regulating the flow of electrons to cytochrome b558.38 Compounding the functional deficit caused by the KKD deletion is the presence of a second mutation in the maternal allele that predicts the nonconservative substitution Arg395 → Trp. While this change may represent a polymorphism, it appears that the substitution of a tryptophan for a strongly basic amino acid such as arginine could lead to some loss of function. Recombinant p67-phox protein carrying this mutation only weakly supports superoxide production in the cell-free NADPH oxidase activation system at a level approximately 15% of normal (Curnutte et al, unpublished results, September 1996). If the Arg395 → Trp substitution is a polymorphism, normal individuals carrying either 1 or 2 alleles encoding this change could have abnormally low levels of NADPH oxidase activity.
Taken together, this series of 6 kindreds shows that a heterogeneous group of mutations are responsible for the p67-phox–deficient form of CGD, a finding consistent with the mutations identified in the 6 individual case reports that have been previously published. Such heterogeneity is also a hallmark of 2 of the other 3 forms of CGD, the X-linked form caused by mutations in the gene encoding gp91-phox and the autosomal recessive type caused by defects in the p22-phox gene. The mutations in the p67-phox gene identified in the patients described here all lead to marked instability of the p67-phox mRNA or protein (or both) and result in undetectable levels of p67-phox, a profound loss of respiratory burst activity, and a relatively severe clinical phenotype.
ACKNOWLEDGMENT
The authors thank Drs Paul Heyworth and Andrew Cross from The Scripps Research Institute for their assistance in measuring NADPH oxidase function in several of the kindreds. The authors also thank Dr Peter Newburger of the University of Massachusetts for performing the p67-phox Northern blots and Dr David Bylund of The Scripps Reference Lab for his help in confirming the mutations in the CGD patients. Finally, the authors thank Isabel Adams of Genentech for her invaluable assistance in the preparation of this manuscript.
Supported in part by Grant No. R01 AI24838 from the National Institutes of Health (to J.T.C.). P.J.P. is a recipient of a grant from the Colombia Agency for Research and Scientific Development (Colciencias).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to John T. Curnutte, MD, PhD, Department of Immunology, Genentech, Inc, 1 DNA Way, South San Francisco, CA 94080.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal