Familial hemophagocytic lymphohistiocytosis (FHL) is a rare but fatal disease in infancy. There are no previous reports on the clonality of T cells in FHL patients. We analyzed here the clonality of β-T cells in 5 FHL patients using an inverse reverse transcriptase-polymerase chain reaction (RT-PCR) of the T-cell receptor variable region gene (TCR V), a joining region gene of the β chain (Jβ)-PCR, a single-strand conformation polymorphism (SSCP), and sequence analysis. A high frequency (15%) of Vβ and V families was observed in 3 of 5 and 4 of 4 patients examined, respectively. In 19 Vβ repertoires, including all highly frequent Vβ, the Jβ-PCR analysis showed restricted usage of the Jβ family, indicating a marked bias to Jβ1 subsets (the mean rate of Jβ1:Jβ2 was 87:13 in 65% of the β-T cells) in widespread β-T cells (in all patients but 1). In all patients, the clonality of specific Vβ-Jβ fragment expanded was confirmed by SSCP and sequence analysis. These results suggest that the existence of clonal expansion and restricted Jβ1 usage of T cells in FHL is genetically associated with the pathogenesis and the immunodysfunction of the disease. These results help to explain some of the abnormal functional behaviors of T cells in FHL and raise new questions regarding the mechanisms responsible for the restricted clonal diversity.
FAMILIAL HEMOPHAGOCYTIC lymphohistiocytosis (FHL) is a rare genetic disorder of the mononuclear phagocyte system characterized by fever, hepatosplenomegaly associated with pancytopenia, hypertriglyceridemia, and hypofibrogenemia.1,2 Despite intensive chemotherapy, most patients with FHL relapse and die of progressive disease.3,4 Only bone marrow transplantation (BMT) has achieved a long remission in some cases.5,6 FHL has been considered a disorder secondary to T-cell dysfunction.7,8Uncontrolled T-cell activation by an abnormal immune response results in a large amount of inflammatory cytokines that promote macrophage infiltration and the formation of a cytokine network.2,9Increased numbers of activated T cells or atypical lymphoid cells are observed in the circulation of affected individuals.4,10High levels of interferon-γ (IFN-γ), soluble interleukin-2 (IL-2) receptor produced by activated T cells, and IL-6 and tumor necrosis factor (TNF) secreted mainly by activated macrophages have been detected in FHL patients.9,11,12 Immunosuppressive drugs, such as cyclosporine A and steroids, are sometimes effective for maintenance therapy in FHL patients.13
The clonal dissemination of T cells has recently been reported in patients with hemophagocytic syndrome. Especially in the majority of patients with Epstein-Barr virus (EBV)-associated hemophagocytic syndrome, infected cells have been shown to proliferate monoclonally.14 15 However, there is no evidence of the clonal origin of T cells in FHL patients. In the present study, we analyzed the clonality of T cells increased in the circulation of 5 FHL patients using an inverse reverse transcriptase-polymerase chain reaction (RT-PCR) for T-cell receptor variable region gene (TCR V), a joining region gene of the β chain (Jβ)-PCR, followed by a single-strand conformation polymorphism (SSCP) and sequence analysis. Our findings suggest that the existence of clonal expansion and restricted Jβ1 usage of T cells in FHL is genetically associated with the pathogenesis and immunodysfunction of the disease.
MATERIALS AND METHODS
Patients.
Samples from a total of 5 patients were available for the study. In the absence of a specific marker for the disease, the diagnoses of all 5 patients were based on the criteria of FHL by the Histiocyte Society and as described elsewhere.1,16,17 Clinical findings, including cytopenia, hypofibrinogenemia, and hypertriglyceridemia, were observed in all patients. They showed the increased histiocytes with hemophagocytosis in peripheral blood, bone marrow, and/or cerebrospinal fluid. Hepatosplenomegaly was also observed in all patients at onset. All of the patients had a family history of affected siblings. No apparent infectious agents, including cytomegalovirus and EBV, were detected in the peripheral blood of all the patients. The natural killer (NK) cell activity and lymphocyte subpopulation at onset in the 5 patients are shown in Table 1. Low NK activity (1% to 9%) was observed in 3 of the 4 patients examined; the NK activity in patient no. 2 was 19% at onset, but it decreased to 1% 1 month after the treatment. An increased number of activated T cells was observed in 4 of the 5 patients examined. Although most of the patients with FHL described in previous reports showed low NK activity or a high number of activated T cells,17,18 these findings were not necessary for the definite diagnosis of FHL.19Chemotherapy, including prednisolone, vincristine, etoposide, cyclophosphamide, methotrexate, 6-mercaptopurine, cytosine arabinoside, or anthracyclines, was administered to these patients. Allogeneic hematopoietic stem cell transplantation (HSCT) was applied for 3 patients. Only the 2 patients who received cord blood stem cell transplantation (CBSCT) have survived, with several relapses; 2 patients died of progressive disease despite chemotherapy, and 1 achieved remission with chemotherapy but died of disease progression after allogeneic BMT.
NK Cell Activity and Lymphocyte Subpopulation of 5 FHL Patients at Onset
| Patient No. . | Age/Sex . | Affected Siblings . | NK Activity (%)* . | CD3 (%) . | CD19 (%) . | CD56 (%) . | CD3/DR (%)† . | CD4 (%) . | CD8 (%) . | Outcome . |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 mo/M | + | NT | 82.5 | 6.8 | 7.2 | 63.8 | 11.4 | Died of PD after BMT (13 mo) | |
| 2 | 18 mo/F | + | 19.0 ()‡ | 77.9 | 10.3 | 12.8 | Died of PD (3.5 mo) | |||
| 3 | 2 mo/M | + | 85.6 | 4.7 | 8.8 | 57.7 | 28.1 | Alive after CBSCT (+23 mo) | ||
| 4 | 17 mo/F | + | 59.4 | 29.2 | 9.1 | 27.3 | 25.3 | Alive after CBSCT (+35 mo) | ||
| 5 | 1 mo/M | + | 67.1 | 15.8 | 14.6 | 0.5 | 47.1 | 22.9 | Died of PD (3 mo) |
| Patient No. . | Age/Sex . | Affected Siblings . | NK Activity (%)* . | CD3 (%) . | CD19 (%) . | CD56 (%) . | CD3/DR (%)† . | CD4 (%) . | CD8 (%) . | Outcome . |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 mo/M | + | NT | 82.5 | 6.8 | 7.2 | 63.8 | 11.4 | Died of PD after BMT (13 mo) | |
| 2 | 18 mo/F | + | 19.0 ()‡ | 77.9 | 10.3 | 12.8 | Died of PD (3.5 mo) | |||
| 3 | 2 mo/M | + | 85.6 | 4.7 | 8.8 | 57.7 | 28.1 | Alive after CBSCT (+23 mo) | ||
| 4 | 17 mo/F | + | 59.4 | 29.2 | 9.1 | 27.3 | 25.3 | Alive after CBSCT (+35 mo) | ||
| 5 | 1 mo/M | + | 67.1 | 15.8 | 14.6 | 0.5 | 47.1 | 22.9 | Died of PD (3 mo) |
Vβ and Vα analysis of T cells in FHL patients.
Blood samples of peripheral blood (4 patients) or bone marrow (1 patient) from the FHL patients were obtained at onset and showed an increased number of atypical lymphoid cells or histiocytes with hemophagocytosis. Mononuclear cells were isolated by Ficoll-Hypaque (Pharmacia, Uppsala, Sweden) density gradient centrifugation, and total cellular RNA was extracted according to a previously described method.20 21
The T-cell receptor β and α chain variable region (TCR Vβ and Vα) repertoires were analyzed by the newly devised RT-PCR method.22 Briefly, double-stranded cDNA was synthesized, followed by circularization and the inverse PCR using 2 constant region primers that are in opposite orientations.23Oligo(dT)-primed double-stranded cDNA was synthesized from 1 μg of total RNA using Moloney murine leukemia virus-derived reverse transcriptase, RNase H, Escherichia coli DNA polymerase I, and E coli DNA ligase, followed by incubation with T4 DNA polymerase for blunt-end formation. The blunt-ended cDNA was circularized with T4 DNA ligase in a volume of 10 μL. The ligated material (5 μL) was used as template for the PCR. The PCR constant (C) primers used were as follows: Cα forward primer, Cα inverse primer, Cβ forward primer, and Cβ inverse primer. After 35 cycles of PCR (denaturation at 95°C for 0.5 minutes, annealing at 62°C for 0.5 minutes, and extension at 72°C for 1 minute), the Klenow fragment of E coli DNA polymerase I was added to ensure full-length DNA synthesis. The sequences of Cβ and Cα primers used are shown in Table 2. The common nucleotide sequences in 2 constant regions of human TCR β chain genes were used.
Primers Used for PCR Assays
| No. . | Primer Orientation . | Sequence . |
|---|---|---|
| 1 | C α forward | 5′-GGG TCG ACG ACC TCA TGT CTA GCA CAG T-3′ |
| 2 | C α inverse | 5′-GCA TGC GGC CGC CCT GCT ATG CTG TGT GTC T-3′ |
| 3* | C β forward | 5′-GGG TCG ACC TGT GCA CCT CCT TCC CAT T-3′ |
| 4* | C β inverse | 5′-GCA TGC GGC CGC ATG GCC ATG GTC AAG AGA-3′ |
| J β 1 Group (anti) | ||
| 5 | J β 1.1 | 5′-AAC TGT GAG TCT GGT GCC TTG-3′ |
| 6 | J β 1.2 | 5′-AAC GGT TAA CCT GGT CCC CGA-3′ |
| 7 | J β 1.3 | 5′-AAC AGT GAG CCA ACT TCC CTC-3′ |
| 8 | J β 1.4 | 5′-GAC AGA GAG CTG GGT TCC ACT-3′ |
| 9 | J β 1.5 | 5′-GAT GGA GAG TCG AGT CCC ATC-3′ |
| 10 | J β 1.6 | 5′-CAC AGT GAG CCT GGT CCC ATT-3′ |
| J β 2 Group (anti) | ||
| 11 | J β 2.1 | 5′-CAC GGT GAG CCG TGT CCC TGG-3′ |
| 12 | J β 2.2 | 5′-TAC GGT CAG CCT AGA GCC TTC-3′ |
| 13 | J β 2.3 | 5′-CAC TGT CAG CCG GGT GCC TGG-3′ |
| 14 | J β 2.4 | 5′-CAC TGA GAG CCG GGT CCC GGC-3′ |
| 15 | J β 2.5 | 5′-CAC CAG GAG CCG CGT GCC TGG-3′ |
| 16 | J β 2.6 | 5′-CAC GGT CAG CCT GCT GCC GGC-3′ |
| 17 | J β 2.7 | 5′-GAC CGT GAG CCT GGT GCC CGG-3′ |
| V β primers (sense) | ||
| 18 | V β 1 | 5′-AAG AGA GAG CAA AAG GAA ACA-3′ |
| 19 | V β 2 | 5′-CAA TGA GGG CTG CAA GGC CAC-3′ |
| 20 | V β 3 | 5′-CGA CAA GAC CCA GGT CTG GG-3′ |
| 21 | V β 4 | 5′-TGA GGC CAC ATA TGA GAG TGG-3′ |
| 22 | V β 5a | 5′-TCA GTG AGA CAC AGA GAA AC-3′ |
| 23 | V β 5b | 5′-AAC TTC CCT GAT CGA TTC TC-3′ |
| 24 | V β 6 | 5′-TCA GGT GTG ATC CAA TTT C-3′ |
| 25 | V β 8 | 5′-CAA CAA CGT TCC GAT AGA TGA T-3′ |
| 26 | V β 12 | 5′-ACT CTG AGA TGT CAC CAG ACT-3′ |
| 27 | V β 13 | 5′-ACA CTG CAG TGT GCC CAG GAT-3′ |
| 28 | V β 15 | 5′-CCT TTG ATG TCA AAG ATA TAA-3′ |
| V α primers (sense) | ||
| 29 | V α 3 | 5′-GAT TAA GAG TCA CGC TTG ACA-3′ |
| 30 | V α 9 | 5′-CAC ATC TCT AGA GAG AGC ATC-3′ |
| No. . | Primer Orientation . | Sequence . |
|---|---|---|
| 1 | C α forward | 5′-GGG TCG ACG ACC TCA TGT CTA GCA CAG T-3′ |
| 2 | C α inverse | 5′-GCA TGC GGC CGC CCT GCT ATG CTG TGT GTC T-3′ |
| 3* | C β forward | 5′-GGG TCG ACC TGT GCA CCT CCT TCC CAT T-3′ |
| 4* | C β inverse | 5′-GCA TGC GGC CGC ATG GCC ATG GTC AAG AGA-3′ |
| J β 1 Group (anti) | ||
| 5 | J β 1.1 | 5′-AAC TGT GAG TCT GGT GCC TTG-3′ |
| 6 | J β 1.2 | 5′-AAC GGT TAA CCT GGT CCC CGA-3′ |
| 7 | J β 1.3 | 5′-AAC AGT GAG CCA ACT TCC CTC-3′ |
| 8 | J β 1.4 | 5′-GAC AGA GAG CTG GGT TCC ACT-3′ |
| 9 | J β 1.5 | 5′-GAT GGA GAG TCG AGT CCC ATC-3′ |
| 10 | J β 1.6 | 5′-CAC AGT GAG CCT GGT CCC ATT-3′ |
| J β 2 Group (anti) | ||
| 11 | J β 2.1 | 5′-CAC GGT GAG CCG TGT CCC TGG-3′ |
| 12 | J β 2.2 | 5′-TAC GGT CAG CCT AGA GCC TTC-3′ |
| 13 | J β 2.3 | 5′-CAC TGT CAG CCG GGT GCC TGG-3′ |
| 14 | J β 2.4 | 5′-CAC TGA GAG CCG GGT CCC GGC-3′ |
| 15 | J β 2.5 | 5′-CAC CAG GAG CCG CGT GCC TGG-3′ |
| 16 | J β 2.6 | 5′-CAC GGT CAG CCT GCT GCC GGC-3′ |
| 17 | J β 2.7 | 5′-GAC CGT GAG CCT GGT GCC CGG-3′ |
| V β primers (sense) | ||
| 18 | V β 1 | 5′-AAG AGA GAG CAA AAG GAA ACA-3′ |
| 19 | V β 2 | 5′-CAA TGA GGG CTG CAA GGC CAC-3′ |
| 20 | V β 3 | 5′-CGA CAA GAC CCA GGT CTG GG-3′ |
| 21 | V β 4 | 5′-TGA GGC CAC ATA TGA GAG TGG-3′ |
| 22 | V β 5a | 5′-TCA GTG AGA CAC AGA GAA AC-3′ |
| 23 | V β 5b | 5′-AAC TTC CCT GAT CGA TTC TC-3′ |
| 24 | V β 6 | 5′-TCA GGT GTG ATC CAA TTT C-3′ |
| 25 | V β 8 | 5′-CAA CAA CGT TCC GAT AGA TGA T-3′ |
| 26 | V β 12 | 5′-ACT CTG AGA TGT CAC CAG ACT-3′ |
| 27 | V β 13 | 5′-ACA CTG CAG TGT GCC CAG GAT-3′ |
| 28 | V β 15 | 5′-CCT TTG ATG TCA AAG ATA TAA-3′ |
| V α primers (sense) | ||
| 29 | V α 3 | 5′-GAT TAA GAG TCA CGC TTG ACA-3′ |
| 30 | V α 9 | 5′-CAC ATC TCT AGA GAG AGC ATC-3′ |
The common nucleotide sequences in 2 constant regions of the human TCR β chain genes were used.39
Two hundred nanograms of each Vβ fragment (from Vβ1 to Vβ20) or Vα fragment (from Vα1 to Vα18, Vα21, and Vα24) was dotted on the filters. Each V-specific fragment was prepared from the series of HBVT/HBVP or HAVT/HAVP plasmids originated from thymus or peripheral T cells.24-26 Fifteen microliters of amplified PCR products was labeled by α-32P-dCTP and hybridized to the filter, including V segments. Using densitometry, a semiquantitative assessment of V gene usage was made from the amount of hybridized products.
In vitro stimulation by superantigen.
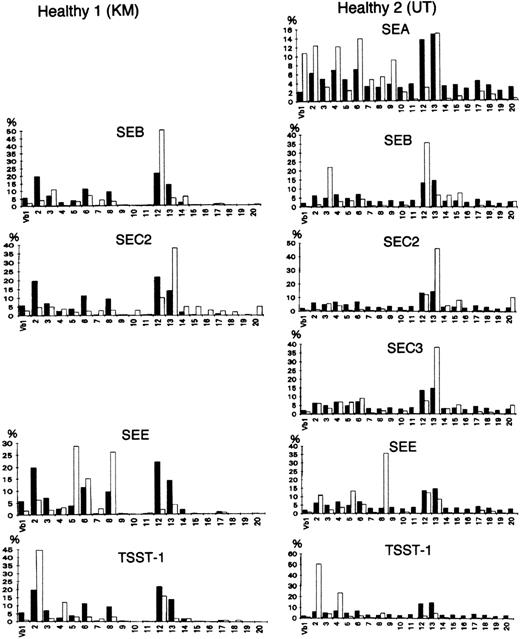
Staphylococcusaureus enterotoxin (SEB, SEA, SEC2, SEC3, and SEE) and toxic shock syndrome toxin 1 (TSST 1) were used as bacterial superantigens to stimulate T cells in a Vβ-specific fashion. Control peripheral T cells were isolated from the blood of 2 healthy donors and analyzed before or after stimulation with bacterial superantigen (1 μg/mL; Toxin Technology, Madison, WI). A solution of the protein (1 μg/mL) was incubated on the well surface for 8 hours at 4°C. Nonadherent protein was removed by extensive washing. This plastic-adherent superantigens were used to stimulate the peripheral blood T cells (5 × l05/mL). Three days later, live cells were collected and RNA was prepared for the analysis of TCR V repertoires. As shown in Fig 1, although the in vitro stimulation with a superantigen appears to have almost no effect on the Vα repertoire (data not shown), the results obtained with the new PCR method suggested that all of the toxins used preferentially stimulated T cells expressing particular Vβs, consistent with the results of previous studies.27
Vβ repertoire stimulated with superantigens in healthy controls. (▪) Healthy donors as the control; (□) stimulated with a superantigen. Peripheral T cells were isolated from the blood of 2 donors and analyzed before or after in vitro stimulation with a bacterial superantigen (1 μg/mL) for 3 days. All of the toxins preferentially stimulated T cells expressing particular Vβs.27
Vβ repertoire stimulated with superantigens in healthy controls. (▪) Healthy donors as the control; (□) stimulated with a superantigen. Peripheral T cells were isolated from the blood of 2 donors and analyzed before or after in vitro stimulation with a bacterial superantigen (1 μg/mL) for 3 days. All of the toxins preferentially stimulated T cells expressing particular Vβs.27
Clonal analysis of Vβ family.
A clonal analysis was then performed using PCR with 13 sets of Jβ primers (Jβ-PCR) and an SSCP and sequence analysis.28,29The sequences of Vβ and Jβ primers used are also shown in Table2.30 Besides the FHL specimens, as control subjects, 3 infants (a 3-month-old healthy infant, a 2-month-old patient with hepatitis, and a 2-year-old patient with diarrhea) were investigated. cDNA was amplified using primers specific to each Vβ family and Jβ primer. The amplified products (Vβ-Jβ) were examined with the same volume of loading solution (0.05% xylene cyanol, 20 mmol/L EDTA, and 95% formamide), incubated at 96°C for 5 minutes, and rapidly chilled on ice. Samples were electrophoresed at a constant voltage of 100 V for 3 hours on a 0.5× MDE gel (AT Biochem, Malvern, PA). The gel was stained with SYBR green (1 μg/mL; FMC Bio Products, Rockland, ME), and the bands were detected and photographed on a UV translluminator.31 Finally, sequence analysis was performed. The Vβ-Jβ amplification fragments were purified by electrophoresis in low melting point agarose. Fragments were ligated into pT7Blue T-Vector (Novagen, Madison, WI). After transformation of Epicurian Coli XL II-Blue (Stratagene, La Jolla, CA), 6 individual colonies were selected for sequencing by ABI PRISM 377 DNA sequencer (Perkin-Elmer, Forster City, CA). Data were analyzed by PC-Gene (IntelliGenetics, Mountain View, CA).
RESULTS
TCR Vβ and Vα repertoire in FHL patients.
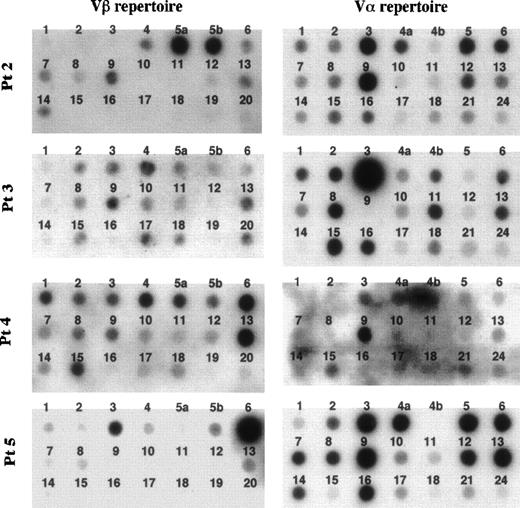
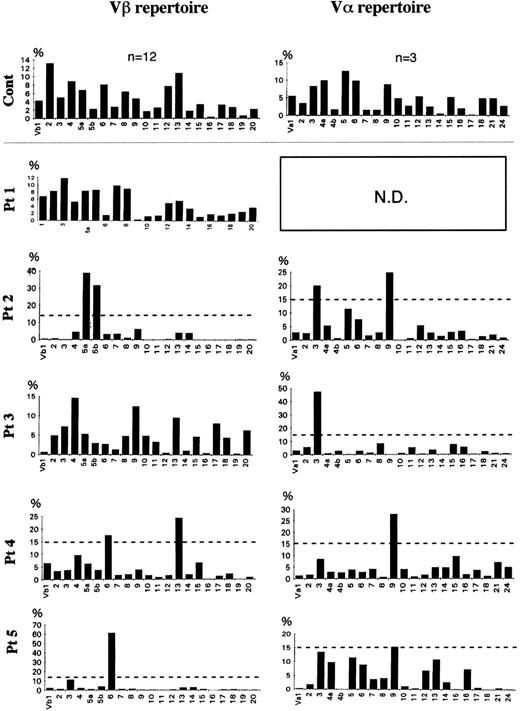
The frequency of the TCR Vβ repertoire in peripheral αβ-T cells of 4 FHL patients (patients no. 1 through 4) and in bone marrow αβ-T cells of patient no. 5 is shown in Figs 2 and 3. Although there was no preferential usage of a Vβ family member in patients no. 1 and 3, high expressions (>15%) of Vβ5a (39%) and 5b (32%) in patient no. 2, of Vβ6 (17.5%) and 13 (24%) in patient no. 4, and of Vβ6 (61%) in patient no. 5 were observed. The TCR Vα repertoire was also analyzed in the 4 FHL patients except patient no. 1. High frequencies (>15%) of Vα3 (20%) and 9 (25%) were detected in patient no. 2, of Vα3 (47%) in patient no. 3, and of Vα9 in patient no. 4 (28%) and patient no. 5 (15.4%). There was no correlation between Vβ and Vα family usage in the FHL patients.
Dot blotting of TCR Vβ and V repertoires of 5 patients with FHL. Fifteen microliters of products that was amplified by an inverse RT-PCR using the set of constant region primers was labeled by -32P-dCTP and hybridized to the Vβ and V filter prepared with 200 ng of each variable gene segment, respectively.
Dot blotting of TCR Vβ and V repertoires of 5 patients with FHL. Fifteen microliters of products that was amplified by an inverse RT-PCR using the set of constant region primers was labeled by -32P-dCTP and hybridized to the Vβ and V filter prepared with 200 ng of each variable gene segment, respectively.
Frequency of TCR Vβ and V family in FHL patients. With densitometry, a semiquantitative assessment of V gene usage was made from the amount of hybridized products. The dotted line shows 15% of β-T cells. TCR V gene segments were regarded as highly expressed when their relative frequency was more than 15%. A highly expressed Vβ family member was observed in 3 of the 5 patients: Vβ5a and 5b of patient no. 2 and Vβ6 of both patients no. 4 and 5. A high frequency of a V family member was detected in all 5 patients: V3 and 9 in patient no. 2, V3 in patient no. 3, and V9 in patients no. 4 and 5.
Frequency of TCR Vβ and V family in FHL patients. With densitometry, a semiquantitative assessment of V gene usage was made from the amount of hybridized products. The dotted line shows 15% of β-T cells. TCR V gene segments were regarded as highly expressed when their relative frequency was more than 15%. A highly expressed Vβ family member was observed in 3 of the 5 patients: Vβ5a and 5b of patient no. 2 and Vβ6 of both patients no. 4 and 5. A high frequency of a V family member was detected in all 5 patients: V3 and 9 in patient no. 2, V3 in patient no. 3, and V9 in patients no. 4 and 5.
Jβ usage in Vβ of FHL patients.
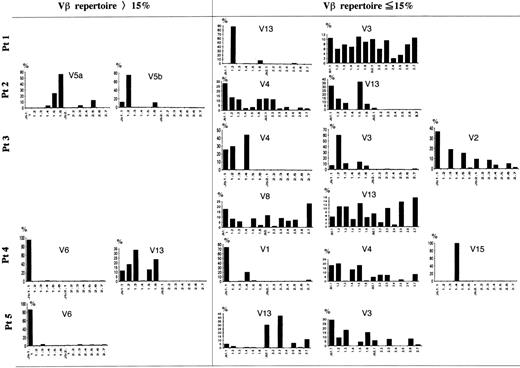
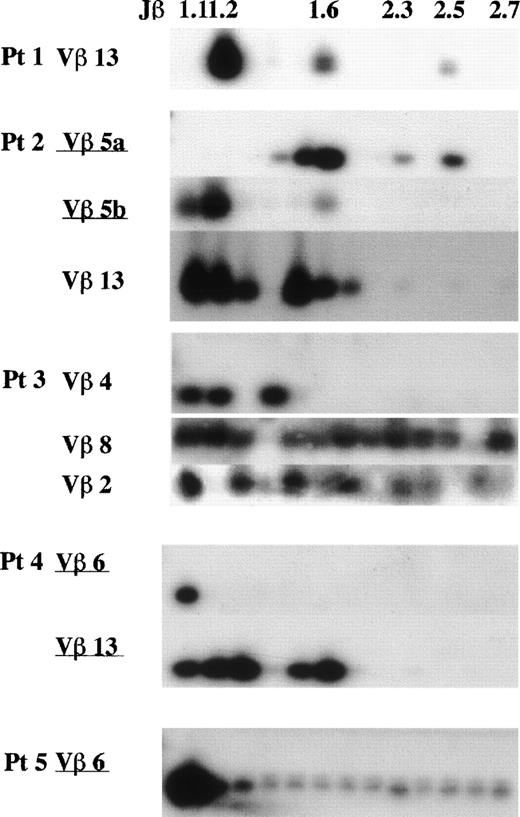
A Jβ-PCR was then performed on the highly expressed Vβ subset(s) (>15%) for each patient using each Vβ specific primer and a set of Jβ primers (Figs 4 and5). In patient no. 2, Vβ5a and 5b used Jβ1.5 (24%) or Jβ1.6 (56%) and Jβ1.2 (75%), respectively. In patient no. 4, Vβ6 exclusively used Jβ1.1 (95%), whereas Vβ13 used several genes of only the Jβ1 subgroup (99%). Vβ6 in patient no. 5 exclusively used Jβ1.1 (86%). These findings show that the highly frequent Vβ families observed in FHL patients use only the Jβ1 subgroup.
Southern blot analysis of PCR-amplified Vβ-Jβ segments in FHL patients. The amplified products were respectively hybridized with each V fragment probe. *Underlining shows a Vβ gene with high frequency (15%). Preferential Jβ usage in highly frequent Vβ families is observed.
Southern blot analysis of PCR-amplified Vβ-Jβ segments in FHL patients. The amplified products were respectively hybridized with each V fragment probe. *Underlining shows a Vβ gene with high frequency (15%). Preferential Jβ usage in highly frequent Vβ families is observed.
Frequency of Jβ usage in Vβ repertoire in FHL patients. Autoradiograms were scanned by computerized densitometry, providing an absolute value for each autoradiographic spot. Each Vβ-Jβ spot is expressed as a percentage of the sum of all of the respective Vβ-Jβ signals detected on the autoradiogram. Some Vβ family members expressed within the normal range were also investigated in each patient. In the TCR V genes with a high frequency (15%), only 1 member of the Jβ1 gene subgroup was highly expressed in 3 patients. Even among the other Vβ family members expressed within the normal range, 1 of Jβ1 with overexpression and Jβ prediposed bias were observed: Vβ13 in patient no. 1, Vβ3 and 4 in patient no. 3, Vβ1 and 15 in patient no. 4, and Vβ13 in patient no. 5.
Frequency of Jβ usage in Vβ repertoire in FHL patients. Autoradiograms were scanned by computerized densitometry, providing an absolute value for each autoradiographic spot. Each Vβ-Jβ spot is expressed as a percentage of the sum of all of the respective Vβ-Jβ signals detected on the autoradiogram. Some Vβ family members expressed within the normal range were also investigated in each patient. In the TCR V genes with a high frequency (15%), only 1 member of the Jβ1 gene subgroup was highly expressed in 3 patients. Even among the other Vβ family members expressed within the normal range, 1 of Jβ1 with overexpression and Jβ prediposed bias were observed: Vβ13 in patient no. 1, Vβ3 and 4 in patient no. 3, Vβ1 and 15 in patient no. 4, and Vβ13 in patient no. 5.
The transplantation of peripheral mononuclear cells in patient no. 1 toscid mice caused an FHL-like physiological disorder. CD4−CD8− αβ-T cells were infiltrated in mice organs and the clonal expansion of Vβ13-Jβ1.2 was recognized, which was described elsewhere.32 Therefore, the clonality of Vβ13 (5.6%) in peripheral αβ-T cells was studied in patient no. 1 and the accumulation of the same Jβ1.2 (88%) was observed (Figs 4 and 5). Several Vβ subsets with a frequency rate less than 15% were investigated in each patient as well at random. These Vβ family members used a variety of Jβ gene families, suggesting polyclonality: Vβ3 (12%) in patient no. 1, Vβ4 (4%) in patient no. 2, Vβ2 (4.9%) and 8 (4.8%) in patient no. 3, Vβ4 (9.6%) in patient no. 4, and Vβ3 (11.6%) in patient no. 5. Polyclonal Vβ13 was recognized in patients no. 2 (4%), 3 (9.5%), and 5 (3%) (Figs 4 and 5).
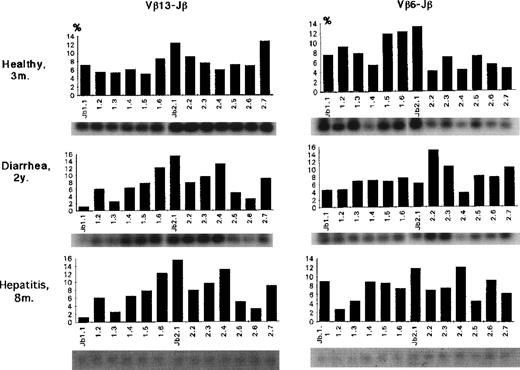
Even in part of those Vβ subsets, a bias of Jβ1 gene usage was observed. Both Vβ1 and Vβ15 in patient no. 4 exclusively used Jβ1.1 (73%) and Jβ1.4 (99%), suggesting clonality in each of these Vβ family members. The Jβ usage of Vβ13 and 6 in 3 control individuals is shown in Fig 6. Compared with FHL patients, both Vβ6 and Vβ13 generally used a variety of 13 Jβ genes with a bias for Jβ2; the average Jβ1:Jβ2 ratios as a whole were 42:58 for Vβ6 and 33:67 for Vβ13. There was no Jβ gene usage greater than 20%.
Jβ usage of Vβ13 or Vβ6 in control infants. Both Vβ6 and Vβ13 generally used a variety of 13 Jβ genes with a bias for Jβ2; the average Jβ1:Jβ2 ratios as a whole were 42:58 for Vβ6 and 33:67 for Vβ13. There was no Jβ gene usage greater than 20%.
Jβ usage of Vβ13 or Vβ6 in control infants. Both Vβ6 and Vβ13 generally used a variety of 13 Jβ genes with a bias for Jβ2; the average Jβ1:Jβ2 ratios as a whole were 42:58 for Vβ6 and 33:67 for Vβ13. There was no Jβ gene usage greater than 20%.
SSCP analysis.
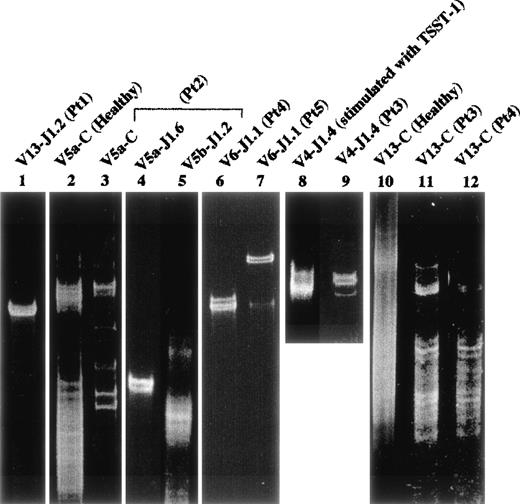
Part of the PCR products amplified by each Vβ and Jβ primer were examined by the SSCP analysis to investigate the clonality of Vβ-Jβ families. On the SSCP gel (Fig 7), there were no distinct bands detectable in the amplified products of each Vβ family of the normal control or in expanded T cells (Vβ4-Jβ1.4) by the in vitro stimulation with TSST-1 superantigen (Fig 7). By contrast, a distinct band(s) was detected in amplified products of several highly frequent Vβ family members in the FHL patients; Vβ5a-Jβ1.6 in patient no. 2 (lane 4) and Vβ6-Jβ1.1 in both patients no. 4 and 5 (lanes 6 and 7). Even in low expressed Vβ repertoires, a clear band(s) suggesting clonality was observed: Vβ13-Jβ1.2 in patient no. 1 (lane 1) and Vβ1 and 15 in patient no. 4 (not shown). These data suggest that some restricted αβ-T–cell clone(s) dominantly proliferates in the peripheral blood or bone marrow of FHL patients.
Clonality by SSCP analysis. Healthy peripheral blood mononucler cells (PBMC) as a control were investigated. Lane 2 (Vβ5a-Cβ) and lane 10 (Vβ13-Cβ); lane 8, Vβ4-Jβ1.4 of healthy PBMC stimulated with TSST1; lane 1, Vβ13-Jβ1.2 in patient no. 1; lanes 3, 4, and 5, Vβ5a-Cβ, Vβ5a-Jβ1.6, and Vβ5b-Jβ1.2 in patient no. 2, respectively; lanes 6 and 12, Vβ6-Jβ1.1 and Vβ13-Cβ in patient no. 4, respectively; lane 7, Vβ6-Jβ1.1 of bone marrow in patient no. 5; lanes 9 and 11, Vβ4-Jβ1.4 and Vβ13-Cβ in patient no. 3, respectively. Smears are shown in lanes 2, 5, 8, 10, 11, and 12. Distinct bands are observed in lanes 3, 4, 6, 7, 9, and 1, suggesting clonal expansion.
Clonality by SSCP analysis. Healthy peripheral blood mononucler cells (PBMC) as a control were investigated. Lane 2 (Vβ5a-Cβ) and lane 10 (Vβ13-Cβ); lane 8, Vβ4-Jβ1.4 of healthy PBMC stimulated with TSST1; lane 1, Vβ13-Jβ1.2 in patient no. 1; lanes 3, 4, and 5, Vβ5a-Cβ, Vβ5a-Jβ1.6, and Vβ5b-Jβ1.2 in patient no. 2, respectively; lanes 6 and 12, Vβ6-Jβ1.1 and Vβ13-Cβ in patient no. 4, respectively; lane 7, Vβ6-Jβ1.1 of bone marrow in patient no. 5; lanes 9 and 11, Vβ4-Jβ1.4 and Vβ13-Cβ in patient no. 3, respectively. Smears are shown in lanes 2, 5, 8, 10, 11, and 12. Distinct bands are observed in lanes 3, 4, 6, 7, 9, and 1, suggesting clonal expansion.
Sequence analysis of TCR β-chain transcript.
To confirm that the SSCP analysis of the predominant Vβ fragment amplified from each patient represented the clonal expansion, the fragments were subcloned into plasmids and sequenced. Six individual colonies were analyzed in each fragment. As summarized in Table 3, the expressed Vβ gene in patient no. 1 was Vβ13.6, and residues comprising the NDβN junction were TSAL. A similar analysis was performed on the dominant Vβ gene for the remaining 4 patients. The sequence identified in 83% and 50% of the recombinant plasmids were identical, which indicates clonally expanded cells.
Structural Features of TCR β-Chain Transcripts Expressed by FHL Clone(s)
| Patient No. . | V β Gene . | J β Gene . | Junctional Sequence . | 6 Subclones Examined3-150 . | ||
|---|---|---|---|---|---|---|
| V β . | ND β N . | J β . | ||||
| 1 | 13.6 | 1.2 | C3-151 ASS | TSAL | TTGTT | 5 |
| TGT GCC AGC AGT | TAC TCG AGA CTG | ACC TAT GGC TAC ACC | ||||
| 2 | 5.1 | 1.6 | C ASS | PAGGA | SPLH | 5 |
| TGC GCC AGC AGC | TTC GCC GGT GGG AAT | TCA CCC CTC CAC | ||||
| 3 | 4.1 | 1.4 | CSV | TGVP | MLACP | 5 |
| TGC AGC GTT | TGG GGG GTT CCG | ATG AAA AAC TGT TTT | ||||
| 4 | 6.4 | 1.1 | CASS | PPGGLA | TGAP | 3 |
| TGT GCC AGC AGC | CCC TTC GAG GGA TTG AAC | ACT GAA GCT TTC | ||||
| 6.4 | 1.1 | CAS | AATGPG | GAP | 2 | |
| TGT GCC AGC | AGG GAC ACG GGT TTT GAG | CAA GCT TTC | ||||
| 5 | 6.5 | 1.1 | CASS | AAAV | TGAP | 5 |
| TGT GCC AGC AGC | CGG GAC AGG GTT | ACT GAA GCT TTC | ||||
| Patient No. . | V β Gene . | J β Gene . | Junctional Sequence . | 6 Subclones Examined3-150 . | ||
|---|---|---|---|---|---|---|
| V β . | ND β N . | J β . | ||||
| 1 | 13.6 | 1.2 | C3-151 ASS | TSAL | TTGTT | 5 |
| TGT GCC AGC AGT | TAC TCG AGA CTG | ACC TAT GGC TAC ACC | ||||
| 2 | 5.1 | 1.6 | C ASS | PAGGA | SPLH | 5 |
| TGC GCC AGC AGC | TTC GCC GGT GGG AAT | TCA CCC CTC CAC | ||||
| 3 | 4.1 | 1.4 | CSV | TGVP | MLACP | 5 |
| TGC AGC GTT | TGG GGG GTT CCG | ATG AAA AAC TGT TTT | ||||
| 4 | 6.4 | 1.1 | CASS | PPGGLA | TGAP | 3 |
| TGT GCC AGC AGC | CCC TTC GAG GGA TTG AAC | ACT GAA GCT TTC | ||||
| 6.4 | 1.1 | CAS | AATGPG | GAP | 2 | |
| TGT GCC AGC | AGG GAC ACG GGT TTT GAG | CAA GCT TTC | ||||
| 5 | 6.5 | 1.1 | CASS | AAAV | TGAP | 5 |
| TGT GCC AGC AGC | CGG GAC AGG GTT | ACT GAA GCT TTC | ||||
There are submembers in some Vβ families. It was indicated that Vβ5a in patient no. 2 is Vβ5.1, that Vβ4 in patient no. 3 is Vβ4.1, and that Vβ6 in patients no. 4 and 5 is Vβ6.4 and Vβ6.5, respectively. In all patients, the junctional sequences expressed by each clone were unique.
Table 4 shows the summarized data. In all 5 FHL patients, the clonality of αβ-T cells was demonstrated. The high frequency of 5 Vβ subsets was found in 3 FHL patients. Three of these five Vβ subsets clonally expanded using 1 of 6 Jβ1 genes. A total of 19 Vβ subsets, including 14 within the normal range, were investigated. Of the 14 Vβ with a frequency rate less than 15%, 4 Vβ were also suggested or confirmed to be a clone(s). Fifteen of the 19 Vβ subsets showed a bias for Jβ1 subsets (>70%). In patient no. 2, 79.3% of the peripheral αβ-T cells use Jβ1 over Jβ2 (89.7:10.3). The marked bias for Jβ1 usage (the mean rate of Jβ1:Jβ2 was 87:13) was to be confirmed by densitometry in all but 1 patient (patient no. 1).
Clonal Expansion and Jβ Bias of β-T Cells in 5 FHL Patients
| Patient No. . | High Frequency (>15%) . | Clonal Expansion Vβ-Jβ4-150 . | Vβ With Jβ1 Bias (>70%)/ Investigated Vβ4-151 . | Comparison Value Jβ1:Jβ2 (αβ-T cells investigated)‡ . | |
|---|---|---|---|---|---|
| Vβ . | Vα . | ||||
| 1 (PB)4-153 | No | Not done | 13-1.2 (88%)4-155 | 13/3, 13 | 48.2:51.8 (23.9%) |
| 2 (PB) | 5a (39%), 5b (32%) | 9 (25%), 3 (20%) | 5a-1.6 (56%) | 4, 5a, 5b, 13/4, 5a, 5b, 13 | 89.7:10.3 (79.3%) |
| 3 (PB) | No | 3 (47%) | 4-1.4 (44%) | 2, 3, 4/2, 3, 4, 8, 13 | 77.4:22.6 (41%) |
| 4 (PB) | 6 (17.5%), 13 (24%) | 9 (28%) | 6-1.1 (95%), 1-1.1 (74%), 15-1.4 (98%) | 1, 4, 6, 13, 15/1, 4, 6, 13, 15 | 94.4:5.5 (64.4%) |
| 5 (BM) | 6 (61%) | 9 (15.4%) | 6-1.1 (86%) | 3, 6/3, 6, 13 | 86.6:13.4 (76%) |
| Patient No. . | High Frequency (>15%) . | Clonal Expansion Vβ-Jβ4-150 . | Vβ With Jβ1 Bias (>70%)/ Investigated Vβ4-151 . | Comparison Value Jβ1:Jβ2 (αβ-T cells investigated)‡ . | |
|---|---|---|---|---|---|
| Vβ . | Vα . | ||||
| 1 (PB)4-153 | No | Not done | 13-1.2 (88%)4-155 | 13/3, 13 | 48.2:51.8 (23.9%) |
| 2 (PB) | 5a (39%), 5b (32%) | 9 (25%), 3 (20%) | 5a-1.6 (56%) | 4, 5a, 5b, 13/4, 5a, 5b, 13 | 89.7:10.3 (79.3%) |
| 3 (PB) | No | 3 (47%) | 4-1.4 (44%) | 2, 3, 4/2, 3, 4, 8, 13 | 77.4:22.6 (41%) |
| 4 (PB) | 6 (17.5%), 13 (24%) | 9 (28%) | 6-1.1 (95%), 1-1.1 (74%), 15-1.4 (98%) | 1, 4, 6, 13, 15/1, 4, 6, 13, 15 | 94.4:5.5 (64.4%) |
| 5 (BM) | 6 (61%) | 9 (15.4%) | 6-1.1 (86%) | 3, 6/3, 6, 13 | 86.6:13.4 (76%) |
The marked bias for Jβ1 usage (the mean rate of Jβ1:Jβ2 was 87:13) was confirmed due to the calculation by densitometory in all but 1 patient (patient no. 1).
By SSCP analysis, all Vβ/Jβ fragments were suggested to be clones. All clones (7/7) used 1 member of the Jβ1 gene family. In Table 3, the sequencing of Vβ-Jβ fragments confirmed to be clones.
Vβ in each patient was investigated at random for the Jβ usage analysis. On the right side, each Vβ family member indicates the bias to Jβ1 usage. Fifteen of 19 Vβ repertoires investigated showed a bias of more than 70% Jβ1 genes usage.
For the Vβ/Jβ fragments investigated, the comparison rate (%) of Jβ1 genes/Jβ2 genes was calculated.
In all patients except patient no. 5, mononuclear cells of peripheral blood were analyzed.
Rate of 1 specific Jβ gene used in 13 types of Jβ genes.
DISCUSSION
The genetic basis of FHL, a rare inherited disorder characterized by multivisceral infiltration by lymphocytes and histiocytes, is still unknown. Although it is now accepted that FHL is a disorder of T-cell dysfuction, clonality of T cells in FHL patients has not been reported. In the present study, we obtained evidence of clonal expansion of αβ-T cells with Jβ1 bias in all 5 FHL patients studied. Immunodysfunction has been speculated in widespread pan-αβ-T cells. Because activated T cells and various cytokines derived from T cells are increased in the circulation of FHL individuals, it can now be speculated that those clonally expanded T cells may produce high levels of inflammatory cytokines followed by the activation of macrophages with hemophagocytosis.
Concentrating on the TCR β chain, a paucity of clonotypic T-cell expansion has been demonstrated in the peripheral blood of healthy individuals by using the PCR method and a subsequent SSCP analysis.31 A number of recent reports have established that oligoclonality and/or clonal expansion is a common feature of the CD8+ T-cell population. Posnett et al33documented that clonal TCR αβ T-cell populations are frequent in normal subjects more than 65 of age, and they showed that these populations are usually clones of CD8+CD28− cells. In addition, Morley et al34 reported that oligoclonal CD8+ T cells are preferentially expanded in the CD57+ subset and that the oligoclonal expansion is a characteristic feature of the normal immune system. In the present study, low NK activity and an increased number of activated T cells (CD3+/HLA-DR+ cells) were observed in 4 of 4 and 4 of 5 FHL patients examined, respectively. In patient no. 1, with an increased number of activated T cells, the clonal T lymphocyte with Vβ13.6-Jβ1.2 was CD4−CD8− (double negative [DN]), and these T cells caused an FHL-like physiological disorder in transplanted scid mice.32 In a previous analysis of normal donors, the expressions of Vβ2, 8, 11, and 13 were reported to be markedly increased in DN αβ-T cells.35 Therefore, it is unlikely that clonally expanded T-cell subsets (Vβ5a, 6, and 6 in patients no. 2, 4, and 5, respectively) are DN. As described above, it appears that expanded T-cell subsets in each FHL patient are not limited to only one common T-cell subclass. It would be worthwhile to determine whether the clonal T cells expand by their own abnormality or by the reaction to some antigens or superantigens. Based on the preferential usage of different Vβ and Jβ genes in each of our patients, we propose that FHL is not a superantigen-mediated disease. In FHL patients who exhibit an excess of HLA DR+CD3+ T cells, it would be worthwhile to check for an abnormal repertoire in separated HLA DR+ and HLA DR− T cells. This study would help to recognize whether possible oligoclonality reflects selected T-cell activation or a truly abnormal repertoire.
In the present study, the highly frequent Vβ family members observed in FHL patients used only the Jβ1 subgroup. In addition, a markedly inverted bias to Jβ1 usage (the mean Jβ1:Jβ2 ratio of 87:13) was observed in many αβ-T cells (65%), suggesting an association with the genetical pathogenesis in FHL. All helper and cytotoxic T cells reported so far have been shown to rearrange and express TCR α and β genes. The use of either of the 2 Cβ regions does not correlate with either the class of T cells or the class of major histocompatibility complex (MHC) molecules that are recognized, although the Cβ2 gene is used more often in the T-cell population as a whole.36 An analysis of the Jβ gene in TCRβ chain message (human T cells) showed that Jβ1 or Jβ2 genes are expressed in a VDJC fragment including Cβ1 or Cβ2, respectively.30 The Jβ2 family was used more commonly than Jβ1 in healthy individuals, indicating the preferential use of Jβ2 over Jβ1 (72%:28%).37 Our results obtained from 3 controls also indicated a similar trend of the Jβ bias. Rosenberg et al37 indicated that this Jβ bias must result from events associated with the rearrangement itself, before subsequent selection pressures are applied to the repertoire. Therefore, the marked inverted bias in only the Jβ1 usage of FHL may be associated with the genetic pathogenesis and/or with thymic-positive selection.
It should be clarified as to whether the clonal T cells and the Jβ1 bias in widespread T cells of FHL patients are responsible for the mechanism of T-cell dysfunction. In severe combined immunodeficiency (SCID) patients, the unusual functional behavior of maternal T cells has been explained as the result of profoundly reduced T-cell receptor (TCR) diversity.38 Those investigators demonstrated the lack of one or several TCRBV segments in SCID patients.38,39 Sottini et al40 documented that TCRBV transcripts were characterized by extremely restricted V-D-J junctional diversity in an SCID patient. The functional alteration of these cells appears to be ascribable to an insufficient TCR diversity. The abnormal inverted Jβ1/Jβ2 usage rate might mean the limitation of the size of the TCR member. Subtle changes in the T-cell functions that may be due to the Jβ bias can genetically influence the regulation of the T-cell network in FHL, resulting in hyperactivation and widespread multiorgan infiltration by lymphocytes and histiocytes.
FHL is generally considered a nonneoplastic disorder, but its treatment requires chemotherapy and subsequent HSCT. Stephan et al8recently recommended immuno-suppressive agents (steroids, antithymocyte globulins, and cyclosporine A) as alternative primary and maintenance therapy. Their effective results also support the key role of T cells in the disease. During the clinical course of our FHL patients, polyclonal Vβ13 T-cell lymphoproliferative disease developed in patient no. 1 with a relapse after an allogeneic BMT, and the clonal change (Vβ5.1-Jβ1.6 to Vβ4) of expanded T cells in patient no. 2 occurred; both patients died. An association with EBV was suggested in both of these relapse cases.41 Patients no. 3 and 4 received CBSCT and are now in remission. In FHL patients with low NK activity and unbalanced regulation of T cells by widespread Jβ bias who are undergoing immunosuppressive therapy, virus infection might induce more aggressive disease. Henter et al1suggested that various viral infections may elicit a bout of FHL disorders in genetically predisposed individuals.
Finally, FHL is often indistinguishable from other types of hemophagocytic syndromes42 in neonates and infants. Clonal T cells may also be present in patients with infection-associated HL. A comparison of the V repertoire and Jβ usage in such patients would be of interest. An unbalanced regulation of T cells by widespread Jβ bias in αβ-T cells may contribute to the pathogenesis of FHL. The association of the Jβ bias of clonal T-cell subpopulations and the genetic pathogenesis of FHL remain topics of ongoing study.
Supported in part by Grants-in-Aid for General Scientific Research from the Ministry of Education, Science, and Culture.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Nobuhiro Kimura, MD, The First Department of Internal Medicine, Fukuoka University School of Medicine, Nanakuma 7-45-1, Jonan-ku, Fukuoka 814-01, Japan.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal