PU.1 is a hematopoietic cell–specific ets family transcription factor. Gene disruption of PU.1 results in a cell autonomous defect in hematopoietic progenitor cells that manifests as abnormal myeloid and B-lymphoid development. Of the myeloid lineages, no mature macrophages develop, and the neutrophils that develop are aberrantly and incompletely matured. One of the documented abnormalities of PU.1 null (deficient) hematopoietic cells is a failure to express receptors for granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage (GM)-CSF, and M-CSF. To elucidate the roles of the myeloid growth factor receptors in myeloid cell differentiation, and to distinguish their role from that of PU.1, we have restored expression of the G- and M-CSF receptors in PU.1-deficient cells using retroviral vectors. We have similarly expressed PU.1 in these cells. Whereas expression of growth factor receptors merely allows a PU.1-deficient cell line to survive and grow in the relevant growth factor, expression of PU.1 enables the development of F4/80+, Mac-1+/CD11b+ macrophages, expression of gp91phox and generation of superoxide, and expression of secondary granule genes for neutrophil collagenase and gelatinase. These studies reinforce the idea that availability of PU.1 is crucial for normal myeloid development and clarify some of the molecular events in developing neutrophils and macrophages that are critically dependent on PU.1.
THE ETS FAMILY transcription factor PU.1 is expressed exclusively in hematopoietic cells. Evidence from PU.1 gene-disrupted mice indicates a pivotal role for PU.1 in myeloid lineage (as well as B-lymphocyte) development. To summarize the PU.1 null phenotype, leukocytes are absent at birth in these mice, but low numbers of neutrophils eventually develop by 2 to 3 days, and T lymphocytes by 5 to 8 days, after birth. Mature monocytes/macrophages and B cells are not detectable in these mice at any age examined, up to 2 weeks.1 The defect in PU.1 null hematopoietic progenitors is cell autonomous and may be due, at least in part, to their failure to express receptors for the myeloid growth factors granulocyte colony-stimulating factor (G-CSF), macrophage (M)-CSF, and GM-CSF.2 Currently there is still debate as to the specific role of these receptors in hematopoietic cell survival, proliferation, and differentiation, and it is not clear whether the absence of receptors can account for the hematopoietic deficits seen in the PU.1 null mouse. It is possible and indeed likely that nonexpression or dysregulation of other key genes that are regulated by PU.1 contributes to the abnormal myeloid development in these gene-disrupted mice.
Because PU.1-deficient cells fail to express myeloid growth factor receptors, they represent a useful vehicle for elucidating the distinct functions served by these receptors in hematopoietic cell expansion and development. Therefore, we reintroduced PU.1, G-CSF receptor, and M-CSF receptor into an established PU.1-deficient myeloid cell line, 503.3 We observed in PU.1 restored myeloid cells the expression of genes and acquisition of functions associated with normal terminal neutrophil maturation that were absent from PU.1-deficient cells. In addition, restoration of PU.1 expression in these cells enabled the development of F4/80+, Mac-1/CD11b+cells (monocytes and macrophages), whereas previously only Gr-1+, CAE+ cells (neutrophils) were generated.3 Furthermore, PU.1 transduction restored the ability of these cells to grow in the presence of G-and M-CSF. Unlike cells transduced with a PU.1-expressing vector, those transduced with the G-CSF receptor showed no detectable change in surface-marker phenotype (other than expression of G-CSF receptor), gene expression, or functional capacity to suggest terminal differentiation or maturation of these cells beyond the point that we have described to occur in PU.1 null neutrophils.3 Similarly, transduction with the M-CSF receptor allowed the cells to use M-CSF for survival and growth with no apparent progression of differentiation along the monocyte/macrophage pathway.
These results support the concept of a permissive but not an inductive or instructive role for growth factor receptors in myeloid cell development. In contrast, PU.1 can regulate the expression of molecules associated with cell differentiation. The results obtained from these studies suggest that normal components of myeloid cells and neutrophils can be restored and distinct differentiation pathways enabled even when PU.1 is reintroduced at a relatively late stage of myeloid development, as shown by the PU.1 null cell line that was used in these experiments.
MATERIALS AND METHODS
The PU.1 null cell line 503.
The myeloid cell line 503 was originally derived from PU.1 null neonatal liver as described3 and was used for PU.1 and G- and M-CSF receptor restoration studies. These cells are maintained in Iscove’s medium supplemented with 20% fetal bovine serum with 100 U/mL recombinant mouse (rm) interleukin-3 (IL-3), 5 ng/mL rmGM-CSF, 5 ng/mL rmG-CSF, and 5,000 U/mL recombinant human (rh) M-CSF. Previous work has shown that the cells will only respond to IL-3,2 3but they are maintained in all factors for experimental consistency and comparison with normal cells.
Construction of retroviral plasmids.
Pfu polymerase (Stratagene, San Diego, CA) was used for high-fidelity polymerase chain reaction (PCR) amplification of cDNAs encoding PU.1 and the G-CSF receptor. The complete coding region of PU.1 (nucleotides 132 to 1000, GenBank accession no. M32370) and the G-CSF receptor coding region (nucleotides 180 to 2691, GenBank accession no. M58288) was amplified from a mixed population of mouse bone marrow myeloid cells obtained after activation in IL-3, G-, and GM-CSF. The 5′ and 3′ amplimers introduced a NotI cloning site at each end and the 5′ primer also includes a Kozak consensus translational initiation sequence.4 PCR fragments were cloned into pCR-script (Stratagene) and verified by sequencing. cDNAs were excised with NotI and cloned into the SrfI site of retroviral vector pBA8b-L1 (Chiron Corp, San Diego, CA). The plasmid pBA8b-L1 was constructed in a 3-fragment ligation using the following 3 fragments: (1) The NdeI-ClaI fragment from pBA-5b (described in Patent Cooperative Treaty [PCT] application no. WO9742338), containing the 3′ long terminal repeat (LTR) and the pUC18 backbone; (2) theClaI-HindIII fragment from pCI-PLAP (the cDNA encoding human Placental Alkaline Phosphatase [PLAP]5 inserted into the pCI cytomegalovirus (CMV) expression plasmid (Invitrogen Inc, San Diego, CA); and (3) theHindIII-NdeI fragment from pBA-6bL1, containing the 5′ LTR and the SV40 promoter. Plasmid pBA-6bL1 is based on pBA-6b (described in example 10 of patent application WO 9742338), where the HIVenv/rev-coding region was deleted via XhoI-ClaI digestion and replaced with the L1 polylinker.
In these vectors, now termed pBA8b-L1.PU.1 (encoding PU.1) and pBA8b-L1.GR (encoding the G-CSF receptor), the Moloney murine leukemia virus LTR drives expression of the gene of interest and the SV40 promoter drives expression of PLAP. The M-CSF receptor expressing vector pMZen (c-fms) was a gift from Dr Larry Rohrschneider (University of Washington, Seattle, WA).6
Retroviral infection of PU.1-deficient cells.
The 293-based amphotropic packaging line 2A-LB (patent application WO no. 9742338) was maintained in Dulbecco’s modified Eagle’s medium (DMEM) plus 10% fetal calf serum (FCS) and 2 mmol/L L-glutamine. Vector-producing cell lines (VPCL) were made by cotransfecting the plasmid pMLG-G encoding vesicular stomatitis virus glycoprotein (VSV-G)7 along with plasmids containing gene of interest into 293 2-3 cells7 to generate transient VSV-G-pseudotyped vectors that were then used to transduce the human 293–based amphotropic packaging cell line, 2A-LB. The resulting pool of vector producing cells were enriched for PLAP-expressing cells using anti-PLAP antibody (Sigma, St Louis, MO) and magnetic beads (Miltenyi Biotec Inc, Auburn, CA). To produce virus-containing supernatants, PLAP+ cells were grown to confluency, fresh medium was added, and supernatants were collected 24 and 48 hours later. Viral titers were determined by infection of HT1080 cells using a series of dilutions of the vector in the presence of 8 μg/mL polybrene and scoring the number of PLAP+ colonies 48 hours later. Briefly, the infected HT1080 cells were fixed in phosphate-buffered saline (PBS) containing 2% formaldehyde and 0.2% glutaraldehyde and stained with the alkaline phosphatase substrate Fast Red TR/Napthol AS-MX phosphate (Sigma) as directed by the manufacturer. Titer (colony-forming units [CFU])/mL was calculated as [No. Red Colonies/μL Vector Added] × Dilution Factor × 1,000. Typical supernatants used contained 0.5 to 1.0 × 106CFU/mL.
To transduce 503 cells, the wells of a tissue-culture plate were first coated with fibronectin (Sigma) by incubating with 10 μg/mL fibronectin solution at 37°C for 2 hours. The fibronectin was aspirated and 1 mL of viral supernatant was added.8 9 The plate was further incubated for 0.5 to 1 hour at 37°C to allow adherence of viral particles to the fibronectin-coated surface. Cells were then added in a minimal volume of media plus polybrene (final concentration 6 to 8 μg/mL), and the plate was centrifuged at 1,000g for 3 hours at room temperature. Afterwards, cells were resuspended and allowed to recover for 24 hours in growth factor–containing medium. The transduction protocol was repeated for a total of 3 times.
Selection of transduced cells and flow cytometric analysis.
Forty-eight hours after the last transduction (5 days after the first transduction), an aliquot of cells was removed and stained with an anti-human alkaline phosphatase (PLAP) antibody (Sigma) followed by a phycoerythrin-conjugated secondary donkey anti-mouse antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) to determine transduction efficiency. Cells were then analyzed for alkaline phosphatase expression by flow cytometry with a Becton Dickinson FACScan and data were analyzed with CellQuest (Becton Dickinson, Franklin Lakes, NJ). For assessment of Gr-1 and Mac-1 expression, cells were incubated with Gr-1PE and Mac-1/CD11bFITC at concentrations recommended by the manufacturer (Pharmingen, San Diego, CA), and washed and analyzed as previously described.1
To isolate PU.1 and G-CSF receptor transduced cells, 107 cells were incubated with 20 μg/mL of anti-PLAP antibody (Sigma) for 30 minutes on ice. This step was followed by 3 washes, then goat anti-mouse antibody-conjugated magnetic beads were added as per manufacturer’s instructions (Miltenyi Biotec, Auburn, CA). Positively stained cells were isolated using a VarioMacs magnetic device as directed by the manufacturer (Miltenyi Biotec). Because the M-CSF receptor-expressing vector did not contain a selectable marker, transduced cells were selected by their ability to survive and expand in 5,000 U/mL rhM-CSF.
Isolation of RNA and reverse transcription (RT)-PCR analysis.
Total RNA was isolated and PCR was performed as described previously.3 PCR primers used herein included the following (listed 5′ → 3′): β actin 5′ GTGGGCCGCTCTAGGCACCAA, 3′ CTCTTTGATGTCACGCACGTATTC; PU.1 (573-873) 5′: TCTGATGGAGAAGCTGATGG, 3′: CTTGACTTTCTTCACCTCGC (these primers are located in exon 4 and exon 5, respectively); c-fms 5′: GCGATGTGTGAGCAATGGCAGT, 3′: AGACCGTTTTGCGTAAGACCTG; murine neutrophil collagenase 5′: CACGATGGTTGCAGAGAAGC, 3′: TCTCCTCCAATACCTTGGCC; murine neutrophil gelatinase 5′: ACGGTTGGTACTGGAAGTTCC, 3′: CCAACTTATCCAGACTCC- TGG; gp91phox.3
Enzyme histochemistry and immunohistochemistry.
Methods for immunostaining of cytospin slides for F4/80 and CD11b were as described.1 Sialoadhesin antibody was obtained from Serotec Inc (Raleigh, NC) and used at 1:25 dilution.
Modified colony-forming assays.
Cells were seeded at 500 cells/mL in Methocult 3234 (Stem Cell Technologies, Vancouver, BC, Canada) which was supplemented with either 10 ng/mL rmG-CSF (R&D Systems, Minneapolis, MN) or 5,000 U/mL rhM-CSF (gift of Dr David Hume, University of Queensland, Brisbane, Australia). Colony formation (50 or more cells) was scored after 7 days.
Western blotting.
Detection of proteins by Western blot was performed as described.3 Polyclonal anti-PU.1 and anti–G-CSF receptor antibodies and secondary antibody were obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
Cytochrome c reduction assay for superoxide generation.
Cells were assayed for their ability to reduce cytochrome c exactly as described.3
RESULTS
Expression of PU.1 in PU.1-deficient cells restores expression of G- and M-CSF receptors and restores response to G- and M-CSF.
PU.1 regulation of the G- and M-CSF receptor promoters in transfection assays has previously been reported,10,11 and we have shown that PU.1-deficient cells fail to express receptors for G- and M-CSF.2 PU.1-deficient hematopoietic cells will grow in the presence of IL-3 but not in G-CSF or M-CSF, further confirming the absence of selective receptor function.2 The PU.1 null myeloid cell line 503 was originally derived from PU.1 null neonatal liver as described.3 The phenotype of this cell line is very similar to cells that can be expanded in short-term (<1 month) cultures of PU.1 null neonatal liver. Most cells are CAE+(90%) and CD18+ (80%), with approximately 40% to 60% Gr-1+, and <3% CD11b+1,3 (and data not shown). Our previous work has shown that these cells express markers consistent with developing neutrophils, but fail to express markers and functions of terminally differentiated neutrophils.3 503 cells exhibit a 10% frequency of colony formation in semisolid media containing IL-3; thus, these cultures consist of a progenitor-type cell in addition to more differentiated, neutrophil-like progeny. Therefore, 503 cells should be useful for the investigation of PU.1-mediated growth factor regulation and myeloid differentiation.
We first tested whether retroviral delivery of PU.1 to PU.1-deficient 503 cells would restore G- and M-CSF receptor expression and function. To assess whether restoration of either G-CSF receptor or M-CSF receptor alone would convey the same biological potential as PU.1 expression, G-CSF– or M-CSF–expressing 503 lines were also produced (called 503-GR and 503-MR, respectively). To deliver PU.1 and the G-CSF receptor, the Moloney murine leukemia virus–based retroviral vector pBA8b-L1 was used. In this vector, PU.1 (or G-CSF receptor) cDNA was under the control of the viral LTR, and the cell-surface–expressed selectable marker, human placental alkaline phosphatase (PLAP), was driven by the SV40 promoter (see Materials and Methods). Delivery of the M-CSF receptor used the pMZen(cfms) vector.6
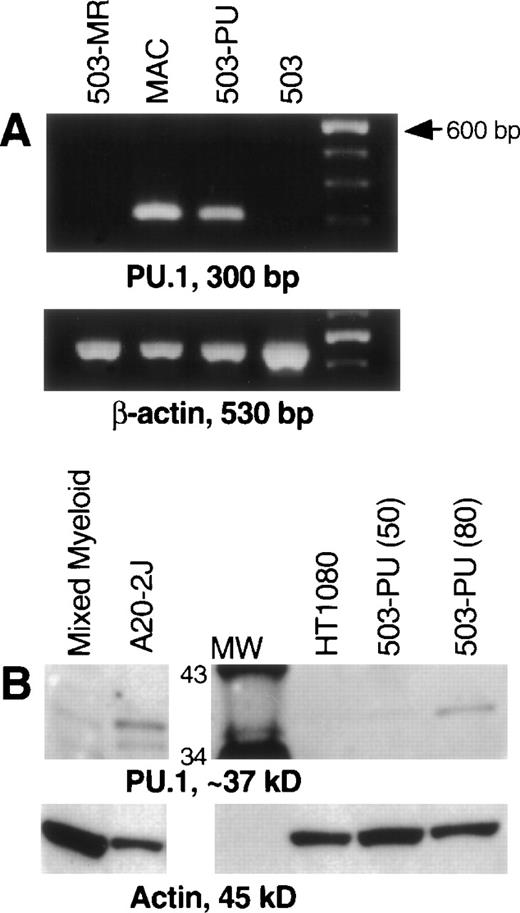
After transduction of the 503 cell line with pBA8b-L1.PU.1, PLAP+ cells were isolated as discussed in Material and Methods. To confirm PU.1 expression, RNA was prepared from these cells, treated with DNase, and subjected to RT-PCR. mRNA for PU.1 was detected in transduced samples (503-PU) but not in the parental 503 cells, or in 503 cells transduced with an M-CSF receptor-expressing retroviral vector (Fig 1A) or a G-CSF receptor-expressing retroviral vector (data not shown). We then analyzed whole-cell lysates for presence of PU.1 protein. Confirmation that PU.1 protein expression was restored can be seen in Fig 1B. The PU.1 protein level in 503-PU cells approached the level of normal mixed myeloid cells, but was less than the B-cell line A20-2J.
PU.1 is detectable in 503 cells transduced with a PU.1-expressing retrovirus. PU.1-deficient 503 cells were transduced with retroviral vectors containing the cDNA for PU.1, G-CSF receptor, or M-CSF receptor. Cells that were positive for the selectable marker PLAP (PU.1 and G-CSF receptor) or positive for growth in M-CSF (M-CSF receptor) were isolated for analysis. (A) RT-PCR showed PU.1 mRNA in normal macrophages (MAC) and PU.1-transduced 503 cells (503-PU), but not in 503 or 503 transduced with M-CSF receptor (503-MR). Primers used were intron-crossing, and controls for DNA amplification in the absence of RT were also included and were negative (not shown). The far righthand lane contains a 100-bp DNA ladder for size determination of PCR products. (B) Whole-cell lysates were prepared and 50 μg (or 80 μg where indicated) of total cellular protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After blotting and probing with a polyclonal anti-PU.1 antibody, PU.1 protein was visible in normal mixed myeloid cells, A20-2J B cells, and 503-PU cells. As expected, nonhematopoietic HT1080 cells were negative for PU.1 expression. The lane labeled MW contains protein size standards (CruzMarker; Santa Cruz Biotechnology). MW, molecular weight.
PU.1 is detectable in 503 cells transduced with a PU.1-expressing retrovirus. PU.1-deficient 503 cells were transduced with retroviral vectors containing the cDNA for PU.1, G-CSF receptor, or M-CSF receptor. Cells that were positive for the selectable marker PLAP (PU.1 and G-CSF receptor) or positive for growth in M-CSF (M-CSF receptor) were isolated for analysis. (A) RT-PCR showed PU.1 mRNA in normal macrophages (MAC) and PU.1-transduced 503 cells (503-PU), but not in 503 or 503 transduced with M-CSF receptor (503-MR). Primers used were intron-crossing, and controls for DNA amplification in the absence of RT were also included and were negative (not shown). The far righthand lane contains a 100-bp DNA ladder for size determination of PCR products. (B) Whole-cell lysates were prepared and 50 μg (or 80 μg where indicated) of total cellular protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After blotting and probing with a polyclonal anti-PU.1 antibody, PU.1 protein was visible in normal mixed myeloid cells, A20-2J B cells, and 503-PU cells. As expected, nonhematopoietic HT1080 cells were negative for PU.1 expression. The lane labeled MW contains protein size standards (CruzMarker; Santa Cruz Biotechnology). MW, molecular weight.
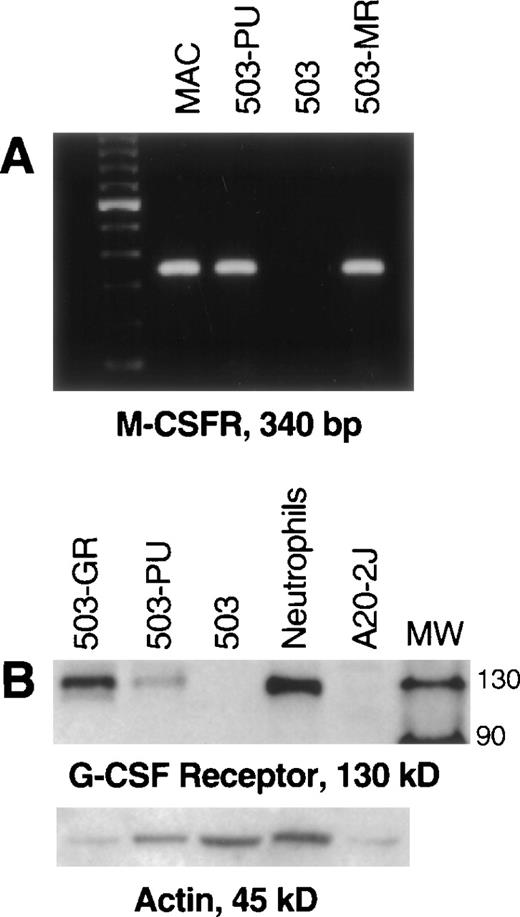
Unlike parental PU.1-deficient 503 cells, pBA8b-L1.PU.1-transduced 503 cells (503-PU) were shown to express M-CSF receptor mRNA (Fig 2A). 503 cells that were transduced with an M-CSF receptor-containing retroviral vector (503-MR) also expressed mRNA for M-CSF receptor (Fig 2A). 503-PU cells also expressed G-CSF receptor protein, as did 503 cells transduced with a G-CSF receptor-expressing retroviral construct (503-GR) (Fig 2B). Parental 503 cells expressed no G-CSF receptor protein, consistent with our previous results2 (Fig 2B). To test whether reintroduction of PU.1 would also restore responsiveness to G- and M-CSF, PLAP+ 503-PU were seeded into methylcellulose media containing either G- or M-CSF. We have shown that cells derived from PU.1 null neonatal liver will form colonies in semisolid media only in the presence of IL-3, but not G- or M-CSF.2 Similarly, parental PU.1-deficient 503 cells formed no colonies in either G- or M-CSF; however, 2 different 503-derived PU.1 transduced clones produced 9.3 ± 3.5 and 20.7 ± 5.6 colonies in G-CSF, and 1.3 ± 0.6 and 4.0 ± 2.7 colonies in M-CSF, per 500 cells seeded in methylcellulose (Table 1).
Transduction of PU.1 into PU.1-deficient cells restores G- and M-CSF receptor expression. (A) PU.1- and M-CSF receptor transduced cells (503-PU, 503-MR) that were selected by PLAP expression or growth in M-CSF, respectively, were analyzed by RT-PCR. M-CSF receptor mRNA was evident in normal macrophages (MAC), 503-PU, and 503-MR. These primers were intron-crossing, and controls for DNA amplification in the absence of RT that were also performed were negative (not shown). The far lefthand lane contains a 100-bp DNA ladder. (B) Whole-cell lysates were prepared and 25 μg of total cellular protein was separated by SDS-PAGE. After blotting and probing with polyclonal anti–G-CSF receptor antibody, G-CSF receptor protein was detectable in G-CSF receptor- and PU.1-transduced 503 cells (503-GR, 503-PU) and normal neutrophils, but not in 503 cells or the B cell line A20-2J. The lane labeled MW contains protein size standards (CruzMarker; Santa Cruz Biotechnology).
Transduction of PU.1 into PU.1-deficient cells restores G- and M-CSF receptor expression. (A) PU.1- and M-CSF receptor transduced cells (503-PU, 503-MR) that were selected by PLAP expression or growth in M-CSF, respectively, were analyzed by RT-PCR. M-CSF receptor mRNA was evident in normal macrophages (MAC), 503-PU, and 503-MR. These primers were intron-crossing, and controls for DNA amplification in the absence of RT that were also performed were negative (not shown). The far lefthand lane contains a 100-bp DNA ladder. (B) Whole-cell lysates were prepared and 25 μg of total cellular protein was separated by SDS-PAGE. After blotting and probing with polyclonal anti–G-CSF receptor antibody, G-CSF receptor protein was detectable in G-CSF receptor- and PU.1-transduced 503 cells (503-GR, 503-PU) and normal neutrophils, but not in 503 cells or the B cell line A20-2J. The lane labeled MW contains protein size standards (CruzMarker; Santa Cruz Biotechnology).
Only PU.1-Transduced 503 Cell Lines and Not Parental PU.1-Deficient 503 Cells Can Form Colonies in Response to G- and M-CSF
| Cell Line/Clone . | G-CSF . | M-CSF . |
|---|---|---|
| 503 | 0 | 0 |
| 503-PU#1 | 9.3 ± 3.5 | 1.3 ± 0.6 |
| 503-PU#2 | 21 ± 5.6 | 4.0 ± 2.7 |
| Cell Line/Clone . | G-CSF . | M-CSF . |
|---|---|---|
| 503 | 0 | 0 |
| 503-PU#1 | 9.3 ± 3.5 | 1.3 ± 0.6 |
| 503-PU#2 | 21 ± 5.6 | 4.0 ± 2.7 |
Data reflect number of colonies formed per 500 cells seeded after 7 days in 10 ng/mL G-CSF or 5,000 U/mL M-CSF. Assays were performed in triplicate and results are reported as mean ± SD.
Thus, we have documented that retroviral delivery of PU.1 into 503 cells results in PU.1 expression and the restoration of expression of functional G- and M-CSF receptors.
Gene expression consistent with neutrophil terminal differentiation is seen after restoration of PU.1 expression to PU.1-deficient myeloid cells.
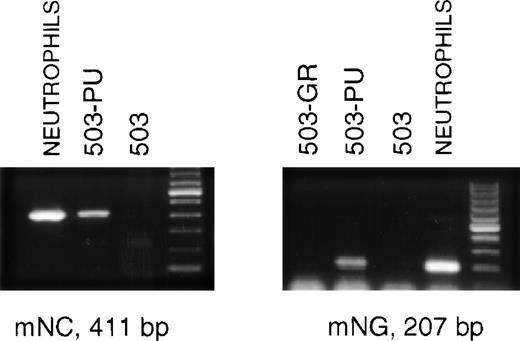
The late stages of neutrophil differentiation are characterized by the appearance of secondary or specific granule gene products. These genes are believed to be coordinately regulated, and are transcribed in a stage-specific manner at the myelocyte stage of development.12-14 We have shown that the PU.1 null neutrophil-like cells that develop, both in vivo and in vitro, do not express detectable mRNA for secondary granule components.3When PLAP+ 503-PU cells were examined using RT-PCR, we were able to detect mRNA for the secondary granule genes neutrophil collagenase and neutrophil gelatinase, whereas no mRNA was found in the parental cell line (Fig 3) or in G-CSF receptor-transduced 503 cells (503-GR) (Fig 3 and data not shown).
503 cells transduced with PU.1 express markers of neutrophil terminal differentiation. RNA was prepared from PLAP+ PU.1-transduced 503 (503-PU) cells and analyzed by RT-PCR. mRNA for secondary granule genes neutrophil collagenase (mNC) and neutrophil gelatinase (mNG) is present in 503-PU as well as normal neutrophils, but not in 503 or 503 transduced with G-CSF receptor (503-GR). The far righthand lane of each gel contains a 100-bp DNA ladder.
503 cells transduced with PU.1 express markers of neutrophil terminal differentiation. RNA was prepared from PLAP+ PU.1-transduced 503 (503-PU) cells and analyzed by RT-PCR. mRNA for secondary granule genes neutrophil collagenase (mNC) and neutrophil gelatinase (mNG) is present in 503-PU as well as normal neutrophils, but not in 503 or 503 transduced with G-CSF receptor (503-GR). The far righthand lane of each gel contains a 100-bp DNA ladder.
PU.1 transduction restores gp91phox expression and superoxide production in PU.1-deficient 503 cells.
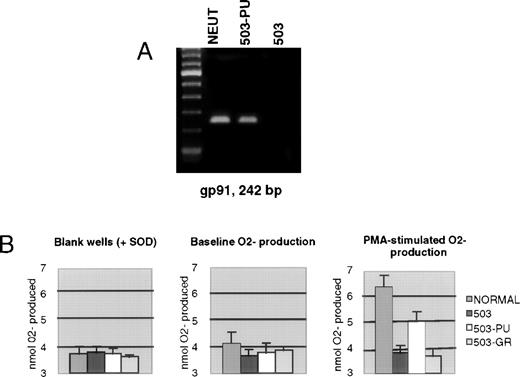
We previously documented that PU.1 null myeloid cells failed to transcribe the gp91phox gene, which encodes a major subunit of the enzyme NADPH oxidase, and also failed to generate the metabolite superoxide (O2−) that is the exclusive product of this enzyme.3 As shown in Fig 4A, 503-PU cells expressed gp91phox mRNA, unlike the parental 503 cell line, or 503-GR cells. To confirm the restoration of NADPH oxidase function, we analyzed the cells for their ability to reduce cytochrome c in response to PMA stimulation. Unlike 503 or 503-GR cells that produced no detectable O2−, 503-PU and normal cells were able to increase their O2−production by roughly 74% and 64%, respectively, in the presence of PMA (Fig 4B).
Expression of gp91phox and associated NADPH oxidase enzyme activity is restored by PU.1 transduction of 503 cells. (A) RT-PCR analysis of cells revealed gp91phox mRNA in normal neutrophils and PU.1-transduced 503 cells (503-PU), but not in 503 as previously documented or in G-CSF receptor-transduced 503 (503-GR). The far lefthand lane contains a 100-bp DNA ladder. (B) An assay for cytochrome c reduction in response to PMA, which measures O2− production, was performed to address the functionality of the enzyme NADPH oxidase, of which gp91phox is a principal subunit. Detectable production of O2− was found in normal neutrophils (NORMAL) and 503-PU, but not in 503 or 503-GR. SOD, superoxide dismutase.
Expression of gp91phox and associated NADPH oxidase enzyme activity is restored by PU.1 transduction of 503 cells. (A) RT-PCR analysis of cells revealed gp91phox mRNA in normal neutrophils and PU.1-transduced 503 cells (503-PU), but not in 503 as previously documented or in G-CSF receptor-transduced 503 (503-GR). The far lefthand lane contains a 100-bp DNA ladder. (B) An assay for cytochrome c reduction in response to PMA, which measures O2− production, was performed to address the functionality of the enzyme NADPH oxidase, of which gp91phox is a principal subunit. Detectable production of O2− was found in normal neutrophils (NORMAL) and 503-PU, but not in 503 or 503-GR. SOD, superoxide dismutase.
G-CSF receptor expression enables PU.1-deficient myeloid cells to survive and proliferate without further differentiation in G-CSF.
Our previous studies have shown that neutrophil development proceeds in the absence of PU.1 and the accompanying lack of detectable G-CSF receptor expression. The number of neutrophils that develop in vivo is severely reduced, however, and the cells that develop in vivo and in vitro are abnormal in their surface marker and functional profile.1-3 As shown above, PU.1 restoration in 503 cells leads to the expression of genes and functions associated with mature neutrophils. To attempt to distinguish the contribution of the G-CSF receptor to neutrophil development from the contribution of PU.1, we transduced PU.1-deficient 503 cells with the retroviral vector pBA8b-L1.GR as discussed above. PLAP+ cells were isolated after transduction and shown to express G-CSF receptor protein (Fig2A). By plating equal numbers of cells initially and counting live versus dead cells on the basis of trypan blue exclusion over the course of 7 days, we were able to document enhanced survival and growth in G-CSF of PLAP+ G-CSF receptor-expressing 503 cells compared to PLAP− cells (Fig 5). However, as shown in Figs 3 and 4, we detected neither mRNA for murine neutrophil gelatinase nor superoxide production in 503-GR cells, which represented no change from the parental PU.1 null cell line, even after 2 weeks of culture in G-CSF. Similarly, no change in the Gr-1 and Mac-1/CD11b surface staining profile of G-CSF–cultured 503-GR cells was detected (data not shown). Therefore, we conclude that G-CSF receptor expression is not sufficient for terminal neutrophil differentiation in the absence of PU.1. Finally, because the G-CSF receptor-expressing retroviral vector was the same vector that was used for PU.1 transduction, it is clear that the phenotypic changes produced by PU.1 transduction were specific to PU.1 expression and not a nonspecific effect of retroviral integration.
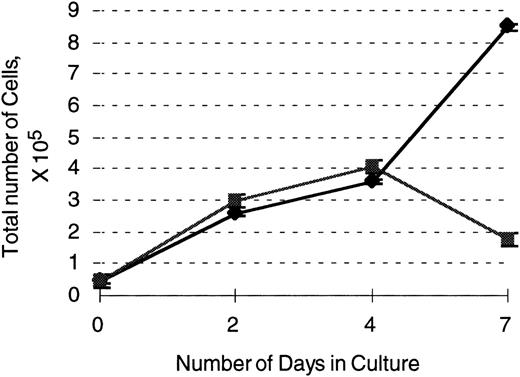
503 cells transduced with the G-CSF receptor display enhanced survival and proliferation in the presence of G-CSF compared with parental 503 cells. After transduction with a G-CSF–expressing retrovirus, an equal number of selectable marker PLAP+(⧫) and PLAP− (▪) 503 cells were plated in 10 ng/mL G-CSF and live cell (on the basis of trypan blue exclusion) counts monitored over 7 days. Note an almost 5-fold greater number of live PLAP+ versus PLAP− cells after 7 days in G-CSF.
503 cells transduced with the G-CSF receptor display enhanced survival and proliferation in the presence of G-CSF compared with parental 503 cells. After transduction with a G-CSF–expressing retrovirus, an equal number of selectable marker PLAP+(⧫) and PLAP− (▪) 503 cells were plated in 10 ng/mL G-CSF and live cell (on the basis of trypan blue exclusion) counts monitored over 7 days. Note an almost 5-fold greater number of live PLAP+ versus PLAP− cells after 7 days in G-CSF.
Macrophage differentiation is partially restored by transduction with a PU.1-expressing retrovirus.
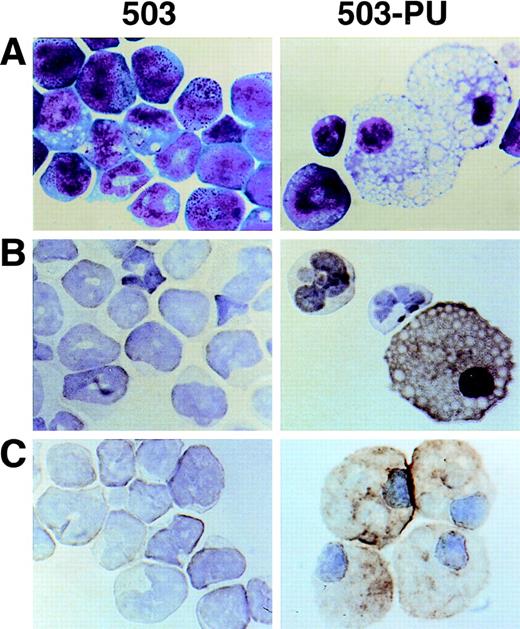
As we have previously reported,3 cell line 503 was derived from the liver of a PU.1 null neonate. Based on their expression of Gr-1 and chloroacetate esterase, 503 cells appear to be of the neutrophil lineage. No cells displaying specific monocyte or macrophage characteristics such as F4/80, M-CSF receptor, scavenger receptor, or mannose receptor expression have been detected in PU.1 null cell cultures2,3 or in PU.1 null mice of any age examined.1 After transduction of 503 cells with PU.1, however, Wright-Giemsa staining showed cells with typical mature macrophage morphology (Fig 6A, compare 503 and 503-PU). Immunohistochemical staining demonstrated the expression of F4/80 and sialoadhesin, two classic markers for this lineage (Fig 6B and C). Flow cytometric analysis of the PU.1-transduced population showed an overall increase in cell size (forward scatter), and both a decrease in Gr-1 expression and an increase in Mac-1/CD11b expression relative to the parental 503 PU.1 null cell line (data not shown). mRNA for macrophage-specific genes including M-CSF receptor (Fig 2A), mannose receptor, and IL-18 was detected in PU.1-transduced but not PU.1-deficient cells (data not shown). Interestingly, scavenger receptor mRNA was still absent from these cells. With the exception of IL-18, the promoters of each of these genes have previously been reported to be regulated by PU.1.11,15 16 Thus, by retroviral delivery of PU.1 into the PU.1-deficient cell line 503, which previously expressed only markers of early myeloid or neutrophil development, we have restored the ability of these cells to express markers associated with mature monocyte and macrophage development.
Macrophage development can be restored by PU.1 transduction of PU.1-deficient 503 cells. (A) PU.1 transduction of 503 cells (503-PU) restored the development of cells with typical macrophage appearance as shown by Wright-Giemsa staining. (B) Immuno-cytochemistry was used to show that 503-PU cells expressed the macrophage marker F4/80, unlike the parental cells (503). Positive-staining cells demonstrate an orange-brown reaction product. Note the presence of F4/80+ 503-PU cells with lobate nuclear morphology of monocytes. (C) An adherent subpopulation 503-PU cells was also positive for the macrophage subset marker sialoadhesin by immunocytochemical staining, whereas no positive cells were found in parental 503 cultures.
Macrophage development can be restored by PU.1 transduction of PU.1-deficient 503 cells. (A) PU.1 transduction of 503 cells (503-PU) restored the development of cells with typical macrophage appearance as shown by Wright-Giemsa staining. (B) Immuno-cytochemistry was used to show that 503-PU cells expressed the macrophage marker F4/80, unlike the parental cells (503). Positive-staining cells demonstrate an orange-brown reaction product. Note the presence of F4/80+ 503-PU cells with lobate nuclear morphology of monocytes. (C) An adherent subpopulation 503-PU cells was also positive for the macrophage subset marker sialoadhesin by immunocytochemical staining, whereas no positive cells were found in parental 503 cultures.
Restoration of M-CSF receptor expression alone permits survival and proliferation, but not macrophage differentiation, of PU.1 null cells.
We have documented that hematopoietic cells deficient in PU.1 express neither M-CSF receptor mRNA nor protein2 (Fig 2A). Recent studies on an independently derived PU.1 null mouse indicated that some macrophage characteristics could be restored when fetal progenitor cells were transduced with PU.1.17 In addition, it has recently been shown that monocyte/macrophage progenitors may be present in our PU.1 null mouse model.18 To determine whether restoration of signaling through the M-CSF receptor could compensate for the absence of PU.1 and allow macrophage differentiation to proceed, we transduced an M-CSF receptor-expressing retrovirus into the PU.1 null myeloid cell line 503. We found that M-CSF receptor expression permitted the cells to use M-CSF for survival and growth, but did not detectably alter the differentiation status of the cells. In contrast to PU.1-complemented 503 cells, M-CSF receptor-restored 503 cells did not express F4/80 or Mac-1/CD11b (data not shown). PU.1-deficient cells transduced with the M-CSF receptor did not express mRNA for macrophage-associated genes such as IL-18 or mannose receptor or the myeloid-associated gene gp91phox, which represented no change from the parental cell line (data not shown).
DISCUSSION
We previously established that myeloid and lymphoid commitment occurs in the absence of PU.1.1,2 Although myeloid commitment was evident in PU.1 null mice, monocytes and macrophages were not produced in vivo or in vitro.1,2 In contrast, neutrophils were produced in the mouse relatively late in development (postnatally, in fact) and remained extremely few in number. Their surface markers, gene expression, and functional profiles were consistent with incompletely or aberrantly matured neutrophils. Further studies documented that PU.1-deficient neutrophils could be expanded in vitro in the presence of IL-3, but not G- and/or GM-CSF.2 Therefore, at least part of the observed developmental abnormality of PU.1 null myeloid cells could be attributable to their failure to express receptors for G-, GM-, and M-CSF.2 The importance of the CSF receptors in normal production and expansion of myeloid lineage-restricted cells is well accepted, although their specific roles are less clear. To determine the contribution of G- and M-CSF receptors and PU.1 to the expression of characteristics associated with late stages of myeloid/neutrophil development, G- and M-CSF receptors and PU.1 were introduced into the PU.1 null 503 myeloid cell line.
As reported previously, both primary PU.1-deficient myeloid cells and the myeloid cell line 503 produce cells that express markers consistent with developing neutrophils, but these cells fail to express markers and functions of mature neutrophils or monocytes/macrophages.3 Restoration of the G-CSF receptor in the 503 cell line did not allow these cells to progress detectably beyond the point of differentiation that they were able to achieve in the absence of PU.1 and the concomitant absence of the effects of G-CSF. It remains a possibility that disruption of PU.1 has resulted in additional downstream effects that preclude normal G-CSF receptor signal transmission or its effects. However, at least a part of the G-CSF receptor pathway was intact, as indicated by the enhanced cell survival and proliferation seen in the presence of G-CSF after receptor restoration. In contrast, PU.1 restoration in 503 cells permitted expression of mRNA for the NADPH oxidase component gp91phox, which was absent from PU.1-deficient neutrophils. Subsequently, the transduced cells were able to reduce cytochrome c in vitro, which is an indication of superoxide generation and restored enzyme activity. Furthermore, we were able to detect mRNA in 503-PU cells for murine neutrophil collagenase and neutrophil gelatinase, terminal maturation-associated secondary granule genes12,13 that were not expressed in PU.1-deficient neutrophils. Importantly, transduction of cell line 503 with PU.1 restored expression of the G-CSF receptor and the ability to survive and proliferate in G-CSF, confirming an essential role for PU.1 in G-CSF receptor expression as has been predicted.2 10Table 2 summarizes the neutrophil characteristics of 503 cells and transduced variants.
Expression of Certain Neutrophil Characteristics Is Restored in PU.1-Transduced But Not in G-CSF Receptor-Transduced or Parental PU.1-Deficient 503 Cells
| . | Cell Lines . | ||
|---|---|---|---|
| 503 . | 503-PU . | 503-GR . | |
| G-CSF receptor protein & response to G-CSF | − | + | + |
| mNC mRNA | − | + | − |
| mNG mRNA | − | + | − |
| gp91phox mRNA | − | + | − |
| Cytochrome c reduction | − | + | − |
| . | Cell Lines . | ||
|---|---|---|---|
| 503 . | 503-PU . | 503-GR . | |
| G-CSF receptor protein & response to G-CSF | − | + | + |
| mNC mRNA | − | + | − |
| mNG mRNA | − | + | − |
| gp91phox mRNA | − | + | − |
| Cytochrome c reduction | − | + | − |
Abbreviations: mNC, murine neutrophil collagenase; mNG, murine neutrophil gelatinase.
Our earlier studies clearly established that neutrophil commitment and some differentiation can take place in the absence of PU.1 and G-CSF receptor signaling, although the resulting cells were not mature and marginally functional. Although G-CSF and its receptor are believed to play an essential role in neutrophil development and function,19 their specific roles are not clear. For example, mice with a targeted disruption of the G-CSF or G-CSF receptor genes were still able to generate neutrophils, albeit in low numbers.20,21 These results suggested that G-CSF-receptor signaling is not necessary for granulocyte lineage commitment or differentiation, but is necessary for lineage expansion. These studies also highlight the point that alternative developmental pathways exist for neutrophils in vivo. Consistent with our original observations, DeKoter et al17 recently described neutrophil development in vitro in their PU.1 knockout system. Thus, these collective results clearly indicate that neutrophil commitment and some development is possible in the absence of PU.1. However, both our previous studies2 3 and the results presented herein show that the late stages of neutrophil development are critically dependent on the presence of PU.1. In contrast to the results obtained in studies with G-CSF or G-CSF receptor null mice, the molecular events allowing neutrophil maturation cannot be circumvented by cytokine receptor signaling in PU.1-deficient cells and PU.1 restoration is required (Table 2). In addition, our current studies put a temporal context on the effects of PU.1 expression; PU.1 reintroduction in later stages of myeloid development can rescue phenotype and functions associated with mature neutrophils.
When M-CSF receptor expression was restored in PU.1-deficient myeloid cells, cell proliferation occurred, but no change in cell phenotype was found that would suggest any progression of macrophage development. These results, similar to what was observed for G-CSF receptor reintroduction, indicated that receptor expression alone could not compensate for the absence of PU.1. In contrast, expression of PU.1 in the same cell line resulted in the development of F4/80+, Mac-1/CD11b+, and large monocyte- and macrophage-like cells. Consistent with previous data that implicated PU.1 in the critical regulation of the M-CSF receptor,11 we found that expression of the M-CSF receptor and response to M-CSF was restored by PU.1 reintroduction. Macrophage characteristics of PU.1 null 503 cells and 503 transduced with PU.1 or M-CSF receptor are summarized in Table 3. Our findings of reintroducing M-CSF receptor at a relatively late stage of myeloid cell development are similar to those obtained from an independently derived PU.1 null mouse model where pluripotent fetal lineage marker-negative hematopoietic cells were targeted for M-CSF receptor delivery. After transduction of PU.1 or M-CSF receptor into these PU.1 null hematopoietic cells before myeloid commitment and/or possibly at very early stages of myeloid commitment, these investigators documented restoration of macrophage development only in the PU.1-transduced cells.17 Thus, our studies show that PU.1 need not be present for commitment of a myeloid lineage that retains the potential for monocyte/macrophage development, but needs to be present for monocyte/macrophage development.
Expression of Certain Macrophage Characteristics Is Restored in PU.1-Transduced But Not in M-CSF Receptor-Transduced or Parental PU.1-Deficient 503 Cells
| . | Cell Lines . | ||
|---|---|---|---|
| 503 . | 503-PU . | 503-MR . | |
| M-CSF R mRNA & response to M-CSF | − | + | + |
| IL-18 mRNA | − | + | − |
| Mannose R mRNA | − | + | − |
| F4/80 protein | − | + | − |
| Mac-1/CD11b protein | − | + | − |
| . | Cell Lines . | ||
|---|---|---|---|
| 503 . | 503-PU . | 503-MR . | |
| M-CSF R mRNA & response to M-CSF | − | + | + |
| IL-18 mRNA | − | + | − |
| Mannose R mRNA | − | + | − |
| F4/80 protein | − | + | − |
| Mac-1/CD11b protein | − | + | − |
Abbreviation: R, receptor.
M-CSF and its receptor have been reported to be crucial for macrophage and osteoclast development.22,23 However, the naturally occurring mutant op/op mouse, which fails to produce measurable M-CSF, was still able to develop detectable macrophages and osteoclasts.24,25 This mutant showed populations of M-CSF–dependent and M-CSF–independent macrophages; the latter were severely and persistently deficient and the former achieved almost normal levels postnatally.26 No M-CSF receptor-deficient mice have yet been described. The hematopoietic cells of the PU.1 null mouse have been shown to lack M-CSF receptor mRNA or protein expression, and no cells with distinct mature monocyte/macrophage characteristics could be identified1,2 in vivo or in vitro. Populations of Moma-2+ER-MP12+ and Moma-2+ERMP-20+ cells have been detected in fetal and neonatal PU.1 null mice18 that may reflect commitment to the monocyte/macrophage lineage. In normal mice, most of these cells were also F4/80+, CD11b+, and Gr-1+.18 Thus, these populations could include early common myeloid and neutrophil precursors as well as monocyte precursor cells. Nevertheless, it is clear that monocyte/macrophage differentiation is blocked early and cells are unable to proceed along the macrophage developmental pathway as a result of the lack of PU.1.
One potential explanation to account for the lack of differentiation after restoration of M-CSF receptor is that PU.1 must be present to turn on a critical molecular switch (or switches) that enables macrophage development in a common macrophage/neutrophil progenitor cell. In this model, the choice of neutrophil development may hinge upon the precise balance of PU.1 and other neutrophil-differentiation-favoring factor(s), such as the transcription factors C/EBPα and C/EBPε, members of the basic leucine zipper class of transcription factors.27-29C/EBPα and C/EBPε gene targeted mice both displayed disruptions in neutrophil development. Loss of C/EBPα resulted in markedly decreased expression of IL-6 and G-CSF receptors,30 whereas the loss of C/EBPε in mice resulted in the generation of atypical neutrophils (hyposegmented), that appeared to express mRNA for the G-CSF receptor, but failed to mature and were not fully functional.31Interestingly,25 ectopic overexpression of C/EBPα in the human leukemic cell line U937 promoted secondary granule gene transcription, which suggests that C/EBPα might have additional effects in neutrophil development.32 A recent study by Iwama et al33 suggests that C/EBPε message level was undetectable in whole fetal PU.1 null liver. This observation is consistent with our previous demonstration that primitive myeloid lineages were reduced and neutrophils and lymphoid cells were undetectable from PU.1 null mice at birth.2 We have found mRNA for both C/EBPα and C/EBPε in PU.1 null myeloid cells isolated from older mice (K.L.A., unpublished data, April 1998); however, further studies need to be performed to quantify levels of mRNA and protein to determine if alterations are present.
Alternatively, molecular events regulating neutrophil versus monocyte/macrophage differentiation in a shared progenitor may not be reliant on PU.1. However, PU.1 appears to be absolutely necessary to allow the monocyte/macrophage differentiation program to proceed in an already-committed macrophage progenitor. Either model is partially supported by the observation that in the absence of PU.1, neutrophils become the principal myeloid lineage that is produced. In contrast to monocytes/macrophages, the need for PU.1 in the neutrophil lineage apparently becomes critical at a relatively late stage of development, because the absence of PU.1 during neutrophil development manifests as a failure of these cells to display characteristics of terminal differentiation and functional maturity.3
The present study has illuminated certain myeloid developmental events that are critically regulated by PU.1. These include expression of genes common to both monocyte/macrophages and neutrophils, such as CD11b and gp91phox, with the restoration of NADPH oxidase function as shown by superoxide generation. Additionally, lineage-specific programs such as restricted expression of G- and M-CSF receptors, neutrophil expression of secondary granule genes, and the monocyte/macrophage expression of F4/80, mannose receptor, and IL-18, are dependent on the presence of PU.1. From these results, coupled with prior observations made in the PU.1 null mouse,1-3 we conclude that PU.1 is indispensable for macrophage differentiation, neutrophil maturation, and myeloid cell proliferation via regulation of CSF receptors. It is still not clear from the present studies whether the developmental characteristics that have been restored in PU.1 null cells by retroviral reintroduction of PU.1 are directly or indirectly mediated by PU.1. Nevertheless, this work defines certain differentiation and development-associated molecular events that are regulated by PU.1 in myeloid cells. Characterization of a PU.1-rescued mouse model will add relevance and further elucidate the role of PU.1 in the developing hematopoietic system.
ACKNOWLEDGMENT
We gratefully acknowledge the technical assistance of Giano Panzarella and of the animal facility personnel at both the Burnham Institute and The Scripps Research Institute, and the secretarial assistance of Bonnie Towle. We also thank Drs Dan Tenen and Dan Link for helpful discussions, and Dr Larry Rohrschneider for providing the pMZen(cfms) vector and packaging line.
Supported by National Institutes of Health (NIH) Grant No. DK49886 (B.E.T.). K.L.A. was supported by an NIH Training Grant [T328HLO7195-22]. This is publication 12368-IMM from The Scripps Research Institute.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Bruce E. Torbett, PhD, Department of Immunology, IMM-7, The Scripps Research Institute, 10550 N Torrey Pines Rd, La Jolla, CA 92037; e-mail: betorbet@scripps.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal