Abstract
Epstein-Barr virus (EBV) infection of humans has been associated with the development of lymphoid malignancies mainly of B-cell lineage, although occasionally T-cell lymphomas have been reported. We describe here the characterization of a novel EBV-like virus (HVMNE) isolated from a simian T-cell lymphotropic virus type I/II (STLV-I/II) seronegative pigtailed macaque (Macaca nemestrina) with a cutaneous T-cell lymphoma. Immunohistochemistry studies on the skin lesions demonstrated that the infiltrating cells were of the CD3+/CD8+ phenotype. Two primary transformed CD8+ T-cell lines were obtained from cultures of peripheral blood mononuclear cells (PBMC) and skin, and, with time, both cell lines became interleukin-2–independent and acquired the constitutive activation of STAT proteins. Polymerase chain reaction analysis of the DNA from the cell lines and tissues from the lymphomatous animal demonstrated the presence of a 536-bp DNA fragment that was 90% identical to EBV polymerase gene sequences, whereas the same DNA was consistently negative for STLV-I/II sequences. Electron microscopy performed on both cell lines, after sodium butyrate treatment, showed the presence of a herpes-like virus that was designated HVMNE according to the existing nomenclature. In situ hybridization studies using EBV Epstein-Barr viral-encoded RNA probes showed viral RNA expression in both CD8+ T-cell lines as well as in the infiltrating CD8+ T cells of skin-tissue biopsies. Phylogenetic analysis of a 465-bp fragment from the polymerase gene of HVMNE placed this virus within theLymphocryptovirus genus and demonstrated that HVMNEis a distinct virus, clearly related to human EBV and other EBV-like herpesviruses found in nonhuman primates.
MYCOSIS FUNGOIDES (MF) is a rare cutaneous T-cell lymphoma (CTCL) that may involve lymph nodes and viscera as well as skin.1,2 The Sézary syndrome (SS) is the erythrodermic variant of MF characterized by the presence of circulating tumor cells. The tumor cells in MF/SS are usually of the CD4+ mature-cell lineage, although CD8+ lineage has been described in a few cases.2 The diagnosis of MF is based on clinical features and histopathological findings that include infiltration of the dermis with lymphocytes with hyperconvoluted nuclei and Pautrier’s microabcesses.1 MF and SS are the most frequent primary lymphomas involving the skin. Genetic predisposition, alteration in cytokine profile, and viruses such as herpesviruses I and II (HSV-I and HSV-II), Epstein-Barr virus (EBV), human herpesvirus 6 (HHV-6), and human T-cell lymphotropic virus type I (HTLV-I) have been suggested as possible causative agents.1
HSV-I/II–specific antigens and DNA have been found in lesions of CTCL.3,4 One report describes the finding of HHV-6 in 1 of 30 patients with CTCL.5 EBV DNA has been found in patients with cutaneous lymphomas6-12 and, in several cases, viral RNA expression has been demonstrated in the neoplastic tissue. Higher incidence of EBV seropositivity in CTCL patients with the consistent emergence of EBV in MF/SS-cultured lymphocytes has also been reported.10 However, a direct causative role of EBV in CTCL has been difficult to prove.13 In contrast, the importance of EBV in the development of B-cell malignancies in human immunodeficiency virus-infected individuals or iatrogenically immune-suppressed patients is more broadly accepted.14
Although humans are the only known natural host for EBV, EBV-like agents have been described in Old World nonhuman primates, including chimpanzee,15-17 baboon,16,18-21 African green monkey,22 gorilla,23 and macaque species.24-29 Although the relative prevalence of these viruses in animals in captivity or in the wild is unknown,30 several studies suggest that it may be as high as that in EBV in humans. In fact, the inability to generate models of EBV-associated lymphomas in Old World monkeys with the human EBV has been ascribed to the presence of cross-reactive immunity against EBV in these species. In contrast, at least 2 New World monkey species, including the cotton-top tamarin (Sanguinus oedipus oedipus)31 and the owl monkey (Aotus trivirgatus),32 develop B-cell lymphoma upon human EBV exposure. We report here the occurrence of a rare case of CD8+ T-cell MF in a pigtailed macaque and the isolation of a novel EBV-like virus from 2 transformed CD8+ T-cell lines obtained from the blood and the skin of the lymphomatous macaque. These findings may help in the development of an animal model for EBV-like virus-induced malignant proliferation of T cells.
MATERIALS AND METHODS
Establishment of macaque blood and skin T-cell lines.
Peripheral blood mononuclear cells (PBMC) were isolated by density-gradient centrifugation on lymphocyte separation medium (LSM; Organon Teknika Corp, Durham, NC) from anticoagulated blood obtained from pigtailed macaque J94356 before death. The cell layers were washed twice in Dulbecco’s phosphate-buffered saline (DPBS; GIBCO BRL, Gaithersburg, MD) and were suspended in RPMI (GIBCO BRL) with 10% heat-inactivated (HI) fetal bovine serum (FBS; GIBCO BRL) with penicillin/streptomycin (500 U/mL and 500 g/mL, respectively; GIBCO BRL) and L-glutamine (2 mmol/L; GIBCO BRL) and stimulated with phytohemagepletrinic (PHA; 5 g/mL; Murex Diagnostics, Norcross, GA). At 72 hours, the cells were washed twice in DPBS and resuspended in fresh RPMI with 10% HI FBS, penicillin/streptomycin, L-glutamine, and recombinant interleukin-2 (IL-2; 20 U/mL; Boehringer Mannheim, Indianapolis, IN). Fresh skin biopsy tissues from diseased areas were minced to release single cells. These were banded on LSM and placed in culture as described above. All cells were incubated at 37°C with 5% CO2; fresh media were added once or twice per week, as needed, to maintain adequate cell growth. After 6 weeks in culture, both the PBMC and skin-derived cells showed evidence of increased proliferation and clustering and the cells have been cultured for more than 1 year. The concentration of IL-2 in the media in both cultures was decreased in a stepwise manner from an initial concentration of 20 U/mL. Each cell line was maintained at a dose of IL-2 until it was clear that cell growth was unimpaired. At the end of 5 months, both J94356PBMC and J94356SKIN cells were growing well, albeit at a slower rate than their IL-2–dependent counterparts.
DNA extraction, polymerase chain reaction (PCR) amplification, and DNA sequence analysis.
Pellets of PBMC obtained by Ficoll gradient separation were incubated for 1 hour at 37°C in a buffer containing 0.5% sodium dodecyl sulfate (SDS), 10 mmol/L Tris (pH 8.0), 1 mmol/L EDTA (pH 8.0), and RNAse (20 g/mL; Boehringer Mannheim) before the addition of Proteinase K (100 μg/mL; Boehringer Mannheim). After an overnight incubation at 37°C, DNA was extracted from the lysates with phenol and chloroform. Frozen tissues, stored at −80°C until processing, were thawed on ice and finely minced with sterile blades. The minced tissue was resuspended in ice-cold DPBS and washed twice before lysis and extraction as detailed above. DNAs were initially amplified with the primer pools DFASA/GDTD1B and VYGA/GDTD1B described by Rose et al,33 corresponding to a conserved region in the herpesvirus family that codes for substrate binding sites within the DNA polymerases.
PCR conditions were optimized using the Invitrogen PCR Optimizer Kit. Optimal amplification was obtained with 500 ng of genomic DNA, 25 pmol of each primer, 300 mmol/L Tris-HCl 5× buffer (pH 10) containing 2.5 mmol/L MgCl2, 75 mmol/L (NH4)2SO4, 10 mmol/L dNTP, and 2.5 U Taq polymerase. The templates were subjected to a maximum of 45 cycles of amplification, each cycle consisting of 15 seconds at 94°C, 30 seconds at 60°C, and 15 seconds at 72°C (Perkin Elmer GeneAmp PCR system 9600 thermal cycler; Perkin Elmer, Branchburg, NJ). An aliquot of these amplification products was electrophoresed in a 1.5% agarose gel, stained with 0.5 μg/mL ethidium bromide, and visualized under UV transillumination. Amplification products of the predicted size (∼536 bp) were detected, and the resulting fragments were cloned into Inv α F′ bacterial cells using the TA cloning kit from Invitrogen (Carlsbad, CA). White colonies were picked and grown in LB broth in the presence of ampicillin at 37°C overnight. Plasmids were isolated using the Promega Miniprep (Promega, Madison, WI) and digested with EcoRI to confirm the presence of the appropriately sized insert, and DNA sequencing was performed using the T7 Sequenase v2.0 chain-termination method from Amersham Life Science (Cleveland, OH) according to the manufacturer’s instructions. Based on the sequence of the DNA fragment first amplified, a primer set, EDR8/97 sense 5′-ATCTCTGTTATTCTACC-3′, MGF2B antisense 5′-CCTGCAGCGTCA-3′, was synthesized. An aliquot of the first PCR products was amplified using these sequence-specific primers. PCR amplification with the primer set was optimal at 5× buffer (pH 8.5) containing 300 mmol/L Tris-HCl, 2.0 mmol/L MgCl2, 75 mmol/L (NH4)2SO4, 10 mmol/L dNTP, and 2.5 U Taq polymerase. The templates were subjected to 45 cycles of amplification as described above. The nested PCR products were analyzed by gel electrophoresis and subsequently cloned into Inv α cells, and sequence analysis was performed.
Other regions of the viral genome were amplified using the primer pairs described by Ino et al25 and included the 2s/2as pair covering IR1 region and the 11s/11as pair covering part of the EBV BRRF-1 region.24 PCR products were analyzed by gel electrophoresis and cloned into Inv α cells, and the DNA sequence was obtained as described above.
Southern blot analysis.
DNA (10 μg) of each sample was digested with Sau3AI orPvu II, electrophoresed in 0.8% agarose gels, and blotted onto Nylon membranes (Nytran plus; Schleicher & Schuell, Keene, NH). The membranes were hybridized overnight at 42°C with the PCR-amplified EBV-like probe labeled using random-primer reaction (Boehringer Mannheim). Hybridization, washing, and detection were performed according to the manufacturer’s instructions.
Phylogenetic analysis.
Phylogenetic analysis of the novel HVMNEpol-gene fragment (465 bp) was performed using sequences from an analogous strain from Macaca arctoides29 (HVMA) and the following related strains published previously: human EBV: V01556; retroperitoneal fibromatosis herpesvirus (RFHV)—Macaca nemestrina: AF005478; retroperitoneal fibromatosis herpesvirus (RFHV)—Macaca mulatta: AF005479; herpesvirus Saimiri (HVS): M31122; Kaposi sarcoma herpesvirus (KSHV): AF005477; equine herpesvirus 2 (EHV-2): and U20824; herpesvirus Papio (HVPA).34Sequences were aligned using ClustalX version 1.63B.35 Genetic distance estimates among all pairs of pol sequences were estimated using the Tajima-Nei model of substitution.36 Phylogenetic associations, using the computer program Mega Version 1.01,37 were ascertained by minimum evolution estimated by neighbor-joining (NJ) in conjunction with the Tajima-Nei model of substitution. Bootstrap analyses, consisting of 100 iterations, were performed to determine the consistency of the data in recapitulating the same tree for NJ analysis. Values of 70% or more were considered strong support for the adjacent node.38
Electrophoretic mobility shift assay (EMSA).
PHA-stimulated human PBMC were cultured in the presence of IL-2 (20 U/mL) for 8 days and used as a control for this assay. In starvation experiments, 1 × 107 stimulated PBMC and IL-2–dependent and –independent J94356PBL were resuspended in 20 mL of RPMI 1640 with 1% FBS after washing with 1× PBS twice and incubated for 21 hours at 37°C in 5% CO2. Protein lysates were prepared in 20 mmol/L HEPES (pH 7.9), 450 mmol/L NaCl, 0.4 mmol/L EDTA, 0.5 mmol/L dithiothreitol (DTT), 25% glycerol, 1 mmol/L Na3VO4, 1 mmol/L AEBSF, 20 μg/mL aprotinin, and 20 μg/mL leupeptin. The binding reaction was performed by preincubating 5 μg of nuclear extracts with 1 μg of poly(dI-dC) in a buffer containing 5.9 mmol/L HEPES (pH 7.9), 30 mmol/L KCl, 5.9 mmol/L Tris (pH 7.4), 0.7 mmol/L DTT, 0.6 mmol/L EDTA, 8.9% glycerol, 0.1 mmol/L Na3VO4, 1 mmol/L AEBSF, 20 μg/mL aprotinin, and 20 μg/mL leupeptin in ice for 20 minutes. A32P-labeled probe (20,000 cpm) corresponding to the MGF binding site in the β-casein gene promoter (5′-TAGATTTCTAGGAATTCG-3′) was added to the reaction mixture and incubated on ice for 30 minutes. For the supershift assay, STAT5 antibody (N-20; Santa Cruz Biotechnology, Santa Cruz, CA) was incubated with cell extracts on ice for 20 minutes after the addition of radiolabeled probe. Complexes were resolved on 4.5% polyacrylamide gels.
Immunohistochemistry and EBV Epstein-Barr viral-encoded RNA (EBER) in situ hybridization.
CD3 immunostaining was performed on formalin-fixed, paraffin-embedded tissue using a rabbit polyclonal anti-CD3 serum (A0452; DAKO, Carpinteria, CA) at 1:50 dilution followed by biotinylated antirabbit secondary antibodies and ABC reagent (Vector Labs, Burlingame, CA) and DAB substrate (Scytek Laboratories, Logan, UT). CD8 immunostaining was performed on tissue sections frozen in OCT using a mouse anti-CD8 monoclonal antibody (clone Leu-2a; Becton Dickinson, San Jose, CA) followed by a biotinylated antimouse Ig (ABC reagent; Vector Labs) and DAB substrate (Scytek Laboratories). Each immunohistochemical analysis included a test tissue assayed either using antibody against an irrelevant antigen (from the appropriate species) or in absence of primary antibody. Cell lines derived from peripheral blood and skin were formalin-fixed, pelleted, and paraffin-embedded. Sections (3 to 5 μm) were cut onto glass slides and hybridized with a cocktail of fluorescein isothiocyanate (FITC)-conjugated probes reactive with the EBER RNAs, which are abundantly transcribed in all cells latently infected with EBV (Novocastra Laboratories, Newcastle, UK). Hybridization was detected using an alkaline-phosphatase–conjugated, anti-FITC antibody (DAKO, High Wycombe, UK) and nitro-blue tetrazolium as chromogenic substrate (DAKO). Subsequently, the EBER in situ hybridization was performed using sections of skin biopsies obtained from monkey skin affected with lymphoma. Hybridization using a poly-dT probe (Novocastra) was used to confirm the integrity of the RNA in all tissue sections.
Na-butyrate induction and electronmicroscopy (EM).
Cells (5 × 105) from the J94356PBMC cell line were incubated in medium containing Na-butyrate (1 μmol/L) for 48 hours, fixed in 0.65% paraformaldehyde and 0.8% gluteraldehyde for 1 to 2 hours, and then incubated in 0.1 mol/L cacodylate buffer (pH 7.2 to 7.4) containing 0.1 mol/L sucrose. The cell pellet was postfixed with 0.5% osmium fixative (pH 7.2 to 7.4) for 1 hour, processed through an acetone gradient to 100% acetone, and then placed sequentially in 1:1 and 1:3 acetone-resin mixes. The cells were embedded into a pure resin capsule and cured in an oven at 60°C for 48 hours. The pellet was sliced with an Ultra Microtome (Leica Microsystems UK, Milton Keynes, UK) at 50 to 60 nm, collected on copper grids, and stained with uranyl acetate and lead citrate. The preparations were analyzed on a Zeiss 109 Transmission Electron Microscope (Cast Leiss, Welwyn Garden City, UK).
RESULTS
MF in animal J94356: Clinical presentation and histopathological findings.
In May 1996, an 18-month-old pigtailed macaque (animal J94356), housed at the Washington Regional Primate Research Center, developed a unilateral eye infection with marked inflammation of the conjunctiva and eyelids, which eventually spread to the opposite eye. Improvement in the clinical manifestations was observed after treatment with antihistamine and antibiotics, but inflammation of both conjunctivas and eyelids recurred 8 days after treatment was discontinued. A small unilateral corneal ulceration was found on examination and antibiotic and antihistamine therapy was reinstated. At this time, an extensive work-up to test for viral infections was performed. A serologic panel of tests was run at a commercial laboratory. Tests were negative for herpes B, HSV-1, measles, and simian immunodeficiency virus and were positive for cytomegalovirus. In August 1996, the ulcerative lesions recurred in the perioral area and new lesions involving the face, trunk, forearms, and legs appeared. At this time, the serology was negative for herpes B and STLV; however, EBV serology was positive and the lesions improved on a 2-week course of acyclovir. Biopsy results of skin lesions examined early in October 1996 were consistent with a multifocal cutaneous lymphoma. The animal was continued on antibiotic therapy with mild improvement of the lesions.
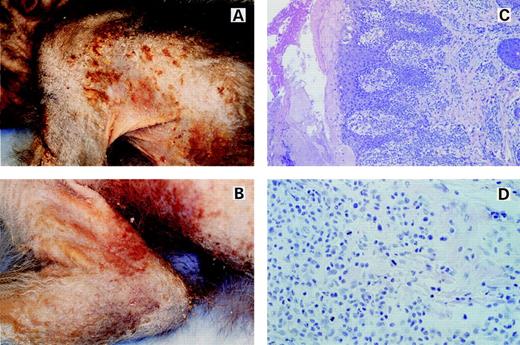
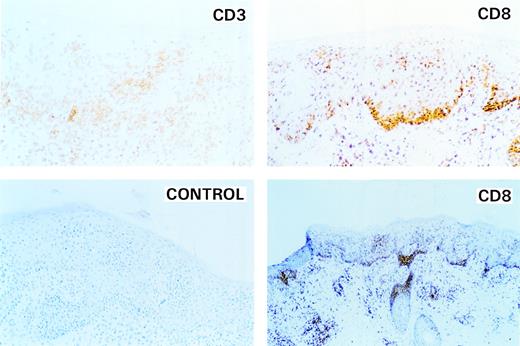
However, in mid-January 1997, the lesions dramatically worsened and, after a course of steady deterioration, the animal was killed in January 1997. At the time of death, animal J94356 displayed an erythematous, hyperkeratotic rash, with alopecia and excoriated lesions with prominent red borders and numerous plaque-like lesions on extensive areas of the skin (Fig 1A and B). Hematoxylin Eosin staining of a skin-lesion sample showed profusely infiltrating mononuclear cells in the dermis and epidermis with prominence at the dermal-epidermal junction in periadnexal tissue and in perivascular tissue (Fig 1C). In the epithelium, the cells were arranged in small solid clusters at the dermal-epidermal interface, resembling Pautrier’s microabcesses. The cells had moderately sized nuclei containing 1 to 2 small nucleoli and up to 2 mitotic figures were present per 400× field (Fig 1D). A few histiocytes and eosinophils were admixed with the neoplastic cells. There were multifocal erosions in the epithelium, which was acanthotic and hyperkeratotic (Fig 1C). Immunohistochemical analysis of the skin lesion showed CD3+ infiltrating T cells both in the dermis (top left panel of Fig 2) and a small portion of the epidermis. Coalescing clusters of CD3+ cells were present along the dermal-epidermal junction and within the epidermis. The lineage of the infiltrating cell population was further characterized and found to be strongly CD8+ (right top and bottom panels of Fig 2) and CD4− (not shown). No staining was observed with an irrelevant antibody (left bottom panel of Fig 2). Thus, clinical features of the skin lesions and the histopathology were indicative of a CTCL with a CD8+ T-cell phenotype.
Macroscopic and histologic characteristics of a skin lesion in the lymphomatous macaque. (A) and (B) show the macroscopic appearance of the lesions present in pigtailed macaque J94356. Note the extensive areas of involvement. The lesions are hyperkeratotic, with alopecia, and with excoriated areas surrounded by prominent borders. (C) shows a low-power magnification of the histology of a skin lesion stained with Hematoxylin and Eosin (HE). Note the profuse mononuclear cell infiltration of the dermis, dermal-epidermal junction, epidermis, periadnexal tissue, and perivascular areas. (D) shows a high-magnification view (400×) of the infiltrating cells stained with HE. Numerous mitotic figures are present on a homogeneous background. The cells have moderately sized nuclei with 1 or 2 nucleoli.
Macroscopic and histologic characteristics of a skin lesion in the lymphomatous macaque. (A) and (B) show the macroscopic appearance of the lesions present in pigtailed macaque J94356. Note the extensive areas of involvement. The lesions are hyperkeratotic, with alopecia, and with excoriated areas surrounded by prominent borders. (C) shows a low-power magnification of the histology of a skin lesion stained with Hematoxylin and Eosin (HE). Note the profuse mononuclear cell infiltration of the dermis, dermal-epidermal junction, epidermis, periadnexal tissue, and perivascular areas. (D) shows a high-magnification view (400×) of the infiltrating cells stained with HE. Numerous mitotic figures are present on a homogeneous background. The cells have moderately sized nuclei with 1 or 2 nucleoli.
Identification of the phenotype of neoplastic cells infiltrating the skin. Top left panel (CD3) shows an area of affected skin stained with monoclonal antibodies against CD3. Clusters of CD3+ cells are present along the dermal-epidermal junction and in the epidermis. Top and lower right panels (high-and low-power magnification, respectively) demonstrate that the infiltrating cells belong to the CD8+ T-cell subgroup. Cells strongly positive for CD8 are present in clusters in the dermis along the dermal-epidermal junction and in the epidermis. Lower left panel is a control for antibody specificity.
Identification of the phenotype of neoplastic cells infiltrating the skin. Top left panel (CD3) shows an area of affected skin stained with monoclonal antibodies against CD3. Clusters of CD3+ cells are present along the dermal-epidermal junction and in the epidermis. Top and lower right panels (high-and low-power magnification, respectively) demonstrate that the infiltrating cells belong to the CD8+ T-cell subgroup. Cells strongly positive for CD8 are present in clusters in the dermis along the dermal-epidermal junction and in the epidermis. Lower left panel is a control for antibody specificity.
Infiltrates of mononuclear cells undergoing mitosis with a few eosinophils and neutrophils were also observed in several organs, including the lungs and lymph nodes (data not shown). Occasional mitoses were present in the enlarged lymphoid follicles and germinal centers. The spleen contained prominent lymphoid nodules and hyaline material in the germinal centers, and the liver had a mild multifocal lymphocytic infiltrate in portal areas and subcapsular tissue. Although staining with CD8 antibodies was not performed in the organs, it is highly likely that these infiltrates were composed of the same CD8+ T cells found in the skin.
Detection of an EBV-like virus in the transformed CD8+T-cell lines from animal J94356.
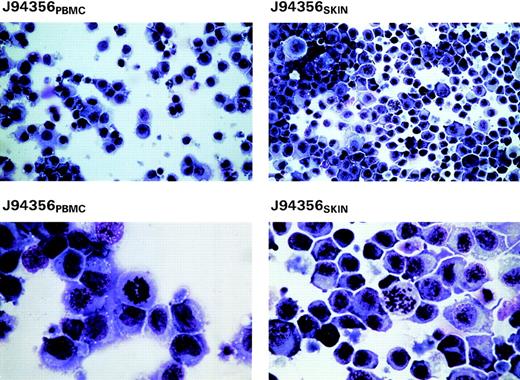
Peripheral mononuclear cells obtained from whole blood at the time of necropsy were cultured with PHA and recombinant IL-2. Skin-derived cells were also placed in culture under similar conditions and, like the PBMC culture, proliferated continuously in response to IL-2. Both cell lines have been in culture for approximately 2 years. Giemsa staining of cytospin preparations of these cells at 2.5 months after initiation of culture of the cells indicated their pleomorphism with myeloid-like features and multiple mitotic figures at higher magnification (lower panels of Fig 3). Cytogenetic analyses of the chromosomes confirmed the simian origin of both cell lines (2n = 42; data not shown).
Giemsa stains of cytospin preparations from the 2 cell lines derived from blood and skin of animal J94356 (J94356PBMC and J94356SKIN, respectively) after 2 months in culture in RPMI with 10% FBS and 20 U/mL of IL-2. High-magnification views (lower panels) show the pleomorphic appearance of these cells with myeloid-like features and presence of mitotic figures.
Giemsa stains of cytospin preparations from the 2 cell lines derived from blood and skin of animal J94356 (J94356PBMC and J94356SKIN, respectively) after 2 months in culture in RPMI with 10% FBS and 20 U/mL of IL-2. High-magnification views (lower panels) show the pleomorphic appearance of these cells with myeloid-like features and presence of mitotic figures.
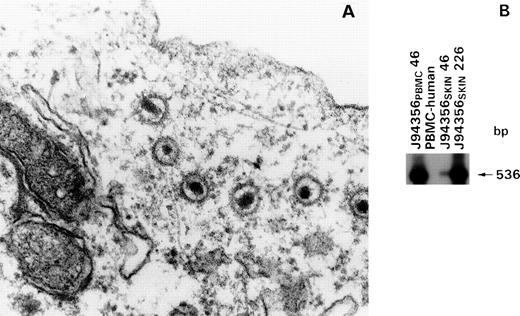
Electron-microscopic analysis of the cell lines under standard culture conditions was negative even after an extensive search for any viral particles, including retroviruses. In addition, the supernatants of these cell lines were consistently negative for reverse transcriptase activity. However, electron microscopic analysis performed 48 hours after treatment with Na-butyrate allowed the detection of large enveloped virions in the cytoplasm of cells derived from the PBMC of animal J94356, consistent with the presence of a herpesvirus (Fig 4A). To assess the genetic composition of this herpesvirus, we used generic DNA primers previously demonstrated to amplify efficiently genomes from several members of the herpesvirus family,33 which amplified a DNA fragment of 536 bp from both CD8+ T-cell lines. This DNA fragment (p536) was molecularly cloned and used as a probe to hybridize the DNA from primary J94356 PBMC cultured for 46 days as well as from the J94356 skin culture at days 46, 226, and thereafter. After PCR amplification, the expected DNA fragment of 536 bp was found in the macaque cell lines but not in human PBMC (Fig 4B), suggesting that this putative herpesvirus persisted in both cell lines originated from animal J94356.
Detection of HVMNE in the cell lines J94356PBMC and J94356SKIN by electron microscopy and DNA PCR. (A) EM of cell line J94356PBMCharvested 48 hours after induction with Na-butyrate (1 μmol/L). Extensive EM search in the same cell line without Na-butyrate induction failed to show viral particles (not shown). (B) Southern blot of PCR products using probe 536. Detection of a 536-bp DNA fragment in the J94356PBMC and J94356SKIN cell lines at 46 days in culture and after 226 days in culture in line J94356SKINby PCR using the generic primer pair DFASA/GDTD1B (described by Rose et al33).
Detection of HVMNE in the cell lines J94356PBMC and J94356SKIN by electron microscopy and DNA PCR. (A) EM of cell line J94356PBMCharvested 48 hours after induction with Na-butyrate (1 μmol/L). Extensive EM search in the same cell line without Na-butyrate induction failed to show viral particles (not shown). (B) Southern blot of PCR products using probe 536. Detection of a 536-bp DNA fragment in the J94356PBMC and J94356SKIN cell lines at 46 days in culture and after 226 days in culture in line J94356SKINby PCR using the generic primer pair DFASA/GDTD1B (described by Rose et al33).
Genetic and phylogenetic characterization of the herpesvirus from animal J94356.
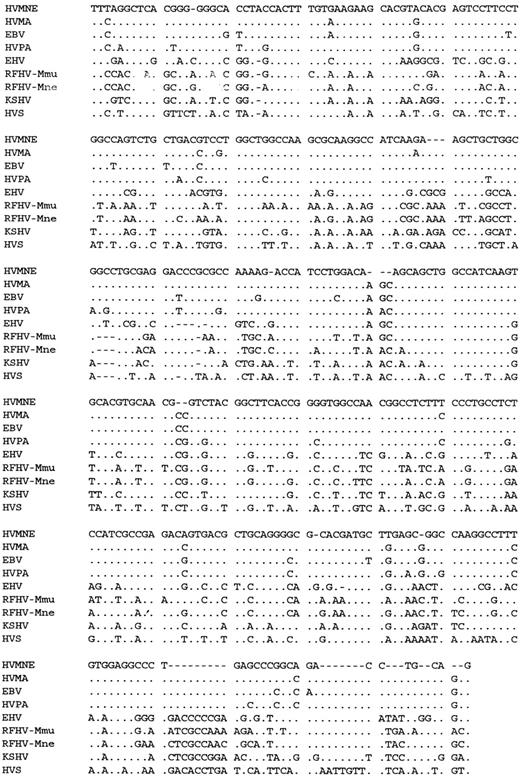
To establish the genetic relationship of the putative viral fragment found in cells from animal J94356 to other known nonhuman primate herpesviruses, we obtained the DNA sequence of plasmid p536 and aligned it to the equivalent polymerase region of the human KSHV and EBV as well as EBV-like viruses or rhadinoviruses from various animal species (Fig 5).29,33,34,39 Both DNA sequence alignment and the phylogenetic analysis by the NJ method (Fig 6) indicated that the herpesvirus in J94356 cells clustered with the human EBV and the nonhuman primate herpesviruses, HVMA and HVPA,34 and was distantly related to the known rhadinoviruses.33,39 40
Alignment of DNA sequence from the polymerase gene of HVMNE and other herpesviruses.
Alignment of DNA sequence from the polymerase gene of HVMNE and other herpesviruses.
Phylogenetic tree based upon 353 bp of the polgene. Shown is the minimum evolution tree estimated by NJ method using distances among all pairs of sequences.36 Numbers on branches indicate the percentage of sequence divergence. Numbers in italics are bootstrap support (ME-NJ) for the adjacent node.
Phylogenetic tree based upon 353 bp of the polgene. Shown is the minimum evolution tree estimated by NJ method using distances among all pairs of sequences.36 Numbers on branches indicate the percentage of sequence divergence. Numbers in italics are bootstrap support (ME-NJ) for the adjacent node.
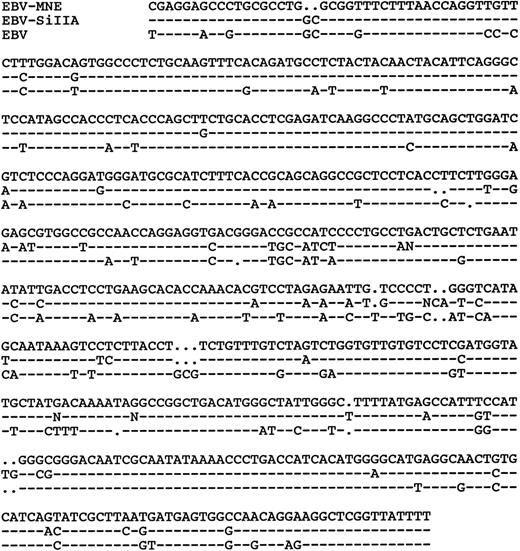
Two other EBV-like viruses, SiIIA and HVMF-1, have been isolated fromMacaca fascicularis.26 41 To assess the genetic relationship of HVMNE to SiIIA, we analyzed the HVMNE DNA with primer sets for the BRRF-1 EBV-equivalent region. As demonstrated in Fig 7, HVMNE appears to be distinct from SiIIA, because the DNA sequence of this region was 85% identical to EBV and 90% identical to the SiIIA strains. In the case of HVMF-1, the DNA sequence from this region is not available; therefore, HVMF-1 and HVMNE DNA were compared by restriction enzyme and Southern blot analyses, which demonstrated differences in both restriction sites used and suggested genetic diversity between HVMNE and HVMF-1 (Fig 8). We therefore designated the virus from animal J94356 as HVMNE for consistency with the common name used for other EBV-like viruses found in nonhuman primates.
Southern blot analysis of HVMNE and HVMF-1–infected cell DNAs. Human PBMC DNA was used as negative control. The molecular-weight complexes are indicated at the left of each panel and the restriction enzyme used is indicated at the bottom of each panel.
Southern blot analysis of HVMNE and HVMF-1–infected cell DNAs. Human PBMC DNA was used as negative control. The molecular-weight complexes are indicated at the left of each panel and the restriction enzyme used is indicated at the bottom of each panel.
JAK/STAT constitutive activation in CD8+ T cells infected with HVMNE correlates with the acquisition of IL-2 independence.
Transforming viruses usually induce growth-factor independence by interfering with intracellular signaling pathways,42-45 and this event occurs also in EBV-transformed B-cell lines.46In addition, in human hematopoietic malignancies, including Sézary syndrome, constitutive activation of JAK/STAT protein has also been described.47-50
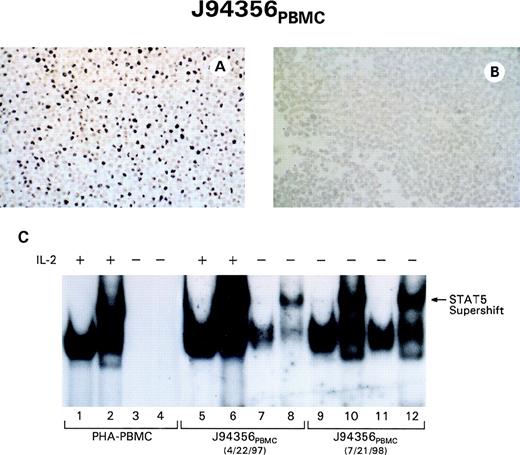
Both T-cell lines from animal J94356 acquired the ability to grow independently from exogenous IL-2, and, to assess whether constitutive activation of STAT5 protein also correlated with the acquisition of IL-2 independence, cell extracts from normal human PHA-stimulated PBMC and the J94356PBMC cell line, at 3 months and 17 months from the start of culture, were analyzed after IL-2 withdrawal. At 3 months from initiation of culture, some degree of constitutive STAT5 binding to the MGF probe could be already observed in the absence of IL-2 and serum (Fig 9C, lanes 7 and 8), and, at 17 months, the cells in culture in absence of exogenous IL-2 displayed constitutive binding of activated STAT5 to the MGF probe (Fig9C, lanes 9 through 12). Constitutive STAT1 and STAT3 binding was also observed when the specific SIE probe was used (data not shown). Thus, as in the case of HTLV-I–infected cells, the transition to IL-2 independence correlates with the acquisition of constitutive activation of STAT proteins.43 44 To assess whether the resident herpesvirus expression was contained in the IL-2–independent cells, viral expression was analyzed using the human EBV EBER probe. As demonstrated in Fig 9A and B, a clear nuclear signal was evident in most HVMNE-infected cells, indicating also a good conservation of this sequence in the HVMNE.
The transition of the J94356PBMC expressing HVMNE cell lines to IL-2 independence is associated with STAT5 activation and HVMNE expression. J94356PBMC cells hybridized with EBER RNA (A) or incubated with hybridization buffer in the absence of probe (B). (C) EMSA on control PBMC (lanes 1 through 4) and the J94356PBMCcell line in the IL-2–dependent (lanes 5 through 8) and IL-2–independent (lanes 9 through 12) status.
The transition of the J94356PBMC expressing HVMNE cell lines to IL-2 independence is associated with STAT5 activation and HVMNE expression. J94356PBMC cells hybridized with EBER RNA (A) or incubated with hybridization buffer in the absence of probe (B). (C) EMSA on control PBMC (lanes 1 through 4) and the J94356PBMCcell line in the IL-2–dependent (lanes 5 through 8) and IL-2–independent (lanes 9 through 12) status.
Detection of EBV-like DNA in the tissues of animal J94356.
To investigate whether HVMNE found in the CD8+T-cell lines from animal J94356 was also present in the infiltrating CD8+ neoplastic T cells from the skin lesion as well as other organs, the DNA from various tissues was analyzed using the same primer set used to amplify the HVMNE polymerase gene from the DNA of the J94356 cell lines.
As demonstrated in Fig 10A, viral DNA sequences were found in the skin but not the PBMC obtained from animal J94356 at the time of CTCL diagnosis (1996). However, at time of death (1997), viral DNA was also found in the uncultured PBMC as well as in multiple skin specimens, lymph nodes, and muscles. The finding of viral DNA in PBMC at time of death is consistent with the clinical finding of an abnormal T-cell count (18,000/mL) in the blood of animal J94356.
Detection of herpesvirus DNA by PCR and of EBV-specific RNA by in situ hybridization in tissues of animal J94356. (A) Southern blot of PCR products using probe p536. Detection of 536-bp and 236-bp DNA fragments in tissues from animal J94356 by PCR using primer pairs DFASA/GDTD1B (top) and VYGA/GDTD1B (bottom) at 2 time points. Lanes 1 and 2, PCR results on DNA obtained from noncultured PBMC and affected skin approximately 7 weeks before death (1996). Lanes 3 through 8, PCR products on DNA obtained from animal J94356 at time of necropsy (time 0). Lane 3, PBMC, noncultured; lane 4, affected skin; lane 5, lymph node; lane 6, muscle; lane 7, normal skin. Lane 8, negative control (DNA from normal human PBMC). EBV EBER in situ hybridization performed on sections of affected skin biopsy at low (B) and high (C) magnification. A large number of cells expressing EBV EBER RNA are found infiltrating the dermis and epidermis in the lymphomatous animal.
Detection of herpesvirus DNA by PCR and of EBV-specific RNA by in situ hybridization in tissues of animal J94356. (A) Southern blot of PCR products using probe p536. Detection of 536-bp and 236-bp DNA fragments in tissues from animal J94356 by PCR using primer pairs DFASA/GDTD1B (top) and VYGA/GDTD1B (bottom) at 2 time points. Lanes 1 and 2, PCR results on DNA obtained from noncultured PBMC and affected skin approximately 7 weeks before death (1996). Lanes 3 through 8, PCR products on DNA obtained from animal J94356 at time of necropsy (time 0). Lane 3, PBMC, noncultured; lane 4, affected skin; lane 5, lymph node; lane 6, muscle; lane 7, normal skin. Lane 8, negative control (DNA from normal human PBMC). EBV EBER in situ hybridization performed on sections of affected skin biopsy at low (B) and high (C) magnification. A large number of cells expressing EBV EBER RNA are found infiltrating the dermis and epidermis in the lymphomatous animal.
To assess whether the HVMNE sequences found in the tissues were also detectable in the CD8+ neoplastic T cells in vivo, the postmortem tissue specimens of animal J94356 were stained with the EBER RNA probe. As demonstrated in the left panels of Fig 10B and 10C, several cells in the skin expressed EBER RNA and the distribution pattern closely mirrored the pattern observed with immunohistochemical analyses using CD8-specific antibody (Fig 2); this suggests that HVMNE was present in the CD8+ T cells that infiltrated the dermis and epidermis of the lymphomatous animal.
DISCUSSION
Herpesviruses have been found in most animal species, and the family Herpesviridae includes the 3 subfamilies Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae. Within the Gammaherpesvirinae subfamily, 2 genuses have been distinguished, the lymphocryptoviruses (Epstein-Barr–like viruses) and rhadinoviruses (Saimiri-ateles–like herpesviruses).51 Only members of the Gammaherpesvirinae subfamily have been associated with human malignancies. EBV causes B-cell lymphoma in immunodeficient individuals and is epidemiologically associated with Burkitt’s lymphoma, Hodgkin’s disease, and nasopharyngeal carcinoma.14Similarly, human herpesvirus 8 has been epidemiologically linked to Kaposi sarcoma40 and rare forms of lymphoma.52-55 EBV induces B-cell lymphoma in previously unexposed New World monkeys but not in Old World primates, presumably because of preexisting cross-immunity against EBV, as demonstrated by their frequent EBV seropositivity.56 However, after simian immunodeficiency virus infection, approximately one third of the infected macaques develop B-cell lymphoma, and this event has been associated with the presence of an EBV-like virus in Macaca fascicularis.57
We describe here the identification and partial characterization of HVMNE from a pigtailed macaque with MF. HVMNEis distinct from the known nonhuman primate EBV-like herpesviruses as demonstrated by the phylogenetic analysis of the polymerase gene and the BRRF-1 region. The finding of HVMNE in the CD8+ infiltrating T cells in vivo as well as in transformed CD8+ T-cell lines in vitro supports the notion that HVMNE might target T cells and might have been involved in the development and/or progression of MF in animal J94356.
EBV has been reported to induce B-cell lymphoma in New World monkeys but not in rabbits,58 whereas other EBV-like strains from nonhuman primates, such as HVMA (Macaca arctoides) and EBV-related strains from cynomolgus monkeys, have been demonstrated to induce B-cell lymphoma in rabbits.24,29 Conversely, the pathogenicity of nonhuman-primate EBV-like viruses has not been assessed in New World monkeys. Interestingly, HSV strains A and B cause lymphoma in New World primates but not in New Zealand white rabbits,58 suggesting that pathogenicity of different viral strains varies in different species. Transfusion of blood from the lymphomatous animal discussed in this study to other pigtailed macaques did not result in lymphoma (data not shown), presumably because of a preexisting infection of EBV-related virus in this species of Old World monkey.30 It would be of interest to assess whether HVMNE induces lymphoma in the rabbit model. If so, HVMNE may provide a model to study viral and cellular determinants involved in oncogenesis. Lastly, the findings reported here may prompt a further investigation on the possible role of human EBV strains in T-cell lymphomas.
ACKNOWLEDGMENT
The authors thank Peter Biberfeld for helpful discussion and Steven Snodgrass for editorial assistance. We thank June Freeland and Ross Blackley, who performed the viral stimulation and EM studies.
Supported in part by National Institutes of Health Grant No. RR00166 (Washington Regional Primate Research Center, Seattle, WA).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to G. Franchini, MD, Chief, Section of Animal Models and Retroviral Vaccines, Basic Research Laboratory, Division of Basic Sciences, National Cancer Institute, National Institutes of Health, 41 Library Dr, Bldg 41, Room D804, Bethesda, MD 20892; e-mail:veffa@helix.nih.gov.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal