Abstract
We determined the breakpoint genes of the translocation t(4;11)(q21;p15) that occurred in a case of adult T-cell acute lymphocytic leukemia (T-ALL). The chromosome 11 breakpoint was mapped to the region between D11S470 and D11S860. The nucleoporin 98 gene (NUP98), which is rearranged in several acute myeloid leukemia translocations, is located within this region. Analysis of somatic cell hybrids segregating the translocation chromosomes showed that the chromosome 11 breakpoint occurs withinNUP98. The fusion partner of NUP98 was identified as theRAP1GDS1 gene using 3′ RACE. RAP1GDS1 codes for smgGDS, a ubiquitously expressed guanine nucleotide exchange factor that stimulates the conversion of the inactive GDP-bound form of several ras family small GTPases to the active GTP-bound form. In theNUP98-RAP1GDS1 fusion transcript (abbreviated asNRG), the 5′ end of the NUP98 gene is joined in frame to the coding region of the RAP1GDS1 gene. This joins the FG repeat-rich region of NUP98 to RAP1GDS1, which largely consists of tandem armadillo repeats. NRG fusion transcripts were detected in the leukemic cells of 2 other adult T-ALL patients. One of these patients had a variant translocation with a more 5′ breakpoint in NUP98. This is the first report of anNUP98 translocation in lymphocytic leukemia and the first time that RAP1GDS1 has been implicated in any human malignancy.
THE STUDY OF GENES at the breakpoints of chromosome translocations has identified a large number of genes involved in the development of human cancer.1-6 We previously reported the translocation t(4;11)(q21;p15) as a t(4;11)(q21;p14-15) in a 21-year-old man with T-cell acute lymphocytic leukemia (T-ALL).7 Somatic cell hybrids containing the der(4) and der(11) chromosomes enabled the localization of the chromosome 11 breakpoint to 11p15.5 in the region between theIGF2 and RRM1 loci.8 Studies using cosmids as fluorescence in situ hybridization (FISH) probes on the patient material further localized the breakpoint region to between the 11p15.5 markers D11S470 and RRM1.9
We describe the identification of the chromosome 4 and 11 breakpoint genes as RAP1GDS1 and NUP98 and show that the (4;11)(q21;p15) translocation is recurrent in T-ALL. This is the first report of RAP1GDS1 involvement in any malignancy. NUP98has previously been shown to be involved in 3 distinct acute myeloid leukemia (AML) translocations. Two translocations involving theHOXA9 and DDX10 genes have been shown to be recurrent, whereas the translocation involving the HOXD13 gene has so far been reported in a single case of therapy-induced AML.10-13
MATERIALS AND METHODS
Patient samples.
Table 1 summarizes the clinical and laboratory features of the patients described here. Patient no. 1, a 21-year-old man, presented with moderate hepato-splenomegaly, a large mediastinal mass, and a white blood cell count of 423 × 109/L, a platelet count of 109 × 109/L, and a hemoglobin level of 7.8 g/dL and was diagnosed as ALL (French-American-British [FAB] L1). The blood film showed 99% blasts. The patient underwent 2 matched bone marrow transplantations but relapsed on both occasions. Patient no. 2, a 25-year-old woman, presented with a white blood cell count of 1.8 × 109/L, a platelet count of 23 × 109/L, and a hemoglobin level of 5.5 g/dL and was diagnosed as ALL (FAB L1). She showed cervical, axillary, and inguinal lymphadenopathy. The blood film showed 87% blasts. Induction of remission was unsuccessful, and the patient died 34 days after presentation. Patient no. 3, a 49-year-old man who presented with a white blood cell count of 169 × 109/L, a platelet count of 116 × 109/L, and a hemoglobin level of 9.3 g/dL, was diagnosed as ALL (FAB L2). The chest x-ray and computerized tomographic (CT) scan showed a thymic mass. The blood film showed 99% blasts. After 4 weeks of induction therapy, the bone marrow showed morphological remission, although thymic enlargement was still evident on the CT scan. Three months later, the marrow showed several foci of primitive cells, which is suggestive of early relapse. A decision was made not to persist with intensive therapy. The patient died 14 months after presentation.
Clinical Features, Cytogenetics, Immunophenotype, and Gene Rearrangements of Patients With a t(4;11)(q21p14-15)
| Patient No. . | Sex/ Age (yr) . | WBC (×109/L) . | Diagnosis . | Survival (mo) . | Karyotype . | Immunophenotype . | Gene Rearrangement . | Reference . |
|---|---|---|---|---|---|---|---|---|
| 1 | M/21 | 423 | ALL L1 | 43 | 46,XY,t(4;11)(q21p15) +2mar (presentation) 48,XY,t(4;11),−7, −7,−9,−9,11p+, 17p+,−20,−21, −21 +9 mar (final relapse) | CD2+, CD3+ (30%), CD4−, CD5+, CD7+, CD8−, CD10+, CD11b (14%), CD14−, CD19−, CD20−, CD33+ (34%), CD34+, CD71+, HLA-DR+ | IgH (R) TCRγ (R) | 7 This report |
| 2 | F/25 | 1.8 | ALL L1 | 1 | 46,XX,t(4;11)(q21p14-15),del(12)(p13), +del(13)(q12q14) | CD2+ (13%), CD3−, CD4−, CD5+ (13%), CD7+, CD8−, CD10+ (14%), CD13+ (5%), CD14−, CD19−, CD33+ (18%), CD34+ (5%) | IgH (G) TCRγ (R) | This report |
| 3 | M/49 | 169 | ALL L2 | 14 | 46,XY,t(4;11)(q21p15), del(5)(q13q31) | CD2−, CD4−, CD5+, CD7+, CD8−, CD10+ (9%), CD19−, CD34+ | IgH (G) TCRγ (R) | This report |
| 4 | M/14 | 1.4 | ALL L2 | 1 | 46,XY,t(4;11)(q21p14), 12p-/46,XY, t(4;11)(q21p14) | CD2−, CD5+, CD10−, CD15+, pan-T+, TdT+ | ND | 21 |
| 5 | M/40 | 127 | ALL | 21 | 46,XY,t(4;11)(q21p15) | T (surface markers not reported) | ND | 22 |
| 6 | F/6 | 49 | ALL L2 | 25+ | 46,XX,t(4;11) (q21p14-15) | CD2+, CD5−, CD7+, CD10−, CD11b+, CD13+, CD14−, CD15−, CD19−, CD20−, CD22−, CD33+, CD36−, HLA-DR+, TdT+ | ND | 23 |
| Patient No. . | Sex/ Age (yr) . | WBC (×109/L) . | Diagnosis . | Survival (mo) . | Karyotype . | Immunophenotype . | Gene Rearrangement . | Reference . |
|---|---|---|---|---|---|---|---|---|
| 1 | M/21 | 423 | ALL L1 | 43 | 46,XY,t(4;11)(q21p15) +2mar (presentation) 48,XY,t(4;11),−7, −7,−9,−9,11p+, 17p+,−20,−21, −21 +9 mar (final relapse) | CD2+, CD3+ (30%), CD4−, CD5+, CD7+, CD8−, CD10+, CD11b (14%), CD14−, CD19−, CD20−, CD33+ (34%), CD34+, CD71+, HLA-DR+ | IgH (R) TCRγ (R) | 7 This report |
| 2 | F/25 | 1.8 | ALL L1 | 1 | 46,XX,t(4;11)(q21p14-15),del(12)(p13), +del(13)(q12q14) | CD2+ (13%), CD3−, CD4−, CD5+ (13%), CD7+, CD8−, CD10+ (14%), CD13+ (5%), CD14−, CD19−, CD33+ (18%), CD34+ (5%) | IgH (G) TCRγ (R) | This report |
| 3 | M/49 | 169 | ALL L2 | 14 | 46,XY,t(4;11)(q21p15), del(5)(q13q31) | CD2−, CD4−, CD5+, CD7+, CD8−, CD10+ (9%), CD19−, CD34+ | IgH (G) TCRγ (R) | This report |
| 4 | M/14 | 1.4 | ALL L2 | 1 | 46,XY,t(4;11)(q21p14), 12p-/46,XY, t(4;11)(q21p14) | CD2−, CD5+, CD10−, CD15+, pan-T+, TdT+ | ND | 21 |
| 5 | M/40 | 127 | ALL | 21 | 46,XY,t(4;11)(q21p15) | T (surface markers not reported) | ND | 22 |
| 6 | F/6 | 49 | ALL L2 | 25+ | 46,XX,t(4;11) (q21p14-15) | CD2+, CD5−, CD7+, CD10−, CD11b+, CD13+, CD14−, CD15−, CD19−, CD20−, CD22−, CD33+, CD36−, HLA-DR+, TdT+ | ND | 23 |
Details of the 3 patients reported in this study (no. 1, 2, and 3) and 3 patients from the literature (no. 4, 5, and 6) are presented. All features except for survival are at presentation. The comparatively long survival of patient no. 1 is at least in part due to the patient having undergone 2 allogeneic transplantations. The survival time of patient no. 5 is unknown but is at least 25 months. Where surface markers are positive in less than 50% of cells, the values are indicated in parentheses. Rearrangements of the heavy chain of Ig (IgH) and of the T-cell receptor γ gene (TCRγ) are indicated as G (germline, no rearrangement) or R (rearranged).
Abbreviations: WBC, white blood cell count; ND, not determined.
Somatic cell hybrid screening.
Human-mouse somatic cell hybrids containing the der(4) and der(11) chromosomes from patient no. 1 were described previously.8Polymerase chain reaction (PCR) was performed on 100 ng of DNA using AmpliTaq Gold (Perkin-Elmer, Foster City, CA), with an initial denaturation at 94°C for 9 minutes followed by 35 cycles of 96°C for 30 seconds, 60°C for 1 minute, and 72°C for 45 seconds. We used published primers for D11S47014and D11S860.15 Primers for NUP98 (Genbank accession no. U41815), exon B (N1428F, 5′GGCATCTTTGTT TGGGAACAACC; N1531R, 5′CAAAGCCCAAAGTGGCTGTCG), and exon C (N1585F 5′CAGGCTGTTCTCCAGCAGCACA; N1681R, 5′CCTTCTTCTTAGGGTCTGACATC) were designed based on the published intron/exon boundaries.12 For exon B, 10 μL of PCR product was digested using 10 U Taq I (New England Biolabs, Beverley, MA) to distinguish the mouse product from the human product.
3′ RACE.
Total RNA was extracted using Trireagent (Sigma, St Louis, MO). One-microgram aliquots of peripheral blood mononuclear cell total RNA were reverse transcribed using Superscript II and the Adapter Primer (AP) from the 3′ RACE kit (Life Technologies, Gaithersburg, MD). The Expand Long Template PCR System (Roche, Mannheim, Germany) was used in all subsequent PCR amplifications. The reverse transcription product was amplified with an NUP98 exon B primer, N1459F (5′ATTGGAGGGCCTCTTGGTACAGGAG), and the Abridged Universal Amplification Primer (AUAP; Life Technologies). Touchdown PCR was performed with an initial step at 94°C for 2 minutes, followed by 10 cycles of 95°C for 30 seconds, 70°C minus 1°C per cycle for 30 seconds, and 68°C for 8 minutes, followed by 25 cycles of 95°C for 30 seconds, 60°C for 30 seconds, and 68°C for 8 minutes plus 20 seconds per cycle. A biotinylated NUP98 exon B oligo, N1491F (5′GGCCCCTGGATTTAATACTACG), internal to the oligo used in the first round, was used to enrich for NUP98 containing sequences using streptavidin-coated magnetic beads (Promega, Madison, WI).16 Second-round PCR of the enriched product was performed using an NUP98 exon B primer, N1511F (5′CGACAGCCACTTTGGGCTTTGGAGC), internal to the previous 2 sense primers and the AUAP with cycling conditions identical to the first-round PCR. Second-round PCR products were electrophoresed in low melting point agarose gels, purified using Wizard PCR Preps (Promega), cloned into pGEM-T (Promega), and sequenced.
PCR and reverse transcription-PCR (RT-PCR) of fusion mRNAs.
Reverse transcription of 1 μg of total RNA with Superscript II and random hexamers was performed according to the manufacturer’s protocol (Life Technologies). One twentieth of the reverse transcription was used for PCR. NUP98 forward primers (N1265F or N1428F) and aRAP1GDS1 reverse primer, R108R (5′TTGAGCCAGGGCTTGAAAGAAGCTG), were used to amplify NRGfusion cDNAs, whereas the primers R 5′UTRF (GGTTCCTCACCCTCGGGGAGC) and N1848R (GGATGGTTCATCGTCATCCAGCC) were used to amplify RGN cDNAs. PCR using AmpliTaq Gold was performed with an initial step at 94°C for 9 minutes, followed by 35 cycles of 94°C for 30 seconds, 65°C for 1 minute, and 72°C for 45 seconds. PCR products were electrophoresed in low melting point agarose gels, purified using Wizard PCR Preps, and sequenced.
Southern analysis of PCR products.
PCR products were electrophoresed through agarose gels and transferred to Hybond N+ membrane (Amersham Pharmacia Biotech, Uppsala, Sweden). Hybridization to end-labeled oligo probes was performed for 16 hours at 42°C in a 20-mL solution of 4× SSPE, 1% sodium dodecyl sulfate (SDS), 1 in 20 dilution of blotto (5% nonfat dried milk powder, 0.02% sodium azide), and 0.1 mg/mL denatured salmon sperm DNA. After washing at 42°C in 2× SSC, 0.1% SDS, the membranes were autoradiographed at −80°C.
NUP98 and RAP1GDS1 probes.
Probes were generated by PCR after reverse transcription of peripheral blood mononuclear cell RNA and gel purified from low melting point agarose gels using Wizard PCR Preps. The identity of the probes was confirmed by sequencing. The 1,084-bp NUP98 cDNA probe was amplified using the primers N301F and N1384R, using AmpliTaq Gold with an initial step at 94°C for 9 minutes followed by 35 cycles of 94°C for 30 seconds, 65°C for 1 minute, and 72°C for 45 seconds. The RAP1GDS1 primers, R11F (5′TCAGTGATACCTTGAAGAAGCTG) and R1673R (5′CTTTCCACAGTAAGTCTCTCTGCTC), were developed from the cDNA sequence (Genbank accession no. X63465). A 1,665-bp RAP1GDS1cDNA probe was amplified using the Expand Long Template PCR System with an initial step at 94°C for 2 minutes, followed by 10 cycles of 94°C for 10 seconds, 63°C for 30 seconds, and 68°C for 2 minutes, followed by 35 cycles of 94°C for 10 seconds, 63°C for 30 seconds, and 68°C for 2 minutes plus 20 seconds per cycle.
Northern analysis.
Ten micrograms of RNA was electrophoresed in a 1% agarose/1.2 mol/L formaldehyde gel, blotted onto Brightstar plus membrane (Ambion, Austin, TX) according to the manufacturer’s protocol, and UV fixed to the membrane. Multiple tissue Northerns were from Clontech (Palo Alto, CA). Hybridization to random nonamer-labeled probe was performed at 42°C in a 20-mL solution of 1 mol/L NaCl, 10% dextran sulphate, 1% SDS, 50% deionized formamide, and 0.2 mg/mL denatured salmon sperm DNA. Membranes were washed to a final stringency of 0.2× SSPE, 1% SDS at 65°C and autoradiographed at −80°C.
RESULTS
Identification of NUP98 as the chromosome 11 breakpoint gene.
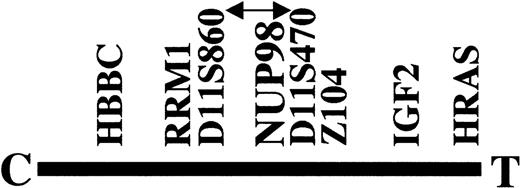
FISH analysis of cosmid probes had narrowed the chromosome 11 breakpoint region of patient no. 1 to between RRM1 andD11S470 on 11p15.5 (Fig1).9 The hybrids containing the der(4) and der(11) chromosomes8 were tested with primers specific toD11S470 and D11S860. D11S470 was on the der(4) chromosome, and D11S860 was on the der(11) chromosome (results not shown). This narrowed the breakpoint to the region between D11S860 and D11S470. The nucleoporin 98 (NUP98) gene maps to a similar region and is located proximal to the region recognized by the cosmid Z104.10 Analysis of the PAC pDJ1173a5 shows that ZNF195, the zinc finger gene within Z104, is distal to D11S470.17NUP98was absent from the PAC sequence, placing NUP98 proximal toD11S470 and therefore within the breakpoint region (Fig 1).
Position of the chromosome 11 breakpoint with respect to the 11p15.5 markers used for FISH and PCR mapping. NUP98 lies within the candidate breakpoint region indicated by the arrowed line. The β chain of hemoglobin (HBBC) and the H-ras oncogene (HRAS) are at the extremities of 11p15.5. C and T denote centromeric and telomeric, respectively.
Position of the chromosome 11 breakpoint with respect to the 11p15.5 markers used for FISH and PCR mapping. NUP98 lies within the candidate breakpoint region indicated by the arrowed line. The β chain of hemoglobin (HBBC) and the H-ras oncogene (HRAS) are at the extremities of 11p15.5. C and T denote centromeric and telomeric, respectively.
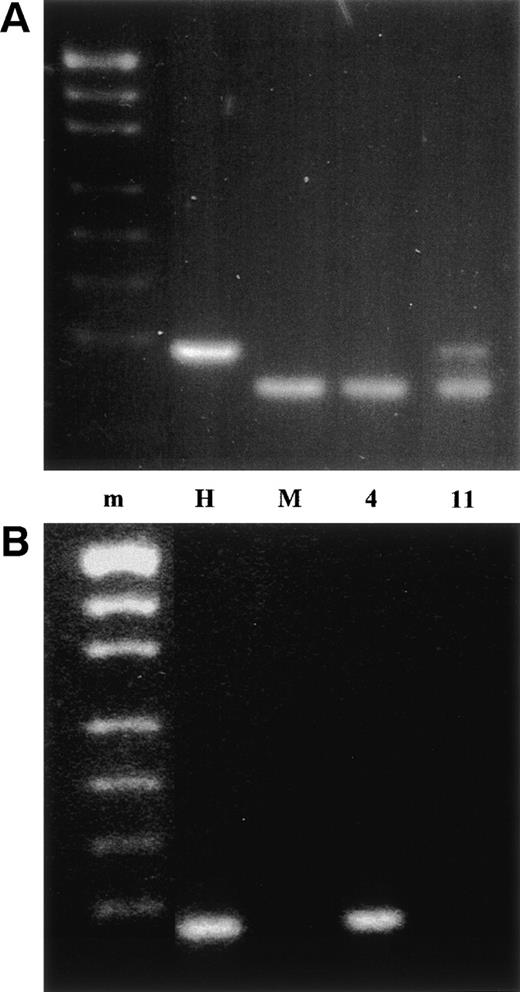
We therefore sought to investigate NUP98 as a candidate breakpoint gene. Five exons, named A through E, have been defined in the NUP98 breakpoint region.12 Most NUP98 breakpoints occur between exons B and C.10-13 The der(4)- and der(11)-containing hybrids derived from patient no. 1 were tested by PCR for the presence of exons B and C. The der(11) hybrid contained exon B and the der(4) hybrid contained exon C (Fig 2). Because exons B and C are on the complementary derivative chromosomes, NUP98 is disrupted between exons B and C in patient no. 1 and is the chromosome 11 breakpoint gene.
PCR analysis of the der(4) and der(11) containing somatic cell hybrids. m is the pUC19/Hpa II molecular weight marker, H is normal human, M is mouse, 4 is the der(4) hybrid, and 11 is the der(11) hybrid. (A) NUP98 exon B PCR product digested withTaq I. Mouse and human NUP98 cDNA sequences are highly conserved and the exon B PCR also amplified mouse NUP98. The mouse and human exon B PCR products were distinguished by a TaqI restriction site, which is present in the mouse product but absent in the human product. (B) NUP98 exon C PCR product.
PCR analysis of the der(4) and der(11) containing somatic cell hybrids. m is the pUC19/Hpa II molecular weight marker, H is normal human, M is mouse, 4 is the der(4) hybrid, and 11 is the der(11) hybrid. (A) NUP98 exon B PCR product digested withTaq I. Mouse and human NUP98 cDNA sequences are highly conserved and the exon B PCR also amplified mouse NUP98. The mouse and human exon B PCR products were distinguished by a TaqI restriction site, which is present in the mouse product but absent in the human product. (B) NUP98 exon C PCR product.
Identification of RAP1GDS1 as the chromosome 4 breakpoint gene by 3′ RACE.
3′ RACE was used to determine the chromosome 4 gene fused toNUP98 in patient no. 1. Experiments were performed in parallel on the presentation sample of patient no. 1 and peripheral blood mononuclear cells from a normal individual. One predominant band was seen in the normal individual. Additional bands were seen in the leukemic presentation sample (results not shown). The bands were sequenced and analyzed using the BLAST algorithm to search the GenBank sequence database.18 The common band was shown to correspond to the normal 4.05-kb NUP98 transcript.
A band that was slightly larger than the normal NUP98 band had the 5′ end of NUP98 fused with the coding region of the guanine nucleotide disassociation stimulator gene, RAP1GDS1.The fusion maintained the reading frame of RAP1GDS1. We hereafter denote this hybrid transcript as NRG (forNUP98-RAP1GDS1). The RAP1GDS1 sequence in NRGstarts at nucleotide 5 of the coding sequence. The methionine and the first G of the codon for aspartic acid are lost. However, the aspartic acid is retained in the fusion protein, because the last base of NUP98 exon B is a G (Fig 3).
Nucleotide and amino acid sequences around the junctions of the (A) NRG and (B) NRG2 fusion transcripts (Genbank accession nos.AF133331 and AF133333, respectively).
Other RACE products that were cloned and sequenced had an identicalNUP98 - RAP1GDS1 junction to NRG but continued into presumed RAP1GDS1 intron/exon splice sites and terminated in either introns of RAP1GDS1 or as yet-unsequenced exons ofRAP1GDS1 (data not shown).
RT-PCR.
RT-PCR of patient no. 1 using primers flanking theNUP98-RAP1GDS1 junction gave a product of the expected size (395 bp), confirming that an NRG fusion mRNA was formed (Fig 4). No bands were seen in the peripheral blood mononuclear cells from normal controls. Two T-ALL patients with a similar karyotype (patients no. 2 and 3; see Table 1) were also tested for the fusion mRNA by RT-PCR (Fig 4). Patient no. 2 was clearly positive, with an RT-PCR product of identical size to that of patient no. 1. Patient no. 3 had a smaller RT-PCR product of 162 bp. Sequencing showed that patient no. 3 had a novel in-frame fusion ofNUP98 to RAP1GDS1 with the NUP98 breakpoint immediately preceding exon A and an RAP1GDS1 junction (nucleotide 5 of the coding sequence) identical to that of patients no. 1 and 2 (Fig 3). This transcript, denoted as NRG2, also maintains the first aspartic acid in the RAP1GDS1 sequence.
RT-PCR analysis of NRG and RGN fusion transcripts in 3 t(4;11)(q21;p15) patients. P1, P2, and P3 are RT-PCR products from peripheral blood mononuclear cells from the patients. C1 and C2 are RT-PCR products from peripheral blood mononuclear cells of normal donors. Samples marked with a minus sign are negative control RT-PCRs without reverse transcriptase. H2O controls are negative control RT-PCRs without target. The lane marked m contains both SPP1/EcoRI and pUC19/Hpa II molecular weight markers. The most prominent bands in the RGN PCR of patient no. 3 are alternative splicings of RGN with and without exon B.
RT-PCR analysis of NRG and RGN fusion transcripts in 3 t(4;11)(q21;p15) patients. P1, P2, and P3 are RT-PCR products from peripheral blood mononuclear cells from the patients. C1 and C2 are RT-PCR products from peripheral blood mononuclear cells of normal donors. Samples marked with a minus sign are negative control RT-PCRs without reverse transcriptase. H2O controls are negative control RT-PCRs without target. The lane marked m contains both SPP1/EcoRI and pUC19/Hpa II molecular weight markers. The most prominent bands in the RGN PCR of patient no. 3 are alternative splicings of RGN with and without exon B.
The complexity of minor bands seen with all 3 patients in theNRG RT-PCR (Fig 4) is a repeatable observation. Whereas some of the faint upper bands in patient no. 3 appear to be the same size as the NRG RT-PCR products in patients no. 1 and 2, they do not contain NUP98 exon B, as shown by hybridization with the N1511F oligo (data not shown), confirming that NRG2 is not just an alternatively spliced version of NRG.
We analyzed expression of the complementary fusion cDNA,RAP1GDS-NUP98 (RGN), by RT-PCR. Primers that could amplify RGN from all 3 patients showed that RGN is only expressed in patient no. 3 (Fig 4).
Some of the RACE products of patient no. 1 showed an insertion of the trinucleotide CAG at the NUP98-RAP1GDS1 junction. The variable insertion of CAG was also seen in RT-PCR products from all 3 patients (data not shown). This insertion is most likely due to alternative splicing of intronic sequence immediately adjacent to an exon. Because there are 2 distinct NUP98 breakpoint regions in our patients, we deduce that it probably comes from the intron adjacent to the firstRAP1GDS1 exon in the translocation. The CAG conforms to the consensus sequence YAG (Y is a pyrimidine) of the 3′ end of an intron.19 Alternative splicing involving a single trinucleotide has previously been reported for the c-kit gene.20
Northern analysis.
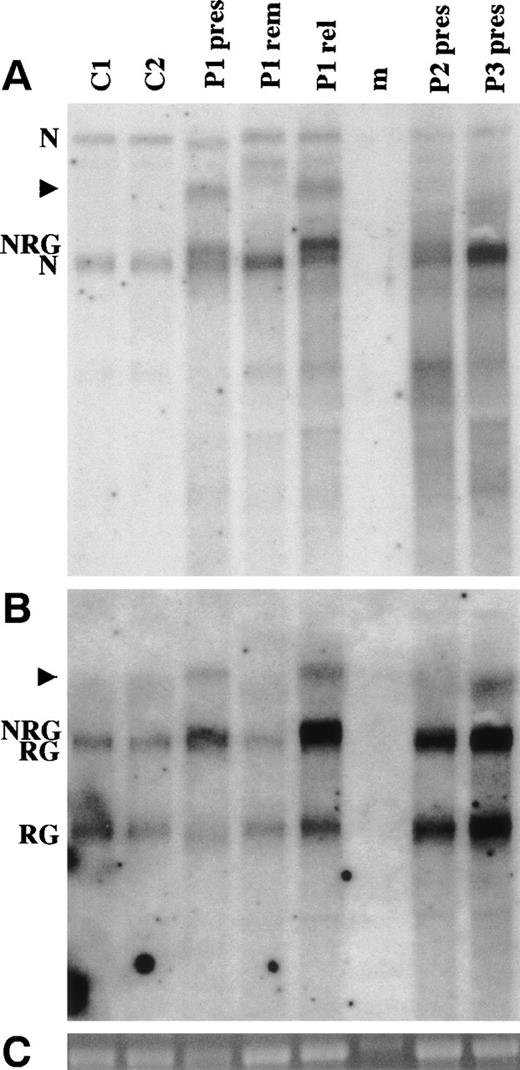
A 1,084-bp NUP98 cDNA probe was used for Northern analysis (Fig 5A). The normal controls show 4.05- and 7.25-kb bands. The 4.4-kb NRG transcript can be seen above the 4.05-kb NUP98 transcript for the presentation samples of patients no. 1 and 2. In patient no. 3, the NRG2 transcript cannot readily be seen as it migrates just above the normalNUP98 band. NRG is not seen in the remission sample from patient no. 1. The relapse specimen from the same patient shows markedly increased NRG expression compared with the endogenousNUP98. The increased NRG expression in the relapse specimen may be related to the addition to the short arm of the previously normal chromosome 11 (Table 1).
Northern analysis of NRG expression. (A) Hybridization using a NUP98 cDNA probe. (B) Hybridization of the same membrane with a RAP1GDS1 cDNA probe. (C) 18S rRNA from the ethidium bromide-stained gel before transfer. RNA was isolated from 2 normal controls (C1 and C2) and from the 3 patients (P1, P2, and P3). pres is a presentation sample, rem is a remission sample, and rel is a relapse sample. Each lane contains 5 μg of total RNA from peripheral blood mononuclear cells, except that P1 rem contains 5 μg of total RNA from bone marrow. The lane marked m is a RNA ladder (Promega). The band in this lane in (C) is marker and not 18S RNA. N indicates theNUP98 4.05- and 7.25-kb bands. The 7.25-kb band is a precursor that also contains the NUP96 coding sequence.47 RG indicates the 2.8- and 4.1-kb RAP1GDS1 bands. NRG indicates the 4.4-kb NRG transcript. NRG2 in patient no. 3 is not indicated, because it is not distinguishable from the 4.05-kbNUP98 and 4.1-kb RAP1GDS1 bands. The arrowheads indicate higher molecular weight transcripts that hybridize with both the NUP98 and RAP1GDS1 probes.
Northern analysis of NRG expression. (A) Hybridization using a NUP98 cDNA probe. (B) Hybridization of the same membrane with a RAP1GDS1 cDNA probe. (C) 18S rRNA from the ethidium bromide-stained gel before transfer. RNA was isolated from 2 normal controls (C1 and C2) and from the 3 patients (P1, P2, and P3). pres is a presentation sample, rem is a remission sample, and rel is a relapse sample. Each lane contains 5 μg of total RNA from peripheral blood mononuclear cells, except that P1 rem contains 5 μg of total RNA from bone marrow. The lane marked m is a RNA ladder (Promega). The band in this lane in (C) is marker and not 18S RNA. N indicates theNUP98 4.05- and 7.25-kb bands. The 7.25-kb band is a precursor that also contains the NUP96 coding sequence.47 RG indicates the 2.8- and 4.1-kb RAP1GDS1 bands. NRG indicates the 4.4-kb NRG transcript. NRG2 in patient no. 3 is not indicated, because it is not distinguishable from the 4.05-kbNUP98 and 4.1-kb RAP1GDS1 bands. The arrowheads indicate higher molecular weight transcripts that hybridize with both the NUP98 and RAP1GDS1 probes.
A second new transcript of approximately 5.8 kb was seen in the presentation and relapse samples of patient no. 1. This band is also present in patient no. 2 but is not discernible on Fig 5A. Patient no. 3 showed a 5.5-kb transcript. The shorter size corresponds approximately to the size difference (233 bp) between NRG andNRG2.
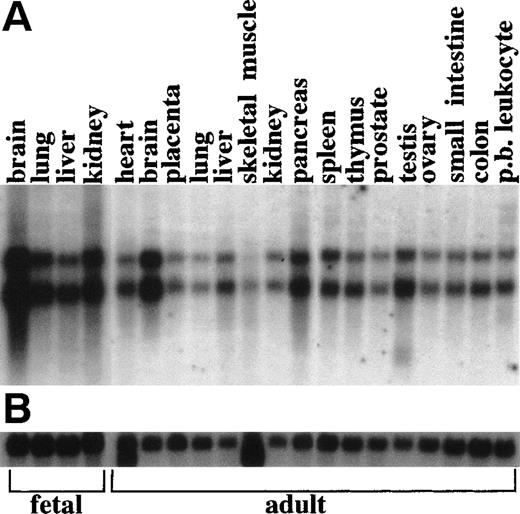
RAP1GDS1 shows 2.8- and 4.1-kb transcripts in all tissues tested (Fig 6). When the patient was Northern probed with the RAP1GDS1 probe, the 4.1-kb transcript was visible as a distinct band slightly lower than the NRG transcript, although the 2 bands are not readily distinguishable after photo-reproduction (Fig 5B). The 5.8- and 5.5-kb bands are present in the patient samples, confirming that they areNRG transcripts. They are probably generated by the same mechanism that generates the upper 4.1-kb RAP1GDS1 transcript.
Multiple tissue Northern analysis of RAP1GDS1. Each lane contains 2 μg of polyA RNA. (A) Hybridization with aRAP1GDS1 cDNA probe shows two predominant bands of 4.1 and 2.8 kb. (B) Hybridization with a β-actin cDNA probe (Clontech).
Multiple tissue Northern analysis of RAP1GDS1. Each lane contains 2 μg of polyA RNA. (A) Hybridization with aRAP1GDS1 cDNA probe shows two predominant bands of 4.1 and 2.8 kb. (B) Hybridization with a β-actin cDNA probe (Clontech).
DISCUSSION
We originally reported a t(4;11)(q21;p14-15) translocation in a patient with T-ALL.7 Molecular analysis then localized the chromosome 11 breakpoint to 11p15.5. 8 Subsequently, 2 further T-ALL patients (no. 2 and 3), karyotyped as t(4;11)(q21;p14-15) and t(4;11)(q21;p15), respectively, were identified by us. Three other patients have been reported with either a t(4;11)(q21;p14-15) or a t(4;11)(q21;p15) as the primary translocation.21-23 The clinical data, cytogenetics, and immunophenotype of all 6 patients are summarized in Table 1.
Whereas different surface markers have been tested in each individual, the following generalizations can be drawn: (1) the cytochemistry and surface markers of all 6 patients are consistent with T-ALL; (2) the leukemic cells are positive for CD7 and CD5 and usually positive for CD2, but are negative for CD4 and CD8; (3) CD10 is often positive in a proportion of the cells; and (4) most express 1 or more of the myeloid markers CD11b, CD13, and CD33 in a proportion of the cells. None of the 6 patients with the primary translocation was an infant. They ranged from 6 to 53 years of age, with a preponderance of younger individuals, as is typical for T-ALL.24 All had a fairly short survival after diagnosis. Patient no. 1, who showed the longest survival, underwent 2 matched allogeneic bone marrow transplants but relapsed with aggressive disease on both occasions.
Four of the 6 patients presented with additional karyotypic rearrangements (Table 1). This may account for some of the differences between their clinical pictures. Interestingly, the 2 patients who presented with a very low white blood cell count both had a 12p deletion.
We identified NUP98 as the chromosome 11 breakpoint gene by PCR analysis of somatic cell hybrids containing the derivative chromosomes of patient no. 1. It was shown that exons B and C of NUP98 were found on the der(11) and der(4) chromosomes, respectively, thereby mapping the breakpoint to the intron between exons B and C. This confirms the previously reported orientation of NUP98 with regard to the centromere.11 Because the principal transcript in the other NUP98 translocations fuses the 5′ end of the NUP98 gene in frame to the 3′ end of a second gene, we used 3′ RACE and identified the RAP1GDS1 gene as the 3′ partner. RAP1GDS1 has previously been mapped to 4q21-25.25
RT-PCR showed that NUP98-RAP1GDS1 (NRG) fusion mRNAs were present in patients no. 1, 2, and 3. Sequencing showed that the same RAP1GDS1 sequence, starting at nucleotide 5 of the coding region, was present in all 3 patients (Fig 3). Patients no. 1 and 2 had an identical fusion mRNA containing the 5′ sequence ofNUP98 up to and including exon B, whereas patient no. 3 lackedNUP98 exons A and B. Breakpoints in NUP98 have been reported to occur between exons B and C10-13 or between exons D and E,12 with a predominance of breakpoints between exons B and C.
The breakpoint in patient no. 3 (in the intron preceding exon A) is the most proximal NUP98 breakpoint reported. The more proximal breakpoint position is consistent with the Northern results in which the NRG2 fusion band is almost identically sized to the 4.05-kbNUP98 transcript. NRG2 is 233 bp shorter thanNRG on account of the missing exons A and B.
The absence of the reciprocal RGN transcript in patients no. 1 and 2 (Fig 4) indicates that NRG is the leukemia-associated transcript. It is unclear why the reciprocal transcript is absent asRAP1GDS1 is universally expressed and RGN is under the control of the RAP1GDS1 promoter. A similar situation has been observed for the BCR-ABL translocation in which the reciprocalABL-BCR transcript is not expressed in all CML patients, although ABL is also universally expressed.26
Nup98 is a component of the nuclear pore complex, involved in the export of RNA and protein from the nucleus.27,28 The previously described fusion partners of NUP98 are functionally diverse.10-13HOXA9 and HOXD13 code for transcription factors required for normal development29,30and DDX10 codes for a putative RNA helicase.31Another nucleoporin gene, NUP214, is also involved in translocations in leukemia. NUP214, also known as CAN, is fused to either the DEK gene or to the SET gene in cases of AML.32 33
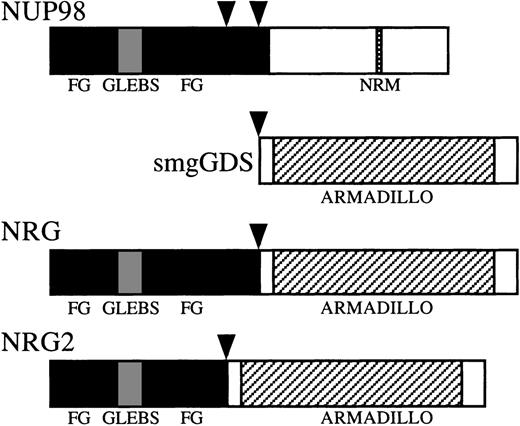
Both nup98 and nup214 contain multiple phenylalanine-glycine (FG) repeats. The FG repeats are presumed contact sites for multiprotein transport complexes that mediate bidirectional transport across the nuclear pores.34 All known NUP98 and NUP214translocations retain the majority of the FG repeats.10-13The FG repeats are also retained in the 3 patients reported here (Fig7). Patient no. 3, who has the most 5′ breakpoint yet reported, still has 30 of the 37 FG repeats.
Schematic representation of the NUP98, smgGDS, NRG, and NRG2 proteins. Vertical arrowheads represent breakpoints in NUP98 and smgGDS. FG, FG (phenylalanine-glycine) repeat-rich areas; GLEBS, GLEBS (Gle2p-binding motif) -like motif48; NRM, nucleoporin RNA binding motif; ARMADILLO, tandem armadillo repeats.
Schematic representation of the NUP98, smgGDS, NRG, and NRG2 proteins. Vertical arrowheads represent breakpoints in NUP98 and smgGDS. FG, FG (phenylalanine-glycine) repeat-rich areas; GLEBS, GLEBS (Gle2p-binding motif) -like motif48; NRM, nucleoporin RNA binding motif; ARMADILLO, tandem armadillo repeats.
The FG repeat-containing nup98 portion of the nup98-hoxa9 fusion protein acts as a potent activator of hoxa9 activity by recruiting the CBP and p300 transcriptional coactivators.35 The CBP/p300 binding activity of the nup98-hoxa9 fusion protein is correlated to its transforming activity. The transforming ability is retained when the FG repeat region from nup98 is exchanged for that of nup214, which directly implicates the FG repeats in the transforming activity. Not all of the FG repeats are required to interact with CBP/p300 or to transform, because a nup98-hoxa9 splice variant with 20 FG repeats still retains transforming ability.35
The entire coding region, except for the initial methionine ofRAP1GDS1, is retained in the NRG and NRG2transcripts. The product rap1gds, usually referred to as smgGDS, has guanine nucleotide exchange factor (GEF) activity.36 GEFs stimulate or inhibit exchange of GDP for GTP at small GTPase proteins to convert the inactive GDP bound form to the active GTP bound form. SmgGDS was first reported as a stimulator of GDP/GTP exchange for rap1a, then called smg p21a.37 SmgGDS also acts on rap1b as well as on other small GTPases, including K-ras, rac1, rac2, rhoA, and ralB.36,38,39 Interestingly, rap1a and K-ras are antagonistic, because the protein smg p21a/rap1a was first identified as Krev-1, which has the ability to revert K-ras–transformed NIH 3T3 fibroblasts.40 However, rap1 is unlikely to be the principal target of smgGDS, because smgGDS cooperates with K-ras in transformation.41 RhoA and rac2 have been reported to be more important targets for smgGDS than rap1a.38
SmgGDS is structurally unique among the GEFs, because it shows no homology to other GEFs and is composed largely of tandem repeats of the 43 amino acid armadillo motif (Fig 7).42 The armadillo motif was originally found in the Drosophila melanogasterarmadillo gene and its vertebrate homologues β-catenin and plakoglobin.43,44 Subsequently, it was identified in a number of other genes that contain tandem repeats of armadillo,42 including importin α.45 It has been suggested that armadillo repeats mediate protein-protein interactions. 42
Determining the cellular location of nrg will be critical in determining its role in malignancy. SmgGDS normally interacts with membrane-bound and cytoplasmic ras superfamily GTPPases. If the nrg hybrid protein is cytoplasmic, its function may involve alterations of signaling via ras family small GTPases. However, by analogy to other armadillo proteins, such as β-catenin and importin α, smgGDS may have an as yet undescribed cytoplasmic-nuclear shuttling capacity. The armadillo repeats of smgGDS may lead it to mimic β-catenin and interact with the transcription factors involved in the wingless signaling pathway.46 Alternatively, the amino terminal end of nup98 might relocate smgGDS to the nuclear pore so that the fusion protein may modify nuclear transport. Because nrg contains an intact smgGDS sequence, it may act as a second GEF for ran in promoting nuclear transport. Finally, nrg may be located in the nucleus, where it may modify transcription, as happens with other nup98 fusion proteins.35 Transcription factors that interact with the armadillo repeats may become coupled to transcription factors that interact with FG repeats.
This report shows that NUP98 can be involved in T-ALL as well as myeloid malignancies. Moreover, the identification of 3 patients with the NRG fusion shows that the t(4;11)(q21;p15) is a recurrent translocation in T-ALL. Whether NRG is capable of causing cellular transformation and hematological malignancy is the subject of further investigation in our laboratory.
ACKNOWLEDGMENT
The authors thank Ed Sage for his support during the duration of this research. Jenny Hardingham, Viki Kalatzis, and Jennie Finch were involved in the preliminary work that led to this investigation. We also thank Alec Morley, Tim Hughes, Luen Bik To, Lesley Snell, and Pam Dyson for access to patient material and information; Peter Little, Marcel Mannens, and Bert Redeker for cosmids; Nick Wickham for reading the manuscript; Peter Aplan, Leonie Ashman, and Sarah Swinburne for discussions; and Tina Bianco for assistance with figure preparation.
Supported by the National Health and Medical Research Council, Anti Cancer Foundation of South Australia, and the Queen Elizabeth Hospital Research Foundation. D.J.H. was supported by the Queen Elizabeth Hospital Research Foundation.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Alexander Dobrovic, PhD, Chief Medical Scientist, Department of Haematology-Oncology, The Queen Elizabeth Hospital, Woodville, SA 5011, Australia; e-mail:adobrovic@medicine.adelaide.edu.au.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal