Peptides corresponding to the fusion site in 210 kD BCR-ABL protein b3a2 (p210b3a2) were previously shown to bind to several HLA class I and II alleles. We have found that b3a2 peptide-specific CD4-positive T-helper cells were able to recognize p210b3a2-positive chronic myelogenous leukemia (CML) blasts in a DR4 restricted manner. Until now, there were no reports of b2a2 breakpoint-specific human T-cell responses. Here we show that repetitive stimulation of T lymphocytes with a 17mer peptide covering the fusion region in p210b2a2 also leads to specific T-cell responses. CD4 and CD4/CD8 double-positive clones obtained from a b2a2 peptide-specific cell line were cytotoxic and proliferative in an HLA-DR2a (DRB5*0101) restricted fashion. Autologous Epstein-Barr virus (EBV) transformed cells, expressing BCR-ABLb2a2 on transfection, and allogeneic HLA-DR matched p210b2a2-positive cells from CML patients were, however, not lysed. BCR-ABL peptide-specific T-cell clones did respond to autologous EBV cells transfected with invariant chain (li) cDNA in which the HLA class II–associated invariant chain peptide (CLIP) was replaced by a BCR-ABL b2a2 fusion oligonucleotide sequence, illustrating the potential of these T cells to recognize an endogenous BCR-ABLb2a2ligand.
THE CLASSICAL t(9;22) translocation in leukemic cells from patients with chronic myelogenous leukemia (CML) results in a fusion gene known as BCR-ABL. The breakpoint on chromosome 9 is nearly always located upstream of abl exon a2,1whereas the breakpoint on chromosome 22 is usually located either between bcr exons b2 and b3 or between exons b3 and b4.2,3The differently fused genes encode principally two variant 210 kD proteins: p210b2a2 or p210b3a2.4 Alternative splicing of BCR-ABL mRNA can result in coexpression of oncoproteins p210b3a2 and/or p210b2a24,5 with p190e1a2.6,7 BCR-ABL oncoprotein-positive cells, which express a novel protein harboring nonself and possibly antigenic peptides at the fusion site, may be targets for appropriate cells of the immune system. The junction region aa sequences in p210BCR-ABL, being well-characterized antigens expressed by leukemic cells, can be considered of potential importance in an immunotherapeutic approach of CML. In approximately two thirds of adult CML patients, the b2a2 protein (in combination with p210b3a2 and/or p190e1a2) is detected.6-13 Thus far, however, only peptides derived from the p210b3a2 protein have been shown to bind to several major histocompatibility complex (MHC) class I14-16 and class II molecules17,18 and peptide-specific human T lymphocytes have been generated only against b3a2 sequences.15-23 Although a 23mer b2a2 peptide appeared to bind with intermediate affinity to HLA DR3,17 no T-cell responses to the b2a2 breakpoint sequence have been described. In this report, we show that in vitro immunization with a 17 aa peptide representing the p210b2a2 fusion region resulted in HLA-DRB5*0101 restricted peptide-specific cytotoxic and/or proliferative CD4+ T-cell clones. In view of the predominant DR positivity of BCR-ABL–positive CML progenitors24,25 and the observation of very low numbers of BCR-ABL transcripts in healthy individuals,26 27 BCR-ABL breakpoint-specific DR restricted T cells may play an important role in CML remission-induction after transplantation and in immunosurveillance. Such T cells can be directed toward the b3a2 and, as shown by our data, the b2a2 breakpoint sequence.
MATERIALS AND METHODS
This study was approved by a local ethical committee. Blood samples were collected from healthy donors after informed consent only.
Preparation of BCR-ABLb2a2-specific T-cell line, P-b2, and clones.
Blood was obtained at regular intervals from a healthy male donor P, blood group A, Rhesus D-negative, with HLA type: A2; B7, B44; Cw5, Cw7; DR2(DRB1*1501/DRB5*0101); DR4(DRB1*0401/DRB4*0101); DQ6, DQ8; DPB*0201, DPB*0401. T-cell line, P-b2, was prepared from peripheral blood mononuclear cells (PBMC) after incubation with 17mer p210b2a2 breakpoint peptide as described earlier19 21 with some modifications. A total of 2 to 3 × 107 PBMC were isolated from whole blood using Ficoll-amidotrizoate centrifugation and, initially, cultured in 20 mL tissue culture flasks. Tissue culture medium (Iscove’s modified Dulbecco’s modified Eagle’s medium [DMEM]; GIBCO-Life Technologies, Paisley, UK), supplemented with 10% autologous heat-inactivated serum, was partially renewed every 5 to 10 days. Final 17mer b2a2 peptide concentration was maintained at 10 μg/mL (approximately 6 nmol/mL). A total of 107 autologous nonirradiated PBMC were added every 9 to 10 days until a proliferative response, as assessed by increasing T-cell numbers, was seen. In case of a peptide-dependent T-cell response, as evidenced in a proliferation assay using autologous irradiated PBMC as antigen-presenting cells (APC), restimulation with 30 Gy irradiated autologous PBMC was continued for at least 40 days. Cultures were restimulated thereafter with allogeneic pooled 30 Gy irradiated PBMC from five healthy donors mixed in a 4:1 ratio with 50 Gy irradiated B-lymphoblastoid cell lines (B-LCL), prepared from Epstein-Barr virus (EBV) transformed B cells from the T-cell donor, and peptide. Peptide-specific T cells were cloned in round bottomed 96-well plates (Costar Co, Cambridge, MA) from T-cell line, P-b2, by limiting dilution using autologous irradiated B-LCL and allogeneic pooled irradiated PBMC as APC/feeder population. The culture medium was supplemented with 10% heat inactivated pooled human serum (HPS) instead of autologous serum. Ten percent T-cell growth factor (TCGF; Lymphocult; Biotest, Dreieich, Germany) was added 48 hours after restimulation with APC and peptide. Clones were maintained in 96-well plates and stimulated weekly with the APC/feeder mixture, peptide, and TCGF as described.
Peptides.
Amino acid sequences of the p210b2a2 and p210b3a2 fusion region were used to design BCR-ABL breakpoint representing 17mer length peptides (b2a2: IPLTINKEEALQRPVAS; b3a2: ATGFKQSSKALQRPVAS),2,3 as well as 15 b2a2 peptide analogs with single aa substitutions to alanine. Peptides were synthesized on an Abimed 422 multiple peptide synthesizer (Abimed, Langenfeld, Germany) at 10 μmol scale. TentagelS AC resins28 were used in combination with Fmoc-protected amino acids applying a sixfold excess amino acid using benzotriazole-1-yl-oxy-tris-(dimethylamino) phosphoniumhexafluorophosphate/N-methylmorpholine activation in N-methylpyrrolidone.29 After cleavage in trifluoroacetic acid (TFA)/water, peptides were isolated and purified by repeated ether precipitations and analyzed by reversed phase high-performance liquid chromatography (HPLC) on a 5C18 Lichrospher RP Select B column (Merck, Darmstadt, Germany), amino acid sequencing and/or mass spectrometry. Peptides were stored at −70°C in aliquots of 1 mL in a concentration of 10 mg/mL in phosphate-buffered saline (PBS: 154 mmol/L NaCl, 1.4 mmol/L Na2HPO4, pH 7.5).
Proliferation assay.
Proliferation tests were performed using a standard3H-thymidine incorporation assay as described.19 Briefly, 105 30 Gy irradiated APC were incubated with 2.5 to 5·104 responder cells (in round-bottomed microtiter wells, Costar Co) in 150 μL culture medium with 15% heat-inactivated serum autologous to the responder cell donor. After 48 hours, 1 μCi 3H-thymidine was added for another 15 hours before harvesting (Microcell harvester; Skatron Co, Tranby, Norway) and counting (1205 Betaplate counter; Wallac Oy, Turku, Finland). All tests were repeated on separate occasions. Results are expressed as the mean (plus the standard deviation) of counts per minute (cpm) from triplicate experiments or as the proliferation index:
Cytotoxicity.
Target cells were labeled with 100 μCi51chromium/106 cells for 1 hour at 37°C. Labeled cells were washed and, when appropriate, pulsed with relevant peptide for 1 hour. The effector cells were mixed at various effector to target ratios with target cells for 4 hours at 37°C, after which cytoxicity was determined by calculating the percent specific lysis from counts of 51Cr released:
For cpmMaximal Release, labeled target cells were incubated in medium with 0.3% Triton X100 detergent (Fluka Chemie AG, Buchs, Germany).
MHC restriction testing.
Functional HLA class I and HLA-DR blocking experiments were performed as described21 using murine monoclonal antibody (MoAb) W6/32 and murine MoAb B8.11.230 in proliferation or MoAb anti-HLA-DR B-D (Becton Dickinson Co, Mountain View, CA) in cytotoxicity assays, respectively. The DQ restricted anti M. Leprae CD4+ clone R3F1031 was used to exclude aspecific blocking by anti-DR. HLA-typed donor-derived fresh PBMC were used in a panel to test the MHC restriction of peptide sensitive T cells. Murine L-cell line LDR2a32 33 expressing DRB5*0101 (kindly provided by Dr J. Bodmer, Imperial Cancer Research Fund, London, UK), HLA-DR cDNA negative murine cells LTKcd323 (kindly provided by Dr K. Moore, DNAX Research Institute of Molecular and Cellular Biology, Palo Alto, CA), and T2 DRB5*0101 transfectants (kindly provided by Dr A. Vogt, Heidelberg, Germany) were used to confirm HLA-DR2a (DRB5*0101) allele restriction.
Flow cytometry.
Double immunostaining of T-cell clones was performed with fluorescein isothiocyanate (FITC)-labeled Moab anti-CD4 (Becton Dickinson) and phycoerythrin (PE)-conjugated anti-CD8 (anti-CD8 α chain, Becton Dickinson) and analyzed by FACscan (Becton Dickinson). CD8 expression was not limited to CD8 α, as verified by staining with a CD8 β chain MoAb (kindly provided by Dr E. Reinhertz, Dana-Farber Cancer Institute and Department of Medicine, Harvard Medical School, Boston, MA).
Peripheral blood cells from four DRB5*0101 and BCR-ABLb2a2reverse transcriptase-polymerase chain reaction (RT-PCR)-positive CML patients in chronic phase (UPN 5831869, 3939610, 3886117, 8033404), stored in N2, were thawed and cultured overnight in Iscove’s medium supplemented with 10% HPS. Cells were ficolled before fluorescence-activated cell sorting (FACS) analysis and all further use. After staining with anti-CD3, CD15, CD20, CD34, CDw65, and anti-HLA-DR fluorescent-labeled MoAbs (Becton Dickinson), the relative contribution of HLA-DR+ myeloid cells in the patients’ cell populations was estimated. Percentage of DR-positive cells varied from 29% to 67%, of which 3.9% to 20% were of nonmyeloid (ie, T or B lymphocytic) origin. Fourteen percent to 50% of cells were CD34-positive, 2% to 17% were CD15 and, 2% to 10% CDw65-positive.
HLA-DR peptide binding assay.
T2-DR2a cells were cultured in RPMI 1640 (GIBCO), supplemented with 2 mmol/L L-glutamine (GIBCO-Life Technologies), 100 U penicillin/100 μg streptomycin/mL (GIBCO), and 10% heat-inactivated fetal calf serum (FCS) (GIBCO). Cells were lysed at a concentration of 108 cells/mL in 50 mmol/L Tris-HCl, pH 8.5, containing 2% Renex (Accurate Chemicals and Scientific Corp, Westbury, NY), 150 mmol/L NaCl, 5 mmol/L EDTA, and 2 mmol/L phenylmethyl sulfonyl fluoride (PMSF). The lysates were cleared of nuclear and other debris by centrifugation at 10,000g for 20 minutes. DR molecules were purified essentially as described.34,35 Purified DR molecules (60 to 600 nmol/L) were incubated at pH 4.5 for 48 hours with 100 fmol (6.7 nmol/L) fluorescent-labeled DR binding class II-associated invariant chain peptide (CLIP, p87-p101)36 in 15 μL (final volume) 100 mmol/L Na-phosphate buffer containing 75 mmol/L NaCl, 1 mmol/L CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-propanesulfonate, Merck, Darmstadt, Germany) and 15% (vol/vol) CH3CN in the presence of a mixture of protease inhibitors (final concentration [f.c.] 1 μmol/L chymostatin, 5 μmol/L leupeptin, 10 μmol/L pepstatin A, 1 mmol/L EDTA, 200 μmol/L pefabloc). Each DR preparation was titrated in the presence of 100 fmol of the reference peptide (CLIP p87-101) to determine the DR concentration necessary to bind 10% to 20% of the total fluorescent signal. All subsequent inhibition assays were then performed at this concentration. Peptides of which the DR binding capacity was to be determined were added to DR molecules simultaneously with the reference peptide. The DR peptide complexes were separated from free peptide by gel filtration on a Synchropak GPC 100 column (250 mm × 4.6 mm; Synchrom Inc, Lafayette, IN). Fluorescent emission was measured at 528 nm on a Jasco FP-920 fluorescence detector (B & L Systems, Zoetermeer, The Netherlands). The percentage of labeled peptide bound was calculated as the amount of fluorescence bound to MHC divided by total fluorescence. Peptides were tested at concentrations ranging from 70 μmol/L to 0.7 nmol/L.The concentration of unlabeled test peptide yielding 50% inhibition of binding (IC50) of FITC-labeled CLIPp 87-101 peptide was deduced from the dose-response curve. Each peptide was tested in three separate experiments.
P210BCR-ABL transfected B-LCL.
B lymphoblastoid cells were obtained by EBV transformation of PBMC. Full-length BCR-ABLb2a2 cDNA in plasmids pCDXX (kindly provided by Dr G. Grosveld, St Jude Children’s Research Hospital, Memphis, TN) was recloned into vector PDR2 (Clontech Laboratories Inc, Palo Alto, CA). Unaltered PDR2 was used as negative control vector (empty vector). PDR2 vectors were transfected into EBV B-LCL by electroporation: cells were washed in serum free Iscove’s medium containing penicillin. Approximately 17 × 106cells in 0.7 mL medium were transferred to a gene pulser cuvet (Bio-Rad Laboratories, Hercules, CA) and 20 μg plasmid was added per cuvet. Electroporation was performed at 960 microfarad and 250 mV. Cells were kept in the cuvet for 10 minutes after electroporation and subsequently cultured at 106/mL in medium supplemented with 20% FCS (Sera Tech Zellbiologische Produkte, Skt Salvator, Germany). After 24 hours, dead cells were removed by ficoll centrifugation and viable cells were cultured in fresh medium under addition of 100 μg/mL hygromycine B (GIBCO Life Technologies Inc). The hygromycine concentration was increased with 100 μg/mL every 3 days to a final concentration of 500 μg/mL, at which the remaining B-LCL appeared hygromycine resistant.
Invariant chain-b2a2 fusion insert-cDNA (Iib2a2) transfected B-LCL.
A cassette vector was constructed from human cDNA coding for Ii, as described earlier,37 in which 33 CLIP-encoding nucleotides were replaced by a BCR-ABL p210b2a2 fusion cDNA sequence. The sequence of the b2a2-oligonucleotide was as follows: 5′-ACCATCAATAAGGAAGAAGCCCTTCAGCGGCCA-3′ (replacing wt CLIP nt 5′-TTCTACGCGTACCGGTGGGGCGACGACTACGTC-3′). The vector-insert was checked by DNA sequencing. The construct Iib2a2 encodes Ii protein with TINKEEALQRP b2a2 fusion peptide (instead of wt CLIP aa KMRMATPLLMQ) within the CLIP aa sequence.
Western blotting.
Laemmli sample buffer solution (60 mmol/L TRIS HCL pH6.8, 10% glycerol, 10 mmol/L EDTA, 2% sodium dodecyl sulfate [SDS], 2% β-mercaptoethanol, 0.03% bromophenol blue, 1 mmol/L Na3VO4) was mixed with an equal volume of 2 × 107/mL cells and boiled for 10 minutes. As p210b3a2, p210b2a2 and p190e1a2-positive control cell lines K562, BV1738, and Tom-138 were used, respectively. A total of 25 μL of cell lysates was subjected to 6.5% polyacrylamide gel electrophoresis (185 V) for 4 hours. Proteins were transferred onto 0.45-μm nitrocellulose membrane (in TRIS 50 mmol/L, glycine 40 mmol/L, 0.04% SDS, and 20% methanol) with 0.1 A at room temperature for 16 hours. Residual membrane antibody binding sites were blocked using 5% nonfat milk powder solution in PBS for 1 hour. Blots were washed in TRIS-buffered saline (TBST, 10 mmol/L TRIS, 150 mmol/L NaCl, 0.05% Tween-20, 1% bovine serum albumin [BSA]) and incubated with mouse anti-Abl MoAb 8E939(kindly provided by Dr Jean Wang, University of California at San Diego, La Jolla, CA), final concentration 2 μg/mL in TBST, for 2 hours. Nonbound 8E9 MoAb was removed by washing with TBST. Secondary incubation of the blots was performed using goat antimouse biotinylated antibody (B7264; Sigma, St Louis, MO), 1:5,000, in TBST for 1 hour. Streptavidin horse-radish peroxidase RPN 1231 (Amersham, Little Chalfont, Buckinghamshire, UK), 1:2,000, was applied for 30 minutes followed by chemiluminescence detection.
T-cell receptor (TCR) analysis.
RNA was extracted from P-b2–derived T-cell clones, Aa3 and Aa14, using RNAzol B (Cinna/Biotecx Laboratories, Houston, TX). Oligo-dT–primed first strand cDNA was reverse transcribed from 2 to 5 μg RNA using the Riboclone cDNA synthesis system (Promega Corp, Madison, WI). cDNA was subjected to PCR amplification with 29 TCRAV and 24 TCRBV family-specific primers as described elsewhere.40 PCR fragments were electrophoresed in agarose and subsequently purified using Whizard columns (Promega Corp). The PCR products were used for direct sequencing using the dideoxy-nucleotide chain termination method41 with the T7 sequencing Kit (Pharmacia LKB, Uppsala, Sweden). DNA sequences were compared with TCR V/J α and β sequences in the Genbank using the PCGENE computer software program (release 6.85; Intelligenetics Inc, Palo Alto, CA).
RESULTS
T-cell line, P-b2, is BCR-ABLb2a2 breakpoint peptide-specific and HLA-DR2 restricted.
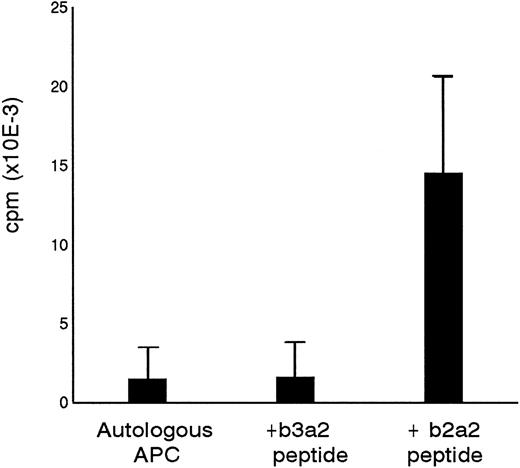
Primary in vitro stimulation of PBMC from a healthy donor with a 17mer b2a2 fusion sequence peptide resulted in a peptide-specific T-cell line designated P-b2. P-b2 proliferated in response to autologous irradiated PBMC incubated with 5 μg/mL b2a2 peptide. P-b2 did not show cross-reactivity to equal length BCR-ABLb3a2 fusion peptide, which also contains the 8 carboxy terminal aa residues encoded by ABL exon a2 (Fig 1). Proliferative responses could be blocked by preincubation of peptide-loaded APC with anti HLA-DR MoAbs (data not shown). A panel of partially HLA-DR–matched APC indicated the HLA-DR2 restriction of the b2a2 peptide-specific T-cell response (Table 1).
Specificity of P-b2 T-cell line for the p210b2a2 fusion region peptide. B3a2 and b2a2 17mer peptides were tested at 5 μg/mL with 105 PBMC as APC and 5 × 104 T cells as effectors.
Specificity of P-b2 T-cell line for the p210b2a2 fusion region peptide. B3a2 and b2a2 17mer peptides were tested at 5 μg/mL with 105 PBMC as APC and 5 × 104 T cells as effectors.
HLA Restriction of BCR-ABLb2a2Peptide-Specific T-Cell Line, P-b2
| APC Code (UPN) . | HLA A . | HLA B . | HLA C(w) . | HLA DR . | HLA DQ . | Response to b2a2 Peptide (cpm) . | Proliferation Index . |
|---|---|---|---|---|---|---|---|
| 9731999 | 2 | 7, 44 | 5, 7 | 2, 4 | 6, 7 | 5,444 ± 1,199 | 8 |
| 8959673 | 3, 31 | 35, 60 | 4, 10 | 4 | 8, 3 | 260 ± 38 | 0 |
| 6056013 | 1, 2 | 8, 60 | 7, 10 | 4, 13 | 6, 8 | 661 ± 443 | 0 |
| 2123634 | 3, 25 | 7, 39 | 7 | 2, 7 | 2, 6 | 8,255 ± 267 | 14 |
| 5024873 | 3, 11 | 7, 55 | 7, 9 | 2, 7 | 2, 6 | 5,472 ± 206 | 13 |
| 7067204 | 1, 24 | 7, 57 | 6, 7 | 2, 7 | 6, 9 | 5,571 ± 777 | 12 |
| 5021880 | 2, 24 | 18, 62 | 5, 9 | 4, 14 | 5, 8 | 261 ± 5 | 0 |
| APC Code (UPN) . | HLA A . | HLA B . | HLA C(w) . | HLA DR . | HLA DQ . | Response to b2a2 Peptide (cpm) . | Proliferation Index . |
|---|---|---|---|---|---|---|---|
| 9731999 | 2 | 7, 44 | 5, 7 | 2, 4 | 6, 7 | 5,444 ± 1,199 | 8 |
| 8959673 | 3, 31 | 35, 60 | 4, 10 | 4 | 8, 3 | 260 ± 38 | 0 |
| 6056013 | 1, 2 | 8, 60 | 7, 10 | 4, 13 | 6, 8 | 661 ± 443 | 0 |
| 2123634 | 3, 25 | 7, 39 | 7 | 2, 7 | 2, 6 | 8,255 ± 267 | 14 |
| 5024873 | 3, 11 | 7, 55 | 7, 9 | 2, 7 | 2, 6 | 5,472 ± 206 | 13 |
| 7067204 | 1, 24 | 7, 57 | 6, 7 | 2, 7 | 6, 9 | 5,571 ± 777 | 12 |
| 5021880 | 2, 24 | 18, 62 | 5, 9 | 4, 14 | 5, 8 | 261 ± 5 | 0 |
DR2 positive cells were DR15/DR51 (DRB1*1501/DRB5*0101) as defined by serology and DNA oligotyping, respectively. B2a2 peptide was used at 5 μg/mL.
Cloning of T-cell line, P-b2.
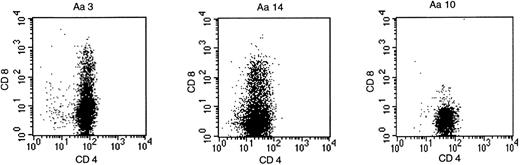
Forty-one peptide-specific T-cell clones were obtained from P-b2 by limiting dilution. FACS analysis showed CD4 positivity in all cases, but several clones expressed CD4/CD8 (Fig2).
BCR-ABLb2a2 fusion peptide-specific T clones were either CD4-single positive, as exemplified by clone Aa10, or as shown for clones Aa3 and Aa14, partially CD4/CD8-double positive.
BCR-ABLb2a2 fusion peptide-specific T clones were either CD4-single positive, as exemplified by clone Aa10, or as shown for clones Aa3 and Aa14, partially CD4/CD8-double positive.
Seven peptide-specific clones, recognizing the b2a2, but not the b3a2 peptide, could be maintained, two of which (Aa3 and Aa14) coexpressed CD4 and CD8. Fine specificity testing of these T-cell clones using altered b2a2 17mer peptides (in which each aa of the b2a2 peptide, other than alanine, was replaced individually by alanine) showed different aa requirements in proliferation assays (Table 2). Substitution however of E8 (E at position 8 of the 17mer peptide) and R13 was not allowed for any of the clones, whereas reduced to absent responses of all clones were seen on replacement of I5 and N6. These results were indicative for a DR2a (DRB5*0101) restriction with I5 and R13serving as anchor residues for binding to this allele.42The ability of the T-cell clones to recognize the b2a2 17 mer in the context of DR2a was confirmed by experiments with the single allele-transfected cells, T2-DR2a and LDR2a (data not shown).
Recognition of b2a2 17mer Peptides With Single Residue Replacement by Alanine
| T-Cell Clones . | 1 I . | 2 P . | 3 L . | 4 T . | 5 I . | 6 N . | 7 K . | 8 E . | 9 E ↑ . | 10 A . | 11 L . | 12 Q . | 13 R . | 14 P . | 15 V . | 16 A . | 17 S . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aa3 | ++ | ++ | ++ | ++ | − | − | − | − | − | ++ | − | ++ | − | − | ++ | ++ | ++ |
| Aa4 | ++ | ++ | ++ | ++ | − | − | + | − | ++ | ++ | ++ | + | − | ++ | ++ | ++ | ++ |
| Aa6 | ++ | ++ | ++ | ++ | + | + | + | − | − | ++ | − | + | − | ++ | ++ | ++ | ++ |
| Aa8 | ++ | ++ | ++ | ++ | + | + | ++ | − | − | ++ | − | + | − | +++ | ++ | ++ | ++ |
| Aa14 | +++ | +++ | ++ | ++ | − | − | − | − | − | ++ | − | + | − | +++ | +++ | ++ | ++ |
| AA15 | ++ | ++ | + | ++ | − | − | + | − | − | ++ | − | + | − | ++ | ++ | ++ | +++ |
| Aa16 | ++ | ++ | ++ | ++ | − | − | − | − | − | ++ | − | + | − | ++ | ++ | ++ | ++ |
| T-Cell Clones . | 1 I . | 2 P . | 3 L . | 4 T . | 5 I . | 6 N . | 7 K . | 8 E . | 9 E ↑ . | 10 A . | 11 L . | 12 Q . | 13 R . | 14 P . | 15 V . | 16 A . | 17 S . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aa3 | ++ | ++ | ++ | ++ | − | − | − | − | − | ++ | − | ++ | − | − | ++ | ++ | ++ |
| Aa4 | ++ | ++ | ++ | ++ | − | − | + | − | ++ | ++ | ++ | + | − | ++ | ++ | ++ | ++ |
| Aa6 | ++ | ++ | ++ | ++ | + | + | + | − | − | ++ | − | + | − | ++ | ++ | ++ | ++ |
| Aa8 | ++ | ++ | ++ | ++ | + | + | ++ | − | − | ++ | − | + | − | +++ | ++ | ++ | ++ |
| Aa14 | +++ | +++ | ++ | ++ | − | − | − | − | − | ++ | − | + | − | +++ | +++ | ++ | ++ |
| AA15 | ++ | ++ | + | ++ | − | − | + | − | − | ++ | − | + | − | ++ | ++ | ++ | +++ |
| Aa16 | ++ | ++ | ++ | ++ | − | − | − | − | − | ++ | − | + | − | ++ | ++ | ++ | ++ |
H-thymidine incorporation responses of P-b2–derived clones to b2a2 17mer peptides with single aa substitutions to alanine. Single letter coded aa represent b2a2 peptides in which the given aa is substituted to alanine. Response (minus the background) <20% of that of the clone to unaltered 17mer peptide: −, 20% to 60%: +, 60% to 120%: ++, >120%: +++. Peptide concentrations, 5 μg/mL.
B2a2 peptide binding to HLA DR2a.
The relative importance of different aa residues within the b2a2 17 mer for T-cell recognition or MHC interaction was evaluated by measuring the effectiveness of the b2a2 peptide with its different alanine substitutions to compete for binding to immunopurified DR2a molecules with a fluorescence-labeled invariant chain control peptide. The results (Table 3) allow, together with the data from Table 2, a designation for the different T-cell clones of putative T-cell contact residues at positions 6, 7, 8, 9, 11, and 14 within the b2a2 peptide and strongly support a role for I5and R13, as so called anchor residues necessary for MHC binding.
Binding of b2a2 17mer Peptide Analogs to HLA DR2a
| aa Sequence . | Relative Binding Capacity IC50(μmol/L) 3-150 . |
|---|---|
| IPLTINKEEALQRPVAS | 5 |
| APLTINKEEALQRPVAS | 7 |
| IALTINKEEALQRPVAS | 9 |
| IPATINKEEALQRPVAS | 8.6 |
| IPLAINKEEALQRPVAS | 4.7 |
| IPLTANKEEALQRPVAS | >70 |
| IPLTIAKEEALQRPVAS | 4 |
| IPLTINAEEALQRPVAS | 4 |
| IPLTINKAEALQRPVAS | 1.2 |
| IPLTINKEAALQRPVAS | 2.7 |
| IPLTINKEEAAQRPVAS | 9 |
| IPLTINKEEALARPVAS | 7 |
| IPLTINKEEALQAPVAS | >70 |
| IPLTINKEEALQRAVAS | 12 |
| IPLTINKEEALQRPAAS | 12 |
| IPLTINKEEALQRPVAA | 15 |
| aa Sequence . | Relative Binding Capacity IC50(μmol/L) 3-150 . |
|---|---|
| IPLTINKEEALQRPVAS | 5 |
| APLTINKEEALQRPVAS | 7 |
| IALTINKEEALQRPVAS | 9 |
| IPATINKEEALQRPVAS | 8.6 |
| IPLAINKEEALQRPVAS | 4.7 |
| IPLTANKEEALQRPVAS | >70 |
| IPLTIAKEEALQRPVAS | 4 |
| IPLTINAEEALQRPVAS | 4 |
| IPLTINKAEALQRPVAS | 1.2 |
| IPLTINKEAALQRPVAS | 2.7 |
| IPLTINKEEAAQRPVAS | 9 |
| IPLTINKEEALARPVAS | 7 |
| IPLTINKEEALQAPVAS | >70 |
| IPLTINKEEALQRAVAS | 12 |
| IPLTINKEEALQRPAAS | 12 |
| IPLTINKEEALQRPVAA | 15 |
IC50 denotes the concentration of b2a2 peptide or an analog thereof required to inhibit the binding of fluorescent labeled CLIP peptide p87-10136 (VSKMRMATPLLMQAL) by 50%. IC50 of unlabeled CLIP peptide was 0.2 μmol/L.
B2a2 fusion peptide-specific T cells fail to lyse autologous B-LCL and allogeneic CML cells expressing p210b2a2.
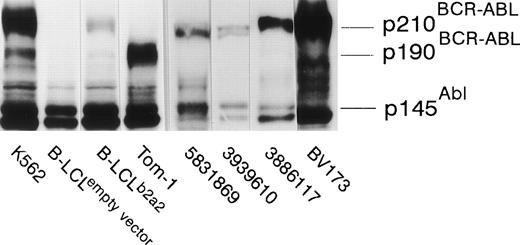
The seven clones were not only proliferative, but displayed b2a2 peptide-specific cytolytic activity against peptide-loaded DR2-positive target cells as well. Autologous B-LCL transfected with full-length BCR-ABLb2a2 cDNA and allogeneic HLA-DR matched BCR-ABLb2a2 RT-PCR-positive cells from CML patients expressed BCR-ABL protein as shown by Western blotting (Fig 3). However, B2a2 peptide-specific clones failed to show lytic or proliferative responses against these cells, unless exogenous b2a2 peptide was added.
EBV-transformed B-LCL, from the anti b2a2 peptide T-cell donor, transfected with full-length BCR-ABLb2a2express p210 BCR-ABL protein, as determined by anti-ABL immunoblot, whereas cells transfected with an empty PDR2 vector do not. K562 and Tom-1 were used as positive controls for p210BCR-ABL and p190BCR-ABL, respectively. Allogeneic HLA-DR–matched BCR-ABLb2a2 RT-PCR-positive leukemic cells from CML patients similarly express p210BCR-ABL as shown for UPN 5831869, 3939610, and 3886117. BV173 was used as a p210b2a2-positive (but p145ABL-negative8) control.
EBV-transformed B-LCL, from the anti b2a2 peptide T-cell donor, transfected with full-length BCR-ABLb2a2express p210 BCR-ABL protein, as determined by anti-ABL immunoblot, whereas cells transfected with an empty PDR2 vector do not. K562 and Tom-1 were used as positive controls for p210BCR-ABL and p190BCR-ABL, respectively. Allogeneic HLA-DR–matched BCR-ABLb2a2 RT-PCR-positive leukemic cells from CML patients similarly express p210BCR-ABL as shown for UPN 5831869, 3939610, and 3886117. BV173 was used as a p210b2a2-positive (but p145ABL-negative8) control.
Iib2a2 cotransfection induces recognition of autologous BCR-ABLb2a2 transfectants by b2a2 peptide-specific T-cell clones.
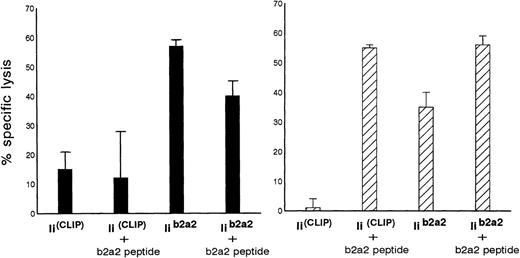
Additional experiments were performed to investigate the possibility of whether the immunogenic fusion region peptide from p210b2a2 could be loaded endogenously and presented in MHC class II molecules of BCR-ABL-transfected cells using a highly efficient endogenous class II loading method. Autologous EBV cells were cotransfected with full-length BCR-ABLb2a2 and invariant chain (Ii) cDNA, in which a CLIP encoding sequence was replaced by a BCR-ABLb2a2 oligonucleotide sequence (Iib2a2). As control APC, identical cells were cotransfected with full-length BCR-ABLb2a2 and with invariant chain without such a CLIP exchange (IiCLIP). As depicted in Fig 4, clones Aa3 and clone Aa14 were both able to kill autologous Iib2a2 transfectants, but not the control Ii transfectants.
Autologous B-LCL transfected with full-length BCR-ABLb2a2 cDNA are lysed by b2a2 fusion peptide-specific clones Aa3 (solid bars) and Aa14 (hatched bars) only after cotransfection with Iib2a2 (invariant chain construct in which part of CLIP was replaced by a b2a2 joining region sequence) and/or incubation with b2a2 peptide.
Autologous B-LCL transfected with full-length BCR-ABLb2a2 cDNA are lysed by b2a2 fusion peptide-specific clones Aa3 (solid bars) and Aa14 (hatched bars) only after cotransfection with Iib2a2 (invariant chain construct in which part of CLIP was replaced by a b2a2 joining region sequence) and/or incubation with b2a2 peptide.
TCR analysis.
The TCR of clones Aa3 and Aa14 was analyzed by RT-PCR and direct sequencing. The results were consistent with a clonal nature of both T-cell populations (Table 4). Differences were seen in their AJ gene usage, N(-D-N) regions and hence complementary determining regions (CDR) 3α and 3β.
TCR Configuration of BCR-ABLb2a2Peptide-Specific Clones
| T-Cell Clone . | TCR AV . | N . | TCR AJ . | TCR BV . | N-D-N . | TCR BJ . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Aa3 | 14S1 | CAY | KGS | QGGSEKLVFG | 57 | 6S5 | CASS | SFWF | SPLHFG | 1S6 |
| Aa14 | 14S1 | CAY | IGSS | NTGKLIFG | 37 | 6S5 | CASS | RGTA | NSPLHFG | 1S6 |
| CDR3α | CDR3β | |||||||||
| T-Cell Clone . | TCR AV . | N . | TCR AJ . | TCR BV . | N-D-N . | TCR BJ . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Aa3 | 14S1 | CAY | KGS | QGGSEKLVFG | 57 | 6S5 | CASS | SFWF | SPLHFG | 1S6 |
| Aa14 | 14S1 | CAY | IGSS | NTGKLIFG | 37 | 6S5 | CASS | RGTA | NSPLHFG | 1S6 |
| CDR3α | CDR3β | |||||||||
DISCUSSION
Although aberrant BCR-ABL fusion genes and BCR-ABL mRNA splicing can occur,4 the majority of patients with CML express p210b3a2 and/or p210b2a2, often together with low levels of pl90e1a2 BCR-ABL proteins.6,7 The immunogenicity of fusion region-derived peptides of p210b3a2 has been demonstrated by our and other groups.15-23 Peptides corresponding to the b3a2 fusion sequence were shown to bind to HLA A2,16A3,14,15 A11 and B8,14 DRB1*0402, DRB1*0301, and DR11.17 HLA A2,16 A3,15,16,20A11,16,20 presumably A24,22 and DRB1*090118 restricted b3a2 peptide-specific cytotoxic T cells have been obtained, as well as HLA DR1,23DRB1*1501,19 DRB1*0401,21DR11,17,20 and DR17(DR317) (and own unpublished observation, July 1996) restricted T-helper cells. Processing of endogenous BCR-ABL proteins by CML cells has not been proven biochemically, albeit that we have observed recognition of HLA-DR4-positive b3a2 CML blasts by HLA-DR4–restricted b3a2 fusion peptide-specific T-helper cells.21 Others have documented lysis of BCR-ABLb3a2 leukemic cells by CD8-positive b3a2 peptide-specific T cells in an MHC class I–restricted16 and unrestricted22 manner. Indirect processing of p210b3a2 has been described in a murine system43 and with mitomycin C–treated CML patient-derived PBMC as APC.23 No reports of BCR-ABL peptides in HLA eluates have been published yet, but a peptide, apparently derived from the BCR protein, was recently eluted from HLA A2.1 molecules.44
The present study shows that it is possible to elicit a T-cell response against a fusion peptide from p210b2a2. Such responses, however, are less easily obtained as those to the b3a2 fusion sequence. The reason for the apparent difference in immunogenicity is unclear. Unlike the b3a2 fusion, the b2a2 sequence lacks a novel amino acid at the fusion site. The glutamic acid encoded by the b2a2 fusion triplet is also present in the original abl sequence (the same holds true for the e1a2 fusion). The b2a2 and e1a2 peptides differ therefore less from their physiologic abl counterpart when compared with the b3a2 fusion. This might contribute to the reduced immunogenicity of b2a2 as compared with b3a2 (the positively charged novel residue lysine at the b3a2 fusion appeared to be essential for recognition by many of our b3a2 peptide-specific clones; ten Bosch et al, unpublished observation, July 1994). Nevertheless, raising the T-cell precursor input by using a modified sensitization protocol, b2a2 peptide-specific DR2a restricted T cells could be obtained from the same donor who responded to b3a2 fusion peptides.19 The latter finding is relevant in view of a potential immunoselective pressure exerted by b3a2-specific T cells, possibly enabling the escape of CML cells expressing b2a2 and e1a2. Several b2a2 peptide-specific T-cell clones were, though clonal by TCR analysis, partially CD4/CD8 double positive. Whether these double-positive T cells, showing a possibly early phenotype, have arisen as a consequence of in vitro differentiation of progenitor cells,45 of expansion of already in vivo present CD4/CD8 double-positive cells, or of certain stimuli that restore the surface coexpression of the CD8 marker46 is unclear. Observations by others of DR2-restricted and cytotoxic CD4/CD8 T cells directed against an EBV BHRF1 peptide47 and CD4/CD8-positive T cells among cutaneous lymphoma infiltrating cells48 suggest an in vivo role for T cells with this phenotype. Fine specificity analyses of b2a2-specific T cells and DR2a binding studies using b2a2 peptides with alanine substitutions were in agreement with proposed motifs within peptide ligands for the DR2a (DRB5*0101) allele.42 The isoleucine at position 5 within the b2a2 17mer and the arginine at position 13 should allow binding to DR51. Moreover, the b2a2 peptide sequence containing a proline at positions 2 and 14 shows a perfect fit with a proposed putative class II processing motif.49 A DR51 restriction was further corroborated by experiments with b2a2 peptide-loaded single allele transfectants. Depending on the T-cell clone, residues at positions 6, 7, 8, 9, 11, and even 14, within the b2a2 peptide appeared to contribute to the TCR contact sites. Clone Aa4 differed from the others in that the glutamic acid at position 9 could be substituted by alanine without affecting the ability of this T cell to respond. Clones, like Aa4, of which the reactivity is not strictly dependent on the presence of the correct amino acid at the fusion site, can still be fusion peptide-specific. As has recently become evident, all amino acids within a given peptide sequence can contribute to the creation of the T-cell epitope.50 51 Our anti b2a2 clones were sensitive to single-residue replacements in the bcr, as well as in the abl part of the intact b2a2 fusion peptide.
TCR analysis by RT-PCR of clones Aa3 and Aa14 confirmed their clonality, despite their partial CD4 and CD8 positivity, and were in agreement with an oligoclonal origin of these cells showing usage of identical AV14S1, BV6S5, and BJ1S6, but different AJ genes and N(-D-N) sequences. Consequently, different CDRs 3α and 3β, which are considered to interact primarily with the antigenic peptide,52 provide an explanation for the different reactivity patterns of the two T-cell clones. We did not observe proliferative or cytotoxic responses of our b2a2 peptide-specific T-cell clones to p210b2a2 expressing autologous B-LCL or HLA-DR–matched CML cells. Nevertheless, it cannot be excluded that the fusion region peptide is processed and presented in the context of HLA-DR2a molecules. Nonresponsiveness of the T-cell clones may be due to an insufficient sensitivity or too low affinity for the antigen. Titration experiments with the 17mer b2a2 peptide and the T2-DR2a cells as target cells showed that relatively high peptide concentrations of 160 ng/mL (88 nmol/L) were required to elicit cytolytic responses by Aa3 and Aa14.
The relative insensitivity of these T cells may limit their responses to APC with artificially elevated numbers of b2a2 peptide MHC complexes, such as exogenous b2a2 peptide-pulsed B-LCL or genetically modified cells carrying the invariant chain construct Iib2a2. Autologous BCR-ABLb2a2-transfected B-LCL were lysed on cotransfection with invariant chain construct Iib2a2 by T-cell clones Aa3 and Aa14. This suggests that the b2a2 epitope for both T-cell clones was presented by HLA-DR2a molecules after endogenous synthesis and optimized processing for MHC class II. Recently, others mentioned the generation of a DRB1*1406 (an allele absent in the Caucasian population, but expressed in approximately 3% of the Japanese population53) restricted b2a2-specific clone, elicited with the same 17mer b2a2 peptide.18 This clone was apparently lost on further culturing, but it illustrates that presentation of b2a2 fusion peptide need not be restricted to HLA-DR51. The present data do not provide proof of processing of endogenous BCR-ABL protein, but demonstrate that the b2a2 fusion region is potentially immunogenic and may serve, if processed properly, as a target for the immune system. Whether immunization with invariant chain-transfectants in which CLIP is exchanged for the b2a2 fusion region (Iib2a2) will yield functionally similar T cells as obtained by peptide immunization or may preferentially induce T cells recognizing epitopes generated by intracellular processing of full-length BCR-ABL protein is currently being evaluated. The quest for BCR-ABL fusion oncoprotein-specific T cells, recognizing endogenously processed peptides in HLA-DR at the tumor cell surface, may perhaps be facilitated by the use of MHC class II tetramers54 to isolate specific T cells with high antigen avidity from donors or CML patients.
Supported by grants from the Dutch Cancer Society, Amsterdam, The Netherlands, and the Fonds National de la Recherche Scientifique Fondation Bekales, Brussels, Belgium.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Onno C. Leeksma, MD, Department of Immunohematology and Bloodbank, Leiden University Medical Center, PO Box 9600, 2300 RC Leiden, The Netherlands.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal