Abstract
About 80% of all cases of Fanconi anemia (FA) can be accounted for by complementation groups A and C. To understand the relationship between these groups, we analyzed the expression pattern of the mouse FA group-A gene (Fanca) during embryogenesis and compared it with the known pattern of the group-C gene (Fancc). Northern analysis of RNA from mouse embryos at embryonic days 7, 11, 15, and 17 showed a predominant 4.5 kb band in all stages. By in situ hybridization, Fanca transcripts were found in the whisker follicles, teeth, brain, retina, kidney, liver, and limbs. There was also stage-specific variation in Fanca expression, particularly within the developing whiskers and the brain. Some tissues known to express Fancc (eg, gut) failed to show Fancaexpression. These observations show that (1) Fanca is under both tissue- and stage-specific regulation in several tissues; (2) the expression pattern of Fanca is consistent with the phenotype of the human disease; and (3) Fanca expression is not necessarily coupled to that of Fancc. The presence of distinct tissue targets for FA genes suggests that some of the variability in the clinical phenotype can be attributed to the complementation group assignment.
PATIENTS WITH FANCONI anemia (FA) suffer from a wide variety of congenital abnormalities. The spectrum of these abnormalities includes growth retardation, skin hyperpigmentation, radial ray deformities, hypogonadism, renal malformations, microcephaly, variable degrees of mental retardation, and microphthalmia.1,2 However, there is extensive phenotypic variability. Also, because many patients develop cytopenias and malignancies,3 FA is regarded as an important genetic model of hematopoietic failure and cancer susceptibility. Cellular manifestations of FA include chromosomal instability, an enhanced sensitivity to bifunctional alkylating agents (eg, mitomycin C [MMC]) and to oxygen, delay in the G2 phase of the cell cycle, and a predisposition to apoptosis.(Kruyt and Youssoufian4 and references therein)
FA is genetically heterogeneous and has been subdivided into at least eight complementation groups.5,6 All subtypes are autosomal recessive. The disease-associated genes for complementation groups C (FANCC),7 A (FANCA),8,9 and G (FANCG)10 have been cloned and shown to encode distinct and novel proteins. In all three cases, the sequences of the predicted polypeptides have failed to suggest a particular function or show any shared motifs. Thus, the ∼63 kD FANCC protein is homologous neither to the ∼163 kD FANCA protein nor to the ∼70 kD FANCG protein (also called XRCC9).
In the absence of such clues from the primary sequences, some of our current understanding about the molecular function of FA gene products has emerged from studies of their cellular localization, protein interactions, and expression patterns. Several studies have shown that FANCC localizes primarily to the cytoplasm under both steady-state and stress conditions.11-14 Other studies have also reported that a fraction of FANCC localizes to the nucleus.15,16Because targeting of FANCC to the nucleus abolishes its ability to correct the hypersensitivity of FA group-C cells to MMC,14its major biological function is most likely performed in the cytoplasm. Consistent with this model is our recent observation that FANCC binds to the microsomal enzyme NADPH cytochrome P450 reductase and regulates its activity, which suggests that FANCC plays a role in detoxification.17 FANCA is localized in both the nucleus and cytoplasm4,15 and functions in a cell compartment distinct from FANCC.4 Forced targeting of FANCA to the cytoplasm abolishes its ability to correct the hypersensitivity of FA group-A cells to MMC.4 Therefore, unlike FANCC, FANCA works in the nucleus. Two reports have also claimed that FANCA interacts physically with FANCC,15,18 but we have been unable to confirm these results.4 Although FANCA has limited homology to a class of peroxidases,19 its precise function remains unknown.
As expected from the pleiotropic nature of FA mutations, bothFANCA and FANCC are thought to be expressed in many tissues, albeit at low levels.4,7-9,12 In situ hybridization studies of the murine Fancc gene has shown that it is expressed initially (embryonic [E] days 8 to 10) in the mesenchyme and its derivatives with osteogenic potential, and at later stages (E 13-19.5) in other cells involved in bone development.20Fancc expression was also observed in the brain, whisker follicles, lung, kidney, gut, and stomach.20 21 We therefore reasoned that a comparison of the expression pattern of Fancc with that of Fancamight clarify the functional relationship between these subgroups and show tissue targets that are differentially sensitive to FA mutations.
MATERIALS AND METHODS
Cloning of murine Fanca cDNA sequences.
A segment of the murine Fanca complementary DNA (cDNA) was amplified by polymerase chain reaction (PCR) from two different cDNA libraries derived from murine erythroleukemia cells and normal murine thymocytes (Youssoufian, unpublished). The following oligonucleotide primers were used for PCR amplification: Fanca1, 5′-CAGACACTGACAGAATGCCAGACTAAG; and Fanca2, 5′-CTGAGGTATTAGCTGCAGCAGAAAAAC. The Fanca1 primer was based on the sequence of the expressed sequence tags (EST) AA146162, and the Fanca2 primer was based on the sequence of the EST AA171025. The PCR-derived fragments were blunt-ended and cloned in pBluescript KS+.
Northern analysis.
A Northern blot containing approximately 2 μg of poly A+RNA per lane from mouse embryos at four different stages of development was probed using conditions recommended by the manufacturer (Clontech, Palo Alto, CA). To improve the detection of Fanca, the 752 bp fragment was labeled simultaneously with both [α-32P]dCTP and [α-32P]dATP.
RNA in situ hybridization.
Embryo collection, sectioning, and in situ hybridization were performed as described.22 Antisense and sense RNA probes of the mouseFanca cDNA corresponding to nucleotides 3491 to 4258 of the human FANCA cDNA were synthesized with T7 or T3 RNA polymerase and labeled with α35S-UTP (1000 Ci/mmol; Amersham, Arlington Heights, IL). Hybridization was done overnight at 50°C. The probe concentration was 0.09 ng/mL. Posthybridization treatments were as follows: (1) one wash in 50% formamide, 2× sodium chloride-sodium citrate (SSC), 20 mmol/L β-mercaptoethanol (FSM) at 50°C for 30 minutes, (2) digestion with 20 μg/mL ribonuclease A in 4× SSC, 20 mmol/L Tris-HCl (pH 7.6), 1 mmol/L EDTA at 37°C for 30 minutes, and (3) one wash in FSM at 58°C for 30 minutes. Slides were dipped in Kodak NTB-2 emulsion (Kodak, Rochester, NY) and exposed for 9 days. Tissues were visualized by fluorescence of Hoechst dye-stained nuclei. Silver grains were visualized with darkfield microscopy. Digital images were captured using a video camera and Adobe Photoshop 3.5 software (Adobe Systems Inc, San Jose, CA) on a Macintosh computer (Apple, Cupertino, CA).
RESULTS
Cloning of mouse Fanca cDNA.
The human FANCA gene consists of 43 exons dispersed over ∼80 kb of genomic DNA.23,24 The structure of the murineFanca gene has not yet been determined. Using sequence information from the EST database, we screened two cDNA libraries from erythroleukemia cells and thymocytes and obtained the same-sized fragment of 742 bp. Sequencing of multiple independent clones showed that the insert corresponds to the human FANCA cDNA residues 3491 to 4258.8 The endpoints of the cDNA fragment fall within exons 35 and 42 of the human FANCAgene.23 24 The sequence also matched with the sequences of EST AA146162 and AA171025 and showed an overall identity of 70% to the nucleotide sequence of the human FANCA cDNA. The labeled insert was then used to isolate a full-length cDNA clone by colony hybridization (complete nucleotide sequence in preparation).
Analysis of Fanca gene expression.
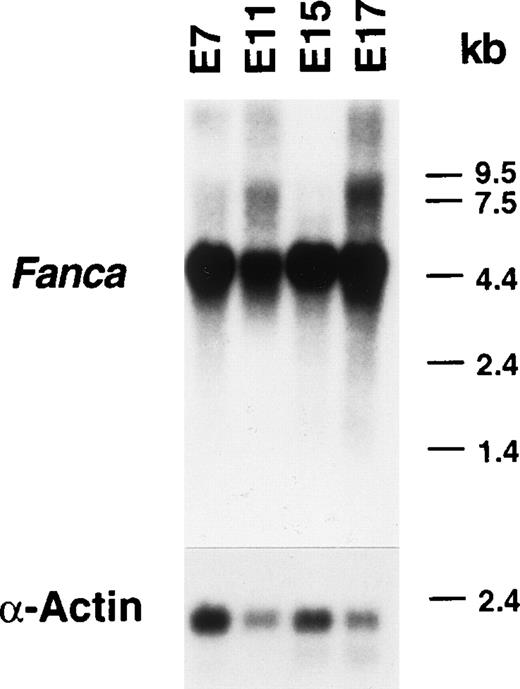
Using the 742 bp fragment as a probe, we analyzed the expression of mouse Fanca during four different stages of embryogenesis. A predominant 4.5 kb band was observed as early as E7, and expression was maintained throughout embryogenesis (Fig1). The same Northern blot probed with a longer cDNA fragment corresponding to the human FANCA cDNA nucleotides 2101-4491 (spanning exons 23-43) showed the same 4.5 kb band, although additional smaller bands were detected at reduced stringency (data not shown). However, as tissue-specific and temporal variations in expression cannot be readily detected by this analysis, we performed in situ hybridization at different stages of mouse development, including E10.5, E13.5, E16.5, and neonatal (P0) stages.
Northern blot of poly A+ RNA from the developing mouse. A 742 bp fragment of mouse Fanca cDNA spanning exons 35 to 42 of the corresponding human FANCA gene and the human -actin cDNA were used as probes. The top blot was exposed for 4 days, whereas the bottom blot was exposed for 6 hours.
Northern blot of poly A+ RNA from the developing mouse. A 742 bp fragment of mouse Fanca cDNA spanning exons 35 to 42 of the corresponding human FANCA gene and the human -actin cDNA were used as probes. The top blot was exposed for 4 days, whereas the bottom blot was exposed for 6 hours.
Expression in the whisker follicles.
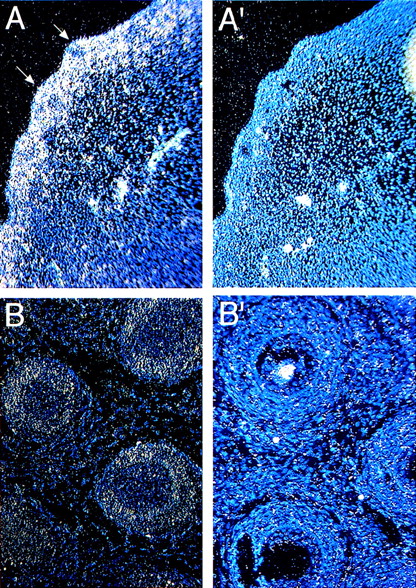
Whiskers or vibrissae are large sensory hairs around the mouth. These structures develop at about E12 with the initial formation of an epithelial placode followed by the formation of dermal papillae (E13-14).25 The papillae surround epithelial placodes, after which the epithelial cells form hair and the hair sheath (E14-17). Hair emerges from the skin at E17.5, which completes the formation of whisker follicles. At E13.4, Fanca is expressed in the forming placode and dermal papilla (arrows in Fig 2A). This expression is maintained at later embryonic stages (E16, not shown), but at the neonatal P0 stage it becomes more restricted to the dermal papilla (Fig 2B).
Expression of Fanca in developing hair follicles (arrows) at (A) E13.5 and (B) P0 stages probed with antisense probes. Controls with sense probes for each panel are shown (A′, B′).
Expression of Fanca in developing hair follicles (arrows) at (A) E13.5 and (B) P0 stages probed with antisense probes. Controls with sense probes for each panel are shown (A′, B′).
Expression in the brain.
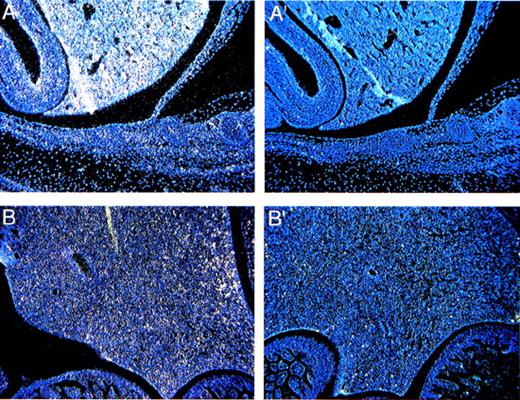
Beginning at E8-9, the neural tube closes and begins to bulge and constrict, which gives rise to the different portions of the brain (fore-, mid-, and hindbrain). The optic vesicles form as secondary outgrowths from the forebrain during embryogenesis and then go on to form the retina during the postnatal period (P0-P16). The forebrain gives rise to the cerebral cortex, olfactory bulb, and hippocampus. Cells in the cerebral cortex proliferate in the ventricular zone, and at E12 begin to migrate within the intermediate zone and give rise to the cortical plate.26 The cerebellum develops between the mid- and hindbrain as early as E13, but most of the proliferative and migratory activities take place postnatally between P0 and P21.27 At E13.5, Fanca expression is limited to the intermediate zone of the developing cerebral cortex (Fig 3A) and the anterior part of the midbrain (Fig 3B), which give rise to the tectum. By the P0 stage,Fanca expression is no longer detectable at these sites (data not shown). Instead, Fanca transcripts now become detectable in the external granular layer of the cerebellum (Fig 3C) and in the inner nuclear layer of the developing retina (Fig 3D) in concert with the postnatal maturation of these organs.
Expression of Fanca in the developing brain at E13.5 (A) within the intermediate zone of the cerebral cortex and (B) the anterior part of the midbrain, and at the P0 stage in (C) the external granular layer of the cerebellum and (D) the inner nuclear layer of the developing retina. Controls are shown as indicated in Fig2.
Expression of Fanca in the developing brain at E13.5 (A) within the intermediate zone of the cerebral cortex and (B) the anterior part of the midbrain, and at the P0 stage in (C) the external granular layer of the cerebellum and (D) the inner nuclear layer of the developing retina. Controls are shown as indicated in Fig2.
Expression in the liver.
The liver arises at E9 from the hepatic diverticulum. By E11.5, hematopoietic foci are found intermingled with the hepatic cords.28Fanca expression is readily detectable at E13.5 in a diffuse pattern (Fig 4A), which makes it difficult to identify the particular lineages of theFanca-positive cells. The expression of Fanca is significantly lower at the P0 stage, although it can still be detected (Fig 4B).
Expression of Fanca in the liver (A) at E13.5 and (B) the P0 stage. Controls are shown as indicated in Fig 2.
Expression of Fanca in the liver (A) at E13.5 and (B) the P0 stage. Controls are shown as indicated in Fig 2.
Expression in the kidney, teeth, and limbs.
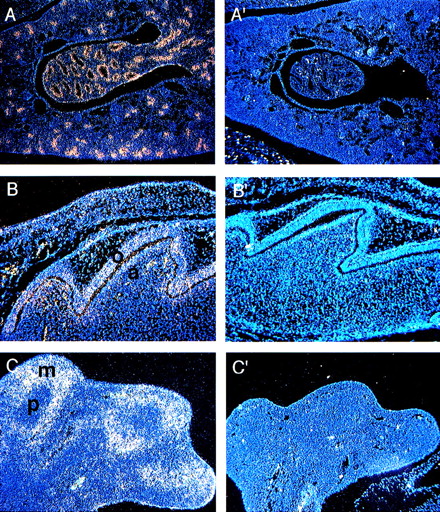
Kidney development begins at E10 as a result of an interaction between two mesodermal derivatives, the ureteric bud, and the metanephric mesenchyme.29 Through the second half of gestation the ureteric bud grows, branches, and makes connections with the maturing nephric tubules that form the renal collecting duct system.Fanca expression appears as early as E16 in the developing ducts (not shown). At P0, there is a high level of expression in the epithelial cells of the developing collecting duct system (Fig 5A).
(A) Expression of Fanca at the P0 stage in (A) the epithelial cells of the developing collecting ducts of the kidney and (B) the developing tooth. Higher level of expression is noted in odontoblasts (o) than ameloblasts (a). (C) Expression of Fancain the hindlimb at E13.5 predominantly in mesenchymal cells (m) rather than in precartilage cells (p). Controls are shown as indicated in Fig2.
(A) Expression of Fanca at the P0 stage in (A) the epithelial cells of the developing collecting ducts of the kidney and (B) the developing tooth. Higher level of expression is noted in odontoblasts (o) than ameloblasts (a). (C) Expression of Fancain the hindlimb at E13.5 predominantly in mesenchymal cells (m) rather than in precartilage cells (p). Controls are shown as indicated in Fig2.
Tooth development occurs through an interaction between the epithelium of the first arch and the underlying ectomesenchyme derived from the neural crest.30 During the perinatal stage the epithelium differentiates into enamel-secreting ameloblasts, whereas the ectomesenchymal cells become the dentin-secreting odontoblasts. Although both cell types contained Fanca transcripts, higher levels were detected in ameloblasts (Fig 5B).
Forelimb buds are first observed at E9.5, whereas hindlimb buds can be observed soon after E10. Cartilage elements begin to form by E12, and by E13.3 mesenchymal cells surround the precartilage localized at the center of each skeletal element. Cartilage is then replaced by bone, and the mesenchymal cells form the muscles and other soft tissues.31 At E13.5, Fanca is expressed in the mesenchymal cells of both the forelimb (data not shown) and hindlimb (Fig 5C), but not in the precartilage.
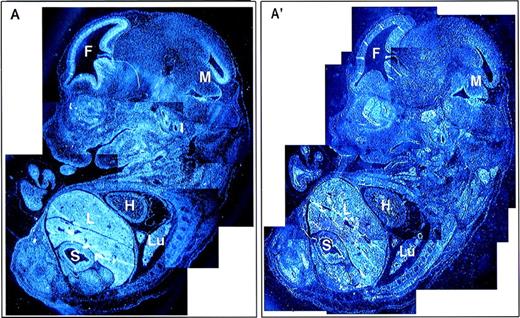
A composite photomicrograph of multiple sections from an E13.5 embryo is shown (Fig 6). In addition to the structures discussed above, some Fanca expression is observed in the heart, but expression in most other tissues, including the developing gut and lung, was very low or negative when individual panels of the composite figure were compared with each other. Finally, there was diffuse but no localized expression at E11.5, including expression in the neural tube (data not shown).
Global expression of Fanca at E13.5 of mouse development. Reconstruction image of a whole embryo at E13.5 from multiple sections visualized with (A) antisense and (A′) sense probes. F, forebrain; H, heart; L, liver; Lu, lung; M, midbrain; S, stomach.
Global expression of Fanca at E13.5 of mouse development. Reconstruction image of a whole embryo at E13.5 from multiple sections visualized with (A) antisense and (A′) sense probes. F, forebrain; H, heart; L, liver; Lu, lung; M, midbrain; S, stomach.
DISCUSSION
Despite the well-established genetic classification of FA into distinct complementation groups5,6 and the identification of molecular etiologies for three subgroups,7-10 the pathogenetic processes involved in this disorder remain obscure. Studies of protein interaction between FANCA and FANCC have yielded conflicting results,4 15 and any epistatic relationship among the FA genes remains speculative. In this context, an understanding of the expression pattern of Fanca and a comparison with the known pattern of Fancc can be instructive. Here we show that the expression of the mouse Fanca gene exhibits tissue-specific and stage-specific regulation during development.
Fanca expression was observed in multiple tissues, including the developing placode and dermal papilla that make up the whiskers and the appendicular skeleton. Expression was noted primarily in cells of mesenchymal origin that give rise to the soft tissues of the forelimb and hindlimb. Such mesenchymal derivatives with osteogenic and hematopoietic potential also contain Fancctranscripts.20,21 Thus, there is concordance between the expression patterns of these two FA genes in mesenchymal derivatives. In addition, Fanca expression was noted in the developing kidney, liver, and brain, indicating a concordance of expression in these tissues as well.20 The expression of Fanca in the brain is particularly noteworthy due to its association with the intermediate zone, which contains migrating neurons that give rise to the cerebral cortex. In this compartment, Fanca may be a useful marker of neuronal migration.
Although many of these findings were reminiscent of the developmental expression pattern of Fancc,20 there were also several tissues that showed seemingly discordant patterns of expression. For example, we found little if any Fancaexpression in the developing lung and gut, organs that were previously identified to contain Fancc transcripts.20 These observations suggest that the expression of Fanca is not necessarily coupled to Fancc during development. In yet other tissues, comparisons between Fanca and Fancc expression were less clear. Fanca was clearly expressed in two cell types that contribute to tooth formation. Although Fancc expression was previously noted in the developing maxilla, the authors did not comment on its expression in the developing teeth.20 A uniform pattern of Fancc expression was also reported previously in many adult tissues, including testes,21 but the pattern of expression in the developing gonads was not stated.20 In the present study, we found little if any specific hybridization in the developing gonads, including the E13.5 and E16 stages. This is somewhat difficult to reconcile with the prevalence of infertility and gonadal abnormalities observed in human patients, features that were also observed in mice nullizygous forFancc mutations.32 33 It may suggest that the gonadal failure is related to hormonal, structural, or other abnormalities rather than to a specific requirement for FA gene products in gonadal tissues.
Beyond the most obvious qualitative differences, however, such comparisons should be made with caution. In situ hybridization is not a quantitative test, and subjective interpretations may account for potential differences between different studies. Also, the analysis of different tissues and stages was less than exhaustive in both cases, and low-levels of expression in some tissues may have been overlooked. Indeed, Fanca expression appeared to be low in most developing tissues. A further lowering of expression in certain tissues, such as the gonads, may place them below the limit of detection by in situ hybridization. Another confounding variable may be the presence of alternatively spliced transcripts that cannot be detected by certain probes. Although it is possible that the probe used in our study cannot detect potential isoforms of Fanca, we believe that this probe is capable of detecting the major Fanca transcript that encodes functional Fanca protein. The relatively large size of the probe spanning eight coding exons, the inability of a longer probe spanning exons 23-43 to detect additional isoforms by high stringency Northern hybridization, and the colinearity of the mouse cDNA sequence with the sequence of the functional human orthologue (data not shown) support this view. Nevertheless, we cannot exclude the presence of additionalFanca-derived transcripts that may encode functional polypeptides.
Taken together, these results show that Fanca expression occurs primarily in tissues of epithelial and mesenchymal origin. Defects in all of these tissues have been implicated in FA patients, confirming again the pleiotropic nature of FA mutations. However, although there are certain similarities in the expression patterns of Fancaand Fancc, there also appear to be distinct domains of expression for each gene during development. At present it is not possible to deduce the complementation group of FA patients on the basis of their clinical or cellular phenotype. The difference in the patterns of expression may provide a useful framework to future studies that attempt to refine genotype-phenotype correlations in FA. Finally, the tissue-specific temporal expression of Fanca during embryogenesis indicates that Fanca-mediated processes are developmentally regulated. We do not yet know the signals that can induce or repress Fanca transcription in these tissues. The 5′ noncoding region of the human FANCA gene is GC-rich and lacks obvious TATA or CAAT sequences,23 suggesting a “housekeeping” role for this gene. This region has not yet been formally tested for promoter activity. It is possible that low levels of Fanca protein could have a housekeeping function in most tissues, whereas higher levels may be required in some tissues during development at certain stages of development. Whether or not this putative promoter contains the elements necessary for tissue- and stage-specific expression remains to be determined.
Supported by grants from the National Institutes of Health (HL52138) and the March-of-Dimes Birth Defects Foundation.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal