Abstract
Chromosomal translocations involving the immunoglobulin heavy chain (IGH) locus at chromosome 14q32 represent a common mechanism of oncogene activation in lymphoid malignancies. In multiple myeloma (MM), variable chromosome partners have been identified by conventional cytogenetics, including the 11q13, 8q24, 18q21, and 6p21 loci. We and others have recently reported a novel, karyotypically undetectable chromosomal translocation t(4;14)(p16.3;q32) in MM-derived cell lines, as well as in primary tumors. The 4p16.3 breakpoints are relatively scattered and located less than 100 kb centromeric of the fibroblast growth factor receptor 3 (FGFR3) gene or within the recently identified WHSC1 gene, both of which are apparently deregulated by the translocation. To assess the frequency of the t(4;14)(p16.3;q32) translocation in MM, we performed a double-color fluorescent in situ hybridization (FISH) analysis of interphase nuclei with differently labeled probes specific for the IGH locus (a pool of plasmid clones specific for the IGH constant regions) or 4p16.3 (yeast artificial chromosome (YAC) 764-H1 spanning the region involved in breakpoints). Thirty MM patients, the MM-derived cell lines KMS-11 and OPM2, and six normal controls were examined. The identification of a t(4;14) translocation, evaluated as the presence of a der(14) chromosome, was based on the colocalization of signals specific for the two probes; a cutoff value of 15% (mean + 3 standard deviation [SD]) derived from the interphase FISH of the normal controls (range, 5% to 11%; mean ± SD, 8.16 ± 2.2) was used for the quantification analysis. In interphase FISH, five patients (one in clinical stage I, two in stage II, one in stage III, and a plasma cell leukemia) were found to be positive (≈15%). FISH metaphases with split or colocalized signals were detected in only two of the translocated cases and confirmed the pattern found in the interphase nuclei. Furthermore, in three of the five cases with the translocation, FISH analysis with the IGH joining probe (JH) showed the presence of the reciprocal product of the translocation [der(4) chromosome]. Overall, our study indicates that the t(4;14)(p16.3;q32) chromosomal translocation is a recurrent event in MM tumors and may contribute towards the detection of this lesion and our understanding of its pathogenetic and clinical implications in MM.
CHROMOSOMAL TRANSLOCATIONS affecting the IGH locus on 14q32 represent the mechanism of activation of a number of proto-oncogenes in distinct types of B-cell lymphoid neoplasms. The best examples are the t(8;14)(q24;q32), t(8;22)(q24;q11), and t(2;8)(p12;q24) translocations leading to the deregulation of c-MYC in Burkitt’s lymphomas; the t(14;18)(q32;q21) involving the BCL-2 gene in follicular lymphomas; the t(11;14)(q13;q32) involving the BCL-1/cyclinD1 locus in mantle cell lymphomas (MCL); the t(3;14)(q27;q32) involving the BCL-6 gene in large cell lymphomas; and the t(9;14)(p13;q32) affecting the PAX-5 gene in plasmacytoid lymphomas.1
Multiple myeloma (MM) is a malignant proliferation of bone marrow plasma cells characterized by a wide spectrum of clinical entities, treatment responses, and survival times.2,3 Conventional cytogenetic analyses are limited and difficult in MM, mainly because of the low proliferation rate of malignant plasma cells; however, a 14q+ marker has been reported in 20% to 40% of tumors,4-6which probably represents translocations in the IGH locus. In almost 30% of cases, this marker is the result of a t(11;14)(q13;q32) chromosomal translocation, although rearrangements of theBCL-1/cyclin D1 regions frequently involved in MCL, rarely occur in MM.7-9 In the remaining cases with a detectable 14q+ marker, the translocation partners have rarely been identified by conventional cytogenetics and mainly involved chromosomes 8q24, 18q21, or 6p21.4-6 10
More recently, we and others using various molecular approaches have demonstrated that translocations involving the IGH locus are a frequent event in MM.11-13 These studies have led to the identification of novel genomic regions involved in MM, mainly as a result of karyotypically undetectable chromosomal translocations. These regions include the 4p16.3, the 6p25, and the 16q23, where the fibroblast growth factor receptor 3 (FGFR3), the interferon regulatory factor 4 (IRF4/MUM1), and the c-maf gene are respectively located.13-16 With regard to the t(4;14)(p16.3;q32) chromosomal translocation, 4p16.3 breakpoints have been reported so far in five MM-derived cell lines and four primary MM tumors and are scattered over a genomic region of approximately 70 kb located less than 100 kb centromeric of the FGFR3gene,17 a member of the tyrosine-kinase receptor family (FGFR1-4), which has been suggested to be a candidate oncogene of the translocation.13,14 It has very recently been reported that some of 4p16.3 breakpoints occur telomeric to or within the 5′ introns of the novel gene WHSC1/MMSET,18,19 a putative transcription factor that is preferentially expressed in rapidly growing embryonic tissues, but also at different levels in a large variety of adult human tissues. The FGFR3 gene is overexpressed in translocated cases, but is absent or barely detectable in cell lines without the translocation,13,14whereas the WHSC1/MMSET gene is expressed at low levels in MM cell lines, but overexpressed in the majority of those carrying the translocation.19 Thus, the t(4;14)(p16.3;q32) in MM may lead to the deregulation of two different potential oncogenes.
The incidence of the t(4;14)(p16.3; q32) chromosomal translocation in MM remains to be established. By using specific probes for some of the 4p16.3 breakpoints, we found rearrangements in only one of 53 primary MM tumors by Southern blot analysis13; however, this approach is conceivably limited because of the dispersion of the 4p14.3 breakpoints and the relatively large amount of repetitive DNA sequences found in their proximity. To assess the incidence of this genetic lesion in MM, we used double-color fluorescent in situ hybridization (FISH) to investigate samples taken from 30 MM patients. This method has the great advantage of detecting chromosomal alterations in interphase nuclei, which has been shown to be very useful in MM patients for whom no abnormal metaphases are generally available.12,20 21 The probes used were a pool of plasmid clones specific for the constant regions of the IGH locus on chromosome 14q32, and a yeast artificial chromosome (YAC) clone 764-H1 spanning the breakpoints on 4p16.3 region. A t(4;14) chromosomal translocation was detected in approximately 15% of the investigated cases.
MATERIALS AND METHODS
Pathological samples and MM cell lines.
The bone marrow aspirates from 30 patients with MM admitted to our Hematology Service were collected during standard diagnostic procedures. The diagnosis and clinical staging were made according to the criteria described by Durie and Salmon.22 All of the patients were at first diagnosis: 12 in stage I, 9 in stage II, 8 in stage III, and one with plasma cell leukemia (PCL). No conventional cytogenetic analyses (G-banding analysis) were performed. Bone marrow cell suspensions were obtained from the bone marrow aspirates after erythrocyte lysis, and aliquots were taken for the FISH and Southern blot analyses. The percentage of malignant plasma cells (ranging from 13% to 86%; Table 1) was assessed by the morphologic analysis of the cytospins from these cell suspensions. The KMS-11 and OPM2 cell lines have been previously described.13 14
DNA preparation and Southern blot analysis.
DNA from the bone marrow cell suspensions was purified by proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation. A total of 10 μg of genomic DNA was digested with BamHI restriction enzyme, electrophorized in a 0.7% agarose gel, and then denaturated, neutralized, and transferred to nylon filters (Amersham International, Amersham, UK). The filters were hybridized to32P-labeled probes according to the manufacturer’s specifications, washed in 0.5 × SSC (NaCl/Na citrate)/1% sodium dodecyl sulfate (SDS) for 1 hour at 60°C, and then autoradiographed using an intensifying screen at −80°C.21IGH gene rearrangement was analyzed using previously described probes.13
FISH studies.
The bone marrow cell suspensions from each case were cultured in RPMI 1640 medium supplemented with 20% fetal calf serum without any mitogen for 24 hours at 37°C in 5% CO2.12 The cultures were treated with colcemid (0.05 mg/mL) for 20 minutes. The cells were harvested using hypotonic potassium chloride, fixed by methanol/glacial acetic acid (3:1)(vol/vol), and then stored at −20°C.
Slides were hybridized in situ with probes labeled with biotin or directly with the fluorochrome Cy3 (Amersham, Little Chalfont, UK) by nick translation, as described23 with minor modifications. Briefly, 200 ng of labeled probe was used for each experiment, and hybridization was performed at 37°C in 2xSSC, 50% (vol/vol) formamide, 10% (wt/vol) dextran sulphate, 5 μg Cot1 DNA (Boehringer, Mannheim, Germany) and 3 μg of sonicated salmon sperm DNA, in a volume of 10 μL. Posthybridization washing was at 60°C in 0.1xSSC (three times). Biotin-labeled DNA was detected using fluorescein isothiocyanate (FITC)-conjugated avidin (green signal) (Vector Laboratories, Burlingame, CA); direct Cy3 was detected as a red signal. The chromosomes were identified by simultaneous 4’,6’-diaminido-2-phenylindole dihydrochloride (DAPI) staining, which produces a Q-banding pattern. Digital images were obtained using a Leica DMR epifluorescence microscope (Leica Imaging Systems Ltd, Cambridge, UK) equipped with a CCD camera (Cohu Inc, San Diego, CA). FITC-avidin, Cy3, and DAPI fluorescence signals were detected using specific filters. The images were recorded, pseudocolored, and merged using QFISH software (Leica Imaging Systems Ltd), and finally edited using Adobe Photoshop, Version 3 (Adobe Systems, Mountain View, CA).
Probes for FISH analysis.
The IGH locus was investigated using a pool of plasmid clones specific for the constant regions Cγ1, Cγ2, Cγ3, Cγ4 (BamHI/HindIII fragments of 7, 4, 7 and 6 kb, respectively), Cα1 and Cα2 (BamHI fragments of approximately 18 kb), and Cμ (HindIII fragment of 10 kb). The JH probe used was the BamHI/HindIII 6.6 kb fragment13 (see scheme in Fig1). The 4p16.3 region was investigated using the YAC clone 764-H1 from the human CEPH2 library (YAC Screening Center, DIBIT, Milan, Italy). This YAC clone (approximately 400 kb in length) hybridizes to theFGFR3 cDNA (see below) and contains sequences specific for the centromeric portion of cosmid clone cl190b4,24 as shown by polymerase chain reaction (PCR) amplification on YAC DNA of a fragment localized between nucleotides 5481-6091 of this clone (GenBank no.Z68276). The breakpoint of the OPM2 cell line has been localized within cosmid clone cl190b4, approximately 17 kb telomeric of this fragment.14 Furthermore, the YAC 764-H1 contains sequences located dowstream of the last exon of the WHSC1gene,18 as shown by PCR amplification of a fragment localized between nucleotides 5035-5600 of the cl27H9 cosmid clone24 (GenBank no. Z49237). Thus, the YAC 764-H1 includes the FGFR3 locus, spans the entire region in which all of the 4p16.3 breakpoints so far reported are located, and includes the entire WHSC1 locus (see scheme in Fig 1). The FGFR3locus was investigated using the 5.3 kb pH9 cDNA clone.13Copy numbers of chromosome 4 were evaluated with the biotin-labeled α-satellite DNA probe (clone D4Z1) (Oncor Inc, Gaithersburg, MD).
Schematic representation of the 14q32.3 (IGHlocus) and 4p16.3 regions. For the IGH locus, the relative location of the variable (V), diversity (D), joining (JH), and constant regions (μ, δ, γ, ɛ, ) is shown. For the 4p16.3 region, a scale representation is given. The location of breakpoints in MM cell lines (OPM2, JIM3, H929, UTMC2, KMS-11) and primary tumors (PCL-1, LB375, LB1017) is indicated by arrows; in case LB278, the breakpoint was localized by Southern blot (see Richelda et al13 and Chesi et al14 for more details). The relative position of the FGFR3 and WHSC1 genes (white box indicates the genomic region spanning the untranslated sequences) on the 4p16.3 region is illustrated (see Chesi et al,14 Thompson et al,17 Stec et al,18 Chesi et al,19and Baxendale et al24 for more details). Cosmid clones encompassing the WHSC1 locus24 are located. The portion of YAC 764-H1 encompassing the 4p16.3 region, as investigated by Southern blot and PCR analyses (see Materials and Methods), is shown by a solid line.
Schematic representation of the 14q32.3 (IGHlocus) and 4p16.3 regions. For the IGH locus, the relative location of the variable (V), diversity (D), joining (JH), and constant regions (μ, δ, γ, ɛ, ) is shown. For the 4p16.3 region, a scale representation is given. The location of breakpoints in MM cell lines (OPM2, JIM3, H929, UTMC2, KMS-11) and primary tumors (PCL-1, LB375, LB1017) is indicated by arrows; in case LB278, the breakpoint was localized by Southern blot (see Richelda et al13 and Chesi et al14 for more details). The relative position of the FGFR3 and WHSC1 genes (white box indicates the genomic region spanning the untranslated sequences) on the 4p16.3 region is illustrated (see Chesi et al,14 Thompson et al,17 Stec et al,18 Chesi et al,19and Baxendale et al24 for more details). Cosmid clones encompassing the WHSC1 locus24 are located. The portion of YAC 764-H1 encompassing the 4p16.3 region, as investigated by Southern blot and PCR analyses (see Materials and Methods), is shown by a solid line.
RESULTS
FISH analysis.
The detection of 4p16.3 rearrangements was based on the analysis of double-color spot distribution in interphase and (when available) metaphase cells. All of the cases were analyzed using a pool ofIGH constant regions and YAC clone 764-H1 as probes. After cohybridization, the interphase nuclei of normal cells have two pairs of nonoverlapping spots (two red signals: homolog chromosomes 4; two green signal: homolog chromosomes 14). Figure 2 shows the pattern of metaphase and interphase FISH of a control sample (Fig 2A and B, respectively). The YAC 764-H1 shows a faint cross-hybridization in metaphase spreads with the short arm of chromosome 2, which is barely or not detectable in interphase nuclei. This cross-hybridization is probably due to the presence of homologous sequences, as two independent and overlapping cosmid clones isolated from a normal human cosmid library (Clontech, Palo Alto, CA), which span the entire FGFR3 locus, show the same pattern of hybridization in FISH metaphases (data not shown).
FISH analyses of normal control and MM-derived cell lines carrying the t(4;14)(p16.3;q32) using the IGH costant regions (green) and the YAC 764-H1 (red) as probes. (A) and (B) Metaphase and interphase nuclei hybridization of normal lymphocytes; arrowheads indicate chromosome 14 and the X indicates chromosome 4. (C) and (D) Metaphase and interphase nuclei hybridization from the KMS-11 cell line. The arrow indicates the colocalized signals on der(14). As shown, the IGH probe recognizes an apparently normal chromosome 14 and an altered, unidentified chromosome; consistent with these findings, there is an additional green signal in the interphase nuclei (D). There are two chromosomes 4 (X) with different long arms; we have previously demonstrated that the longest is the der(4) on the basis of the absence of the FGFR3 locus.13 (E) and (F) Metaphase and interphase nuclei hybridization from the OPM2 cell line. There are two der(14) chromosomes with colocalized signals (arrows). The IGHprobe hybridized to a putative isochromosome and an unidentified, structurally altered chromosome. Three chromosomes 4 were detected (X). Consistent with this pattern, the OPM2 interphase nuclei show five green and five red spots, two of which are overlapping (yellow signal) and indicated by arrows. The FGFR3 locus was found to be localized on both der(14) (data not shown).
FISH analyses of normal control and MM-derived cell lines carrying the t(4;14)(p16.3;q32) using the IGH costant regions (green) and the YAC 764-H1 (red) as probes. (A) and (B) Metaphase and interphase nuclei hybridization of normal lymphocytes; arrowheads indicate chromosome 14 and the X indicates chromosome 4. (C) and (D) Metaphase and interphase nuclei hybridization from the KMS-11 cell line. The arrow indicates the colocalized signals on der(14). As shown, the IGH probe recognizes an apparently normal chromosome 14 and an altered, unidentified chromosome; consistent with these findings, there is an additional green signal in the interphase nuclei (D). There are two chromosomes 4 (X) with different long arms; we have previously demonstrated that the longest is the der(4) on the basis of the absence of the FGFR3 locus.13 (E) and (F) Metaphase and interphase nuclei hybridization from the OPM2 cell line. There are two der(14) chromosomes with colocalized signals (arrows). The IGHprobe hybridized to a putative isochromosome and an unidentified, structurally altered chromosome. Three chromosomes 4 were detected (X). Consistent with this pattern, the OPM2 interphase nuclei show five green and five red spots, two of which are overlapping (yellow signal) and indicated by arrows. The FGFR3 locus was found to be localized on both der(14) (data not shown).
In the presence of a t(4;14) chromosomal translocation, colocalized overlapping red and green (yellow) signals [der(14) chromosome] should be found in interphase nuclei. Separate red and green signals are also found as normal chromosomes 4 and 14, respectively. In addition, a third red signal can be found representing sequences recognized by YAC 764-H1 retained on der(4), whereas no split of the IGH signal should be expected, given the nature of our probe (the sequences between the JH and the constant region involved in switching recombinations are generally deleted).25 The detection of the translocation in interphase FISH was based on evidence of colocalization. The additional red signal was not considered a restrictive finding because breakpoints may occur outside the region recognized by the YAC; portions of chromosome 4p16.3 may be deleted during translocation; one copy of chromosome 4 may be lost; the signal may be barely detectable over the background for technical reasons; or cross-hybridization signals may be present. We therefore evaluated the background of false-positive interphase nuclei showing coincidentally colocalized signals: under our exprimental conditions, 5% to 11% of false-positive interphase nuclei were observed (mean ± SD, 8.16 ± 2.2) among the 200 nuclei taken from six normal controls (phytohemagglutinin [PHA]-stimulated lymphocytes from three healthy donors and bone marrow samples from three patients affected by thrombocytopenia). On the basis of this finding, we considered the mean value plus 3 SD (15%) as the cutoff point for the quantitative analysis of our samples.12,20 21 Two hundred interphase nuclei and all of the detectable metaphases were evaluated for each pathological sample. A percentage of interphase nuclei with colocalized signals above the cutoff value, with or without an additional red signal, was considered as identifying samples with translocation.
The MM-derived cell lines KMS-11 and OPM2 carrying a t(4;14)(q16.3;q32) were evaluated as positive controls (Fig 2C through F); the 4p16.3 breakpoints in these cell lines respectively define the telomeric and centromeric boundaries of the breakpoint region13,14 (see scheme in Fig 1). Colocalization of red and green signals was observed at the telomeric end of a der(14) chromosome in both MM cell lines (Fig2C and E); in the OPM2 cell line, two apparently similar der(14) were detected (Fig 2E) together with three chromosomes 4, presumably as a result of chromosome duplication of both reciprocals of the translocation (see below). In line with these findings, interphase FISH detected one pair of colocalized signals in KMS-11 and two pairs in OPM2, (Fig 2D and F). Furthermore, as a consequence of the split of the 4p16.3 region hybridizing with YAC 764-H1, one (in KMS-11) or two (in OPM2) additional red signals were also detected (Fig 2D and F). TheFGFR3 gene is translocated to the der(14) chromosome in both cell lines as observed using the FGFR3 cDNA as a probe in metaphase FISH13,14 (data not shown). Although the switch-mediated translocation in the OPM2 cell line involves an Sγ region,14 the IGH probe was not split on der(4), which is consistent with the deletion events during switching recombinations. For KMS-11, no IGH split was expected because the breakpoint on 14q32 has been reported to occur within the Sμ region.14 Furthermore, der(4) chromosomes (one in KMS-11 and two in OPM2) hybridized with the JH probe as a consequence of the t(4;14) reciprocal chromosomal translocation (data not shown). Finally, the IGH probe hybridized to apparently abnormal chromosomes in both cell line metaphases: an unidentified putative isochromosome in OPM2 and a structurally altered, unidentified chromosome in OPM2 and KMS11 (Fig 2C and E).
FISH analysis of MM primary tumors.
Interphase FISH analysis was successfully performed in all of the 30 patients. In five patients (nos. 9, 13, 18, 24, and 30), the percentage of nuclei with clearly colocalized green and red signals was far above the cutoff value derived from normal controls (Table 1) and in accordance with the percentage of plasma cells in the pathological samples. In cases no. 9 and 30, we found a typical pattern of translocation in interphase nuclei: a colocalized red and green signal together with an extra red signal, which was probably the result of the split of the 4p16.3 region (Fig 3A and C). In two cases (nos. 13 and 24), no extra red signal was detected (Fig 3D, data not shown), whereas the remaining case (no. 18) showed colocalized red and green signals together with a red signal and the absence of a second green signal (Fig 3E).
FISH Analysis of t(4;14)(p16.3;q32) in MM
| Patient No. . | Sex/ Age . | Ig Type . | Stage . | PC% . | Interphase* FISH (%) . | Metaphase† FISH . |
|---|---|---|---|---|---|---|
| 1 | M/67 | Aλ | IA | 15 | 8 | 0/6 |
| 2 | M/54 | Gκ | IA | 20 | 7 | 0/7 |
| 3 | F/66 | Gκ | IA | 24 | 11 | 0/20‡ |
| 4 | M/72 | Gκ | IA | 21 | 9 | 0/17 |
| 5 | F/78 | Gκ | IA | 16 | 3 | 0/37 |
| 6 | M/68 | Gκ | IA | 18 | 13 | 0/43‡ |
| 7 | M/65 | Gκ | IA | 13 | 7 | 0/47‡ |
| 8 | M/61 | Gκ | IA | 19 | 11 | 0/41 |
| 9 | F/56 | Gκ | IA | 22 | 0/37 | |
| 10 | F/80 | Gκ | IA | 28 | 6 | 0/46‡ |
| 11 | F/72 | Gκ | IA | 22 | 9 | 0/40 |
| 12 | M/70 | Gκ | IA | 14 | 7 | 0/37 |
| 13 | M/66 | Aκ | IIA | 28 | 0/7 | |
| 14 | F/71 | κ | IIA | 42 | 12 | 0/43 |
| 15 | M/67 | Gλ | IIA | 56 | 8 | 0/27 |
| 16 | M/63 | Gκ | IIA | 39 | 11 | 0/13 |
| 17 | M/36 | Dκ | IIA | 50 | 13 | 0/42 |
| 18 | F/66 | Aλ | IIA | 38 | 13/28 | |
| 19 | M/77 | Gκ | IIA | 52 | 6 | 0/19 |
| 20 | F/64 | Aκ | IIA | 39 | 10 | 0/16 |
| 21 | M/84 | Gκ | IIA | 36 | 8 | 0/23 |
| 22 | M/60 | Aκ | IIIA | 45 | 12 | 0/37 |
| 23 | M/67 | Gκ | IIIB | 60 | 10 | 0/27‡ |
| 24 | M/54 | Aκ | IIIA | 45 | 0/47 | |
| 25 | M/76 | Aλ | IIIA | 49 | 5 | 0/2 |
| 26 | M/79 | Gκ | IIIB | 56 | 10 | 0/19 |
| 27 | M/57 | Gκ | IIIB | 46 | 13 | 0/17 |
| 28 | M/66 | Gκ | IIIA | 54 | 12 | 0/18‡ |
| 29 | F/71 | λ | IIIA | 63 | 9 | 0/14 |
| 30 | M/65 | Aκ | PCL | 86 | 20/23 |
| Patient No. . | Sex/ Age . | Ig Type . | Stage . | PC% . | Interphase* FISH (%) . | Metaphase† FISH . |
|---|---|---|---|---|---|---|
| 1 | M/67 | Aλ | IA | 15 | 8 | 0/6 |
| 2 | M/54 | Gκ | IA | 20 | 7 | 0/7 |
| 3 | F/66 | Gκ | IA | 24 | 11 | 0/20‡ |
| 4 | M/72 | Gκ | IA | 21 | 9 | 0/17 |
| 5 | F/78 | Gκ | IA | 16 | 3 | 0/37 |
| 6 | M/68 | Gκ | IA | 18 | 13 | 0/43‡ |
| 7 | M/65 | Gκ | IA | 13 | 7 | 0/47‡ |
| 8 | M/61 | Gκ | IA | 19 | 11 | 0/41 |
| 9 | F/56 | Gκ | IA | 22 | 0/37 | |
| 10 | F/80 | Gκ | IA | 28 | 6 | 0/46‡ |
| 11 | F/72 | Gκ | IA | 22 | 9 | 0/40 |
| 12 | M/70 | Gκ | IA | 14 | 7 | 0/37 |
| 13 | M/66 | Aκ | IIA | 28 | 0/7 | |
| 14 | F/71 | κ | IIA | 42 | 12 | 0/43 |
| 15 | M/67 | Gλ | IIA | 56 | 8 | 0/27 |
| 16 | M/63 | Gκ | IIA | 39 | 11 | 0/13 |
| 17 | M/36 | Dκ | IIA | 50 | 13 | 0/42 |
| 18 | F/66 | Aλ | IIA | 38 | 13/28 | |
| 19 | M/77 | Gκ | IIA | 52 | 6 | 0/19 |
| 20 | F/64 | Aκ | IIA | 39 | 10 | 0/16 |
| 21 | M/84 | Gκ | IIA | 36 | 8 | 0/23 |
| 22 | M/60 | Aκ | IIIA | 45 | 12 | 0/37 |
| 23 | M/67 | Gκ | IIIB | 60 | 10 | 0/27‡ |
| 24 | M/54 | Aκ | IIIA | 45 | 0/47 | |
| 25 | M/76 | Aλ | IIIA | 49 | 5 | 0/2 |
| 26 | M/79 | Gκ | IIIB | 56 | 10 | 0/19 |
| 27 | M/57 | Gκ | IIIB | 46 | 13 | 0/17 |
| 28 | M/66 | Gκ | IIIA | 54 | 12 | 0/18‡ |
| 29 | F/71 | λ | IIIA | 63 | 9 | 0/14 |
| 30 | M/65 | Aκ | PCL | 86 | 20/23 |
Percentage of nuclei with colocalized signals (IGH constant and YAC746-H1 probes). Values above the 15% cutoff are underlined.
Number of metaphases with evidence of a t(4;14) out of the total number of metaphases observed in each sample.
Patients with structurally altered or an abnormal number of chromosomes 14 (see text).
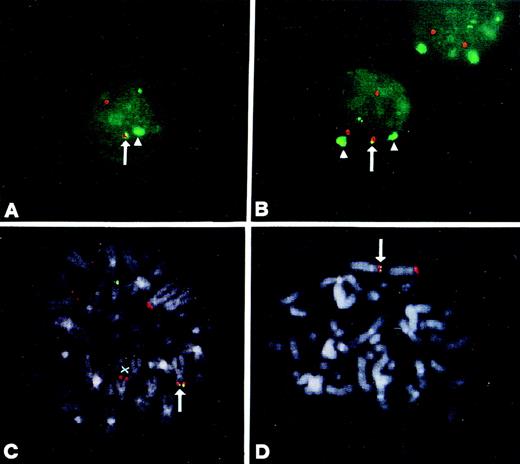
FISH analysis of MM patients using the IGHconstant regions (green) and the YAC 764-H1 (red) as probes. (A) and (B) Interphase and metaphase nuclei of patient no. 30. The colocalized signals at der(14q) are indicated. YAC 764-H1 recognized two chromosomes 4 on metaphase spreads indicated by X. In one chromosome 4, the signal is faint and consistent with a der(4) chromosome (see text). (C) and (D) Interphase nuclei from cases 9 and 24, respectively. (E) and (F) Interphase and metaphase nuclei of patient no. 18. In this case, the IGH probe is colocalized with YAC 764-H1 on the tip of one chromosome 4p (F); the other chromosome 14q32 is apparently absent, but the second chromosome 4 can be seen (indicated by X).
FISH analysis of MM patients using the IGHconstant regions (green) and the YAC 764-H1 (red) as probes. (A) and (B) Interphase and metaphase nuclei of patient no. 30. The colocalized signals at der(14q) are indicated. YAC 764-H1 recognized two chromosomes 4 on metaphase spreads indicated by X. In one chromosome 4, the signal is faint and consistent with a der(4) chromosome (see text). (C) and (D) Interphase nuclei from cases 9 and 24, respectively. (E) and (F) Interphase and metaphase nuclei of patient no. 18. In this case, the IGH probe is colocalized with YAC 764-H1 on the tip of one chromosome 4p (F); the other chromosome 14q32 is apparently absent, but the second chromosome 4 can be seen (indicated by X).
FISH metaphases were detected in all of these five patients. In cases no. 9, 13, and 24, none of the observed metaphases showed any split or colocalized signals, suggesting that they may be derived from normal hematopoietic cells. In case no. 30, almost all of the observed metaphases had a hybridization pattern consistent with a t(4;14)(p16.3;q32) chromosomal translocation (Fig 3B), thus confirming the finding in the interphase nuclei (Fig 3A). One of the chromosomes 4p showed a faint signal accounting for the der(4) (see below), as is also supported by the absence of the FGFR3 locus that was found to be translocated to the der(14) chromosome when dual color FISH was performed with the IGH and FGFR3 cDNA probes (data not shown). This case also apparently had an abnormally long arm in the putative nontranslocated chromosome 4 (Fig 3B). The hybridization pattern was different in case no. 18, in so far as the IGHprobe was colocalized with YAC 764-H1 on 4p16.3 (Fig 3E). Moreover, this case did not show the other chromosome 14 homolog, and the other chromosome 4 was apparently normal: this pattern was consistent with that observed in the interphase nuclei. Furthermore, we were able to demonstrate that the FGFR3 locus was still retained on the 4p chromosome involved in the recombination (data not shown).
FISH metaphases were found in all of the remaining 25 cases (see Table1), but no split or colocalized signals were detected. However, cases no. 3, 6, 7, and 10 showed a few metaphases with the localization of the IGH probe on structurally altered, unidentified chromosomes, and a similar pattern was observed in all of the metaphases found in case no. 23. In case no. 28, four chromosomes 14 were observed in three metaphases.
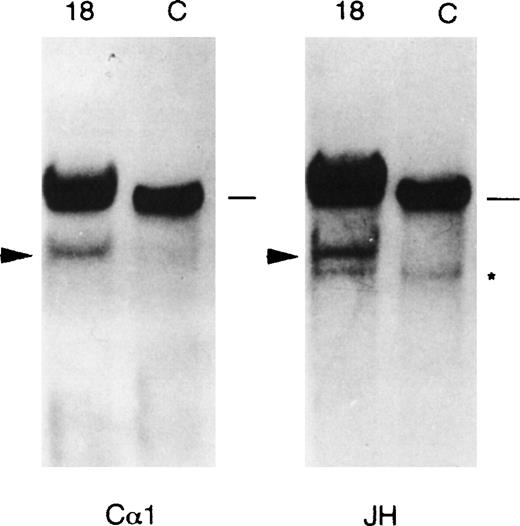
The recently reported evidence of IGH-WHSC1/MMSET hybrid transcripts originating from both the der(14) and der(4) chromosomes in MM cell lines carrying the translocation18,19 prompted us to investigate the presence of the der(4) chromosome in positive cases. FISH analyses were possible in cases no. 9, 18, 24, and 30, but not in case no.13, for which no material was available. We used the YAC 764-H1 probe (red) and the JH probe (green); in the cases in which metaphases with split or colocalized signals had not been previously observed (see Fig 3C and D), the cohybridization was performed together with the α-satellite DNA probe specific for chromosome 4 (green). As shown in Fig 4A and B, in both nos. 9 and 24, one JH signal and one YAC signal were colocalized, thus indicating the presence of a der(4) chromosome. In case no. 24 (Fig 4A), a second green signal (JH probe) was observed together with only one additional red signal (YAC 764-H1 probe): this pattern was similar to that shown in Fig 3D for the same case. Interestingly, in the interphase nuclei with colocalized signals, the centromeric cromosome 4 DNA probe detected only one signal (Fig 4A). Taken together, these findings suggest that, in this case, the two red signals represented the two reciprocal products of translocation, der(14) and der(4), whereas the nontranslocated chromosome 4 was lost. In case no. 9, a typical pattern of translocation was observed (similar to that shown in Fig 3C), together with two signals dectected by the centromeric chromosome 4 DNA probe that indicated the presence of a normal number of chromosomes 4. Finally, in both cases no. 9 and 24, Southern blot analysis detected two rearranged JH fragments in BamHI digests, thus confirming the presence of the two JH signals in the interphase nuclei (data not shown). In case no. 30, FISH metaphases showed that the JH probe was colocalized with the YAC probe in the putative der(4), but not in the der(14) (Fig 4C). In case no. 18, the JH probe was colocalized with the YAC (Fig 4D) and the IGH constant regions probes (see above; data not shown) in the telomeric region of 4p. On the basis of the absence of a second IGH locus and the expression of the IgA heavy chain in this case (see Table 1), it can be argued that the translocated 14q32 region is also apparently able to control the transcription of a functional IGH gene. To add more information to this point, we performed a Southern blot analysis of this case by subsequently hybridizing BamHI-digested DNA with IGHprobes, following the approach previously described.13 As shown in Fig 5, a single rearranged fragment was detected with the JH probe, which was comigrating with a single Cα-rearranged fragment, thus suggesting that these sequences are linked on the genome and capable of producing a functional IgA allele.
FISH analysis of MM patients. (A) and (B) Interphase nuclei of patients 24 and 9, respectively, hybridized with the JH (green), YAC 764-H1 (red), and the -satellite chromosome 4 (green) probes. The colocalized JH/YAC signals are indicated by an arrow, and the signals specific for centromeric chromosome 4 sequences by an arrow head. (C) Metaphase from case no. 30 hybridized with the JH (green) and YAC 764-H1 (red) probes. Colocalized signals at the putative der(4) chromosome are indicated by an arrow, and der(14) by X. (D) Metaphase from case no. 18 hybridized with the JH (green) and YAC 764-H1 (red) probes. The colocalized signals on the telomeric region of chromosome 4p are indicated by an arrow.
FISH analysis of MM patients. (A) and (B) Interphase nuclei of patients 24 and 9, respectively, hybridized with the JH (green), YAC 764-H1 (red), and the -satellite chromosome 4 (green) probes. The colocalized JH/YAC signals are indicated by an arrow, and the signals specific for centromeric chromosome 4 sequences by an arrow head. (C) Metaphase from case no. 30 hybridized with the JH (green) and YAC 764-H1 (red) probes. Colocalized signals at the putative der(4) chromosome are indicated by an arrow, and der(14) by X. (D) Metaphase from case no. 18 hybridized with the JH (green) and YAC 764-H1 (red) probes. The colocalized signals on the telomeric region of chromosome 4p are indicated by an arrow.
IGH rearrangement analysis of case no. 18 by Southern blotting. DNA was digested with BamHI restriction enzyme and the same filter was subsequently hybridized with the JH and C1 probe. Comigrating JH- and C-rearranged fragments are indicated by an arrowhead; germ line bands (approximately 18 kb in length) are indicated by a dash. The asterisk shows a JH cross-hybridizing band of approximately 10 kb detectable under our experimental conditions.
IGH rearrangement analysis of case no. 18 by Southern blotting. DNA was digested with BamHI restriction enzyme and the same filter was subsequently hybridized with the JH and C1 probe. Comigrating JH- and C-rearranged fragments are indicated by an arrowhead; germ line bands (approximately 18 kb in length) are indicated by a dash. The asterisk shows a JH cross-hybridizing band of approximately 10 kb detectable under our experimental conditions.
Finally, concerning the clinical staging of this relatively limited number of positive cases, we note that patient no. 9 was in stage I (indolent phase), patients no. 13 and 18 were in stage II, and patient no. 24 in stage III. Patient no. 30 was affected by PCL and died 2 months after diagnosis.
DISCUSSION
We and others have recently reported the molecular identification of a novel t(4;14)(p16.3;q32) translocation in a number of MM-derived cell lines and primary MM tumors.11,12 This translocation had not previously been shown by conventional cytogenetics, probably because of the involvement of the telomeric region of both chromosome partners. The available data indicate that 4p16.3 breakpoints are dispersed over a region of approximately 70 kb located less than 100 kb centromeric of the FGFR3 locus,13,14 and so it is difficult to detect t(4;14) using conventional approaches. Southern blot analysis with specific 4p16.3 breakpoint probes previously allowed us to detect only one rearranged case in 53 MM tumors.13The aim of this study was to assess the incidence of the novel t(4;14)(p16.3;q32) chromosomal translocation in MM by double-color FISH analysis.
The incidence of t(4;14) was about 15%, which is lower than that suggested by others (25%) on the basis of the combined cytogenetic and molecular analyses of MM-derived cell lines.14 As far as our FISH approach is concerned, the use of a pool of plasmid clones specific for the constant IGH regions as probes was based on the evidence that 14q32 translocations in MM involve the switch regions,11-16 and so these probes should detect the majority, if not all of the breakpoints on 14q32. The YAC clone 764-H1 encompasses the 4p16.3 region in which the FGFR3 gene, all of the breakpoints identified so far, and the WHSC1/MMSET gene are located. However, as discussed above, we did not consider the presence of the extra YAC 764-H1 signal, der(4), to be a requisite for the detection of the translocation in interphase nuclei. Our results were primarily based on the presence of colocalized signals using a cutoff value of 15% for quantification analysis; this was derived from the interphase FISH analysis of six normal controls in which the number of nuclei with coincidentially colocalized signals ranged from 5% to 11%. Similar relatively high rates of false-positive results have been reported in interphase FISH analyses of other types of chromosomal translocation using the colocalization of two different probes.26 It is therefore conceivable that the frequency of translocated cases found in our study may be underestimated because of the level of sensitivity of our assay. It should also be pointed out that the percentage of malignant plasma cells in bone marrow aspirates from MM patients is generally relatively low (less than 50% in 23 cases of our series), and it is generally accepted that distinct subclones harboring specific genetic lesions may exist in the neoplastic population.27 Finally, a very recent abstract has been published, which reports the detection by FISH analysis of this translocation in about 10% of primary MM tumors,28and so this novel lesion may account for about 15% to 20% of the cases of MM. The identification of the t(4;14)(p16.3;q32) chromosomal translocation and the evaluation of its relative incidence further supports the evidence of the complex heterogeneity of genetic lesions in MM.6,11,12,20,21 29
In our study, FISH metaphases with split or colocalized signals were detected in only two of the five cases with positive interphase nuclei. In one case (no. 30), the translocation pattern was similar to that found in the MM cell lines with the FGFR3 gene localized on der(14), thus implying that the 4p16.3 breakpoint is centromeric of the gene and the JH probe on der(4). In the other case (no. 18), theIGH probe hybridized to the 4p16.3 region where the FGFR3gene was still retained. We demonstrated by FISH that the JH region is also localized on 4p16.3, and by Southern blot that it is apparently linked to a Cα region. These findings are consistent with the presence of a monoclonal IgA-type component and, because of the apparent loss of one IGH allele, this suggests that a singleIGH locus may control the expressed IgH chain and the target gene of the translocation. With regard to the mechanisms of chromosomal rearrangement in this case, our results suggest that the breakpoint at 14q32 may be located centromeric of the IGH locus, as has been reported by others who have used FISH in B-cell malignancies and MM12,30 and telomeric of the FGFR3 gene on 4p16.3. Alternatively, they may suggest that tumor no. 18 represents an example of an insertion of a portion of the IGH locus at 4p16.3, either centromeric or telomeric to FGFR3. Unfortunately, no further material was available from this case to perform additional FISH experiments with telomeric 4p and 14q probes that may have clarified this point. In the U266 MM cell line carrying a t(11;14)(q13;q32) chromosomal translocation not detectable by conventional cytogenetics, we have recently found that the IGH Cα1 region is translocated on the der(11) as a result of a complex chromosome rearrangement.31 Our data therefore suggest that different patterns of t(4;14) translocation may exist.
In two more cases with a t(4;14) translocation, we also found evidence of the presence of the chromosome der(4) by using the JH probe in interphase FISH. Interestingly, one case (no. 24) with only two signals specific for the 4p16.3 had lost one copy of normal 4, and so these signals probably represented the two reciprocal products of the translocation. With regard to our FISH assay, it is important to note that, in this case as well as in case no. 18 (see above) with an apparent t(4;14) translocation, a third 4p16.3 signal (YAC 764-H1) was not detected as a result of distinct mechanisms. An interesting finding of our study is the detection of metaphases involving chromosome 14q32 in six cases with no evidence of a t(4;14) translocation, which supports the hypothesis that this locus is frequently involved in MM.11-13 Finally, the presence in almost all of the cases of an appreciable number of FISH metaphases without split, or colocalized signals, or any apparently abnormal chromosomes detected by the IGH or 4p16.3 probes, suggest that these may be metaphases derived from normal hematopoietic cells32 and further support the use of interphase FISH as a valid approach for investigating chromosomal abnormalities in MM.
One important question arising from the investigation of the t(4;14)(p16.3;q32) in MM is related to the nature of the target gene of the translocation. As previously discussed, the FGFR3 gene is located less than 100 kb telomeric of the 4p16.3 breakpoints, is overexpressed in translocated cases, but is absent or barely detectable in MM cell lines without the translocation. The FGFR3 gene is a member of the tyrosine-kinase receptor family (FGFR1-4) whose ability to bind a large repertoire of related mitogenic fibroblast growth factors (FGFs) leads to the activation of complex signaling pathways that regulate cell proliferation, differentiation, and migration in many different tissues.33,34 Point mutations in distinct domains of the FGFR3 gene, which lead to its ligand-independent activation, are associated with specific types of autosomal dominant human skeletal disorders.35,36 Interestingly, such mutations have been found in the deregulated FGFR3 gene of the KMS-11 and OPM2 cell lines.13,14,37 Furthermore, lymphoid tumors have provided several examples of oncogenes deregulated byIGH-mediated translocations occurring at a relatively long distance from the gene, such as cyclin D1, c-myc, and PAX-51; at the distance of the FGFR3 gene from the breakpoint, it is therefore conceivable that IGHregulatory elements such as IGH transcriptional enhancers and locus control regions38 may contribute to gene deregulation. Taken together, this evidence suggests that FGFR3may represent the target gene of the t(4;14)(p16.3;q32) translocation in MM.
The characterization of a novel gene (WHSC1) has very recently been reported, which has been proposed as a candidate for the Wolf-Hirschhorn malformation syndrome that is associated with a hemizygous deletion of the 4p16.3 region.18 This gene (also named MMSET19) contains several functional domains, is expressed in early development (particularly in rapidly growing tissues), and is thought to play a role in transcriptional regulation. It extends about 120 kb on the genome and its 5′ untranslated region lies approximately 50 kb centromeric of the FGFR3.Interestingly, three of the 4p16.3 breakpoints reported so far occur within this gene: two in intron 4 (cell lines NCI-H929 and JIM3) and one in intron 5 (OPM2), and other breakpoints located 10 to 30 kb upstream may involve the putative 5′ region and/or transcription regulatory regions of the gene. This gene is overexpressed in MM cell lines with the translocation, and it is worth noting thatIGH/MMSET fusion transcripts generated from both the der(14) and der(4) have also been found in such cell lines.19 In this context, our study demonstrates the presence of der(4)-containing JH sequences in fresh MM tumors carrying the t(4;14) translocation and suggests that fusion mRNA transcripts may occur in these cases. Overall, these findings suggest that the t(4;14) translocation in MM may lead to different pathogenetic pathways and indicate the need for the careful identification and characterization of the 4p16.3 breakpoints in MM. Our study and the FISH assay used in it may make a useful contribution toward this end.
ACKNOWLEDGMENT
We are grateful to Dr K. Alitalo for providing the FGFR3 cDNA pH9 clone; Dr R. Dalla-Favera for providing the IGH Cγ plasmids; and the YAC Screening Center, DIBIT-Milan, for providing the YAC clone 764-H1.
P.F. and S.F. contributed equally to this work.
Supported by grants from the Associazione Italiana Ricerca sul Cancro (AIRC) to A.N. and from the Ministero della Sanità to Ospedale Maggiore IRCCS “Ricerca Corrente 1996.”
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Antonino Neri, MD, Servizio Ematologia, Centro “G.Marcora”, Istituto Scienze Mediche, UniversitàMilano, Ospedale Maggiore di Milano, IRCCS, Via Francesco Sforza 35, 20122 Milano, Italy; e-mail: filobus@imiucca.csi.unimi.it.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal