Abstract
Microsatellite instability (MSI) and p53 mutations have been reported to occur in a significant proportion of patients with therapy-related acute myeloid leukemia (AML). MSH2 is one of the genes involved in DNA mismatch repair to maintain fidelity of genomic replication, and defects of MSH2 are directly involved in MSI in hereditary nonpolyposis colorectal tumors and other human tumors. We have examined the expression of MSH2 protein by Western blotting in 43 adult leukemia samples, including 42 AML and 1 acute lymphoblastic leukemia (ALL) using the antibody MSH2 (Ab-1) (Calbiochem, La Jolla, CA). Abnormal expression of MSH2 protein was found in 14 of 43 (32.6%) cases; a control antibody to actin was always positive. Of the 14 patients that had abnormal expression of MSH2, 2 had therapy-related acute leukemia and 9 were elderly patients (>60 years of age). Expression of MSH2 mRNA was further examined by reverse transcriptase-polymerase chain reaction (RT-PCR). Deletion of MSH2 mRNA was found in 1 of 14 cases with deficient MSH2 protein expression. This group of patients was also screened for loss of heterozygosity (LOH) at the MSH2 locus using a panel 4 microsatellite markers (D2S367, D2S288, D2S391, and D2S2294). LOH was found in 5 of 11 cases examined. There was no evidence of LOH in 14 patients with normal MSH2 expression who were examined using the same markers. Functional evidence for defective DNA mismatch repair in leukemic cells lacking MSH2 as manifest by MSI was found in 7 of 11 cases studied. Mutations of the p53 gene in these 43 samples were also investigated by direct sequencing of full-length p53 cDNA. Mutations of p53 were found in 6 of 43 cases, including 5 of the 14 (35.7%) cases that did not express MSH2 protein. In contrast, mutation of p53 was only found in 1 of 29 (3.4%) cases with normal MSH2 protein expression (χ2 = 5.720, P < .02). These results suggest that abnormalities of DNA mismatch repair due to defective MSH2 expression could play a key role in leukemogenesis, in particular in AML arising in elderly patients or secondary to previous chemotherapy.
THE MISMATCH REPAIR GENES play a role in maintaining the genetic stability of DNA. Several highly conserved mismatch repair genes from bacteria to humans have been found, including MSH2, MLH1, PMS1, and PMS2. In bacteria, these genes are responsible for correcting mismatched DNA basepairs that arise as a result of misincorporation errors during DNA replication.1Human MSH2 is a homologue of the bacterial MutS protein and has been cloned and mapped to chromosome 2p22-2p21. The genomic MSH2 locus covers approximately 73 kb and contains 16 exons, which encode a 100-kD protein including 934 amino acids.2,3 Human MSH2 was found to be capable of binding to mismatched nucleotides, initiating the mismatch repair process.4,5 Loss of this function may lead to a mutator phenotype, which has been proposed to lead to the accumulation of the mutations in oncogenes or tumor-suppressor genes.6 A consequence of the mutator phenotype is the production of multiple replication errors in simple repetitive DNA sequences, resulting in microsatellite instability (MSI). This phenotype has been observed in tumors from hereditary nonpolyposis colon cancer (HNPCC) patients and several forms of sporadic tumors.2,7-13 A direct role for MSH2 in mutation avoidance, MSI, and tumorigenesis in human cells has been established.5,7,14 15
P53 is another gene critical to the DNA repair process. One central function of the p53 protein is to bind DNA and regulate genes that control cell cycle and cell death. P53 has been called the guardian of the genome,16 because it can induce cell cycle arrest at G1 and allow cells to repair DNA damage induced by ionizing radiation, UV light, or chemotherapeutic drugs before proceeding to DNA replication.17,18 P53 is one of the most frequently mutated genes in human cancers.19,20 Point mutations and/or p53 gene deletions occur in approximately 50% of solid tumors.21-24 In acute leukemia, both p53 mutations and MSI are relatively infrequent. P53 mutations have been reported in 5% to 10% of patients with acute leukemia, but when present are associated with a low complete remission rate, early relapse, and poor survival.25-28 More recently, a high incidence (39%) of p53 mutations has been reported in therapy-related acute myeloid leukemia (t-AML) in which a high incidence of MSI has also been reported.29
Currently, little is known about the role of MSH2 in leukemogenesis; however, MSH2 knock-out mice have an increased propensity to develop lymphoma.30 Also, abnormal expression of MSH2 protein has been reported in some leukemic cell lines.31 The aim of this study was to investigate the expression of MSH2 protein in acute leukemia and its relationship to other biological features of these cells. We have found a high incidence of abnormal expression of MSH2 protein that was associated with both MSI and p53 mutations. These results suggest that abnormalities of DNA mismatch repair could play a role in leukemogenesis in some patients.
MATERIALS AND METHODS
Patients.
Blood samples were obtained at diagnosis from 43 patients, including 42 with AML and 1 patient with t-ALL. The latter case was examined because of the published evidence for mismatch repair defects in therapy-related leukemia.29 The diagnosis of AML was made using the French-American-British (FAB) criteria after conventional cytochemical stains and surface marker analysis. Blast cells from peripheral blood were separated by Ficoll-Hypaque sedimentation and samples were depleted of T cells by Dynabeads M-450 Pan-T (CD2; Dynal, Oslo, Norway).
Western blotting.
Total protein was isolated from AML cells using the lysis buffer (0.9% NaCl, 20 mmol/L Tris-HCl, pH 7.6, 0.1% triton X-100, 1 mmol/L phenylmethylsulphonyl fluoride, and 0.01% leupeptin). The lysates were collected by micro-centrifugation (4,000g for 10 minutes). The protein concentration was determined using the Bio-Rad protein assay following the manufacturer’s protocol (Bio-Rad, Richmond, CA). Thirty-microgram aliquots of each lysate were analyzed by 7.5% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis with Laemmli buffer system. The gels were run at 150 V for 45 minutes in a mini-protein II slab cell (Bio-Rad). Separated proteins were electroblotted on to pure nitrocellulose membranes and the blot was blocked overnight at 4°C in PBS-T (phosphate-buffered saline [PBS], pH 7.4, with 0.3% Tween-20) containing 8% fat-free dried milk powder. The blot was then incubated with a monoclonal antibody to the amino terminal of hMSH2 (Ab-1; Oncogene Science, Cambridge, MA) diluted 1:1,000 in PBS-T containing 8% fat-free dried milk powder for 2 hours at room temperature. The blot was subsequently washed with PBS-T and incubated with a second antibody (goat antimouse IgG linked to horseradish peroxidase conjugate, diluted 1:2,000 in PBS-T-8% milk powder) for 1 hour at room temperature. The blot was washed with PBS-T, incubated with SuperSignal CL-HRP Substrate System (Pierce Chemical Co, Rockford, IL) for 1 minute, and finally exposed to Hyperfilm-ECL. Reprobing of actin was performed by incubating the membrane in stripping buffer (100 mmol/L 2-mercaptoethanol, 2% SDS, and 62.5 mmol/L Tris-HCl) at 50°C for 30 minutes with occasional agitation, washing the membrane in a large volume of PBS-T, blocking the membrane overnight in PBS-T-8% milk powder at 4°C, and then following the steps to perform immunodetection with an anti-actin antibody (N350; Amersham Life Science, Buckinghamshire, UK) as described above.
RNA extraction and reverse transcriptase-polymerase chain reaction (RT-PCR).
Total RNA was extracted from AML cells by the Micro RNA isolation Kit (Stratagene, La Jolla, CA) according to the manufacturer’s protocol. RT-PCR was performed as follows. Two micrograms of total RNA was reverse transcribed in a total volume of 50 μL using 10 U/μL of Moloney’s murine leukemia virus (M-MLV) reverse transcriptase (GIBCO BRL, Gaithersburg, MD), 1 U/μL RNasin (RNase Block 1), 2.5 μmol/L oligo[d(T)15], 1 mmol/L of each dNTP, 10 mmol/L dithiothreitol (DTT), 1× first-strand buffer freshly diluted from 5 × stock (GIBCO BRL) at 37°C for 60 minutes. Samples were heated at 96°C for 3 minutes to terminate the action of the reverse transcriptase. Aliquots (5 μL) of the RT products were subsequently used for PCR amplification. The RT product (5 μL) was brought to a volume of 25 μL containing 1 mmol/L MgCl2, 0.12 mmol/L of each dNTP, 1× buffer (20 mmol/L Tris-HCl, pH 8.4, 50 mmol/L KCl), 0.5 U of Taq polymerase (Flowgen, Staffordshire, UK), and 1 μmol/L of both the upstream and downstream PCR primers. Amplification was performed in a Techne Thermal Cycler, PHC-3 (Techne Ltd, Cambridge, UK), after an initial denaturation at 94°C for 3 minutes. Thirty-five cycles of PCR were performed using the following temperature and time profile: denaturation at 94°C for 1 minute, primer annealing at 60°C for 1 minute, primer extension at 72°C for 1 minute, and a final extension of 72°C for 5 minutes. The primers for MSH2 and p53 are shown in Table 1. The PCR reaction products were analyzed by 1.5% agarose gel electrophoresis in 0.5× TBE at 150 V for 60 minutes after staining with 0.5 μg/mL ethidium bromide.
The Primers for PCR, RT-PCR, and Sequencing
| Primer Name . | Primer Sequences 5′ → 3′ . | Size (bp) . |
|---|---|---|
| RT-PCR of MSH2 (exon 13 plus parts of exon 12 and 14) | ||
| Sense | ATC CAG GCA TGC TTG TGT TG | 547 |
| Antisense | CTT CAC CTG ATA AAG CAT AG | |
| GAPDH | ||
| Sense | CCA CCC ATG GCA AAT TCC ATG GCA | 600 |
| Antisense | CCA CCT GGACTG GAC GGC AGA TCT | |
| RT-PCR of p53 (full-length) | ||
| Fragment 1 (exons 2, 3, and 4 plus parts of exon 1 and 5) | GAC ACG CTT CCC TGG ATT GGC GCA AAA CAT CTT GTT GAG GGC A | 454 |
| Fragment 2 (exons 5 and 6 plus parts of exons 4 and 7) | GTT TCC GTC TGG GCT TCT TGC A GGT ACA GTC AGA GCC AAC CTC | 368 |
| Fragment 3 (exons 6, 7, 8, and 9 plus parts of exons 5 and 10) | TGG CCC CTC CTC AGC ATC TTA CAA GGC CTC ATT CAG CTC TC | 482 |
| Fragment 4 (exons 9, 10, and 11 plus a part of exon 8) | CGG CGC ACA GAG GAA GAG AAT C CGC ACA CCT ATT GCA AGC AAG GG | 444 |
| PCR of MSH2 (exon 13) | ||
| Sense | AGAAAGAAGTTTAAAATCTTGC | 282 |
| Antisense | CTATCTTCAAGGGACTAGG |
| Primer Name . | Primer Sequences 5′ → 3′ . | Size (bp) . |
|---|---|---|
| RT-PCR of MSH2 (exon 13 plus parts of exon 12 and 14) | ||
| Sense | ATC CAG GCA TGC TTG TGT TG | 547 |
| Antisense | CTT CAC CTG ATA AAG CAT AG | |
| GAPDH | ||
| Sense | CCA CCC ATG GCA AAT TCC ATG GCA | 600 |
| Antisense | CCA CCT GGACTG GAC GGC AGA TCT | |
| RT-PCR of p53 (full-length) | ||
| Fragment 1 (exons 2, 3, and 4 plus parts of exon 1 and 5) | GAC ACG CTT CCC TGG ATT GGC GCA AAA CAT CTT GTT GAG GGC A | 454 |
| Fragment 2 (exons 5 and 6 plus parts of exons 4 and 7) | GTT TCC GTC TGG GCT TCT TGC A GGT ACA GTC AGA GCC AAC CTC | 368 |
| Fragment 3 (exons 6, 7, 8, and 9 plus parts of exons 5 and 10) | TGG CCC CTC CTC AGC ATC TTA CAA GGC CTC ATT CAG CTC TC | 482 |
| Fragment 4 (exons 9, 10, and 11 plus a part of exon 8) | CGG CGC ACA GAG GAA GAG AAT C CGC ACA CCT ATT GCA AGC AAG GG | 444 |
| PCR of MSH2 (exon 13) | ||
| Sense | AGAAAGAAGTTTAAAATCTTGC | 282 |
| Antisense | CTATCTTCAAGGGACTAGG |
Direct sequencing.
The RT-PCR products were purified by Chroma Spin Columns (Clontech, Palo Alto, CA) before sequencing. The PCR products were sequenced by both forward and reverse primers (Table 1) in each sample. Approximately 180 ng of purified RT-PCR products was added to the sequencing reaction. The final reaction volumes were 20 μL and included 8.0 μL of Terminator Ready Reaction Mix (Perkin Elmer, Foster City, CA), 3.2 pmol of forward or reverse primer, and dH2O. The cycle sequencing was performed for 25 cycles in a PTC-100TM Programmable Thermal Controller (MJ Research, Inc, Watertown, MA) using the following program: 96°C for 30 seconds, 50°C for 15 seconds, and 60°C for 4 minutes. The extension products were purified by an ethanol precipitation protocol. The purified products were dissolved in 6 μL of loading buffer by combining 5 μL of deionized formamide and 1 μL of 25 mmol/L EDTA (pH 8.0) containing 50 mg/mL of dextran blue. The samples were heated at 90°C for 2 minutes and then cooled on ice until ready to load. Samples (1.5 μL) were then loaded on to a 4.2% polyacrylamide gel containing 8.3 mol/L urea. Sequence analysis was performed on a 377 automated DNA sequencer (PE Applied Biosystems, Foster City, CA).
DNA extraction, loss of heterozygosity (LOH), and MSI analysis.
High molecular weight DNA was prepared from cryopreserved peripheral blood cells from the patients. T cells were depleted by Dynabeads M-450 Pan-T (CD2) and were used as controls for the LOH and MSI analyses. DNA was extracted using a DNA extraction kit (Stratagene) following the manufacturer’s instructions. Matched leukemic and T-cell control DNA were investigated using a panel of 11 microsatellite markers for MSI and 4 markers located in or near the 2p21-22 locus for LOH (Table 2). Primers were synthesized by Cruachem (Glasgow, Scotland) and by Perkin Elmer (Warrington, UK), and the forward primer of each pair was fluorescently labeled. The PCR reaction was set up in a 25 μL of volume including 1 mmol/L MgCl2, 0.12 mmol/L of each dNTP, 1× buffer (20 mmol/L Tris-HCl, pH 8.4, 50 mmol/L KCl), 100 ng of genomic DNA, 1 U of AmpliTaq Gold DNA polymerase (PE Applied Biosystems), and 1 μmol/L of both the upstream and downstream PCR primers. Amplification was performed in an MJ Research Thermal Controller after an initial denaturation at 95°C for 5 minutes. A total of 30 cycles were performed using the following temperature and time profile: denaturation at 95°C for 40 seconds, primer annealing at 55°C for 40 seconds, primer extension at 72°C for 40 seconds, and a final extension of 72°C for 1.5 minutes. The PCR products were analyzed on a 6% polyacrylamide gel (Scotlab, Coatbridge, UK) in 1× TBE buffer in the 377 automated DNA sequencer (PE Applied Biosystems). One microliter of each PCR reaction was combined with 4 μL of formamide, 0.5 μL of dextran blue, and 0.5 μL of a fluorescent size marker (GS-350; PE Applied Biosystems). The mixed samples were heated to 90°C for 2 minutes and then cooled on ice until ready to load. The gel was run for 6 hours at 40 W and 40°C. Data analyses are performed using ABI GeneScan and ABI Genotyper software (PE Applied Biosystems) for automatic sizing of fragments.
The Characteristics of Microsatellite Markers for LOH and MSI Analysis
| Name* . | Chromosomal Location . | Repeat Motif . | Primer Sequence 5′-3′ . | Size (bp) . |
|---|---|---|---|---|
| D2S367 | 2p21-22 | (CA)n | Supplied by Perkin Elmer | 300-330 |
| D2S288 | 2p21-22 | (CA)n | AGGGCCTTGCTCTGGATT | 266-298 |
| GGCCAGTGATTGTTCCCC | ||||
| D2S391 | 2p21-22 | (CA)n | Supplied by Perkin Elmer | 125-155 |
| D2S2294 | 2p21-22 | (CA)n | GGCCCATGACTGTAATGC | 211-243 |
| CTGAATGGACACACTGGTTAAG | ||||
| TNF | 6p21.3 | (GT)n | GCCTCTAGATTTCATCCAGCCACA | 95-123 |
| CCTCTCTCCCCTGCAACACACA | ||||
| D6S264 | 6q26 | (CA)n | AGCTGACTTTATGCTGTTCCT | 102-130 |
| TTTTCCATGCCCTTCTATCA | ||||
| APC | 5q | (CA)n | CCCAATTGTATAGATTTAGAAGTC | 155-192 |
| ATCAGAGTATCAGAATTTCT | ||||
| Rb1 | 13q14 | CTTT(T) 4-5 bp repeat | CTCCTCCCTACTTACTTGT | 255-310 |
| AATTAACAAGGTGTGGTGGTACAC | ||||
| P53 | 17p | (CA)n | ACTGCCACTCCTTGCCCCATTC | 105-135 |
| AGGGATACTATTCAGCCCGAGGTG | ||||
| D7S636 | 7q36 | (CA)n | GGAGTGACTGGGCAGGAA | 115-170 |
| AGCTTGTGTGGGGTTTCA | ||||
| LPL | 8p22 | (CA)n(GT)n | CAGTGGGTTATTTGTGGGATA | 105-140 |
| TAGAGCACACTATCCAGGTGA | ||||
| DCC | 18q21 | (TA)n | TCCCTCTAGAAATTGTGTG | 106-160 |
| TGACTTTATCTCATTGGAG | ||||
| D5S107 | 5q12 | (CA)9AA(CA)19(GT)7 | GATCCACTTTAACCCAAATAC | 115-155 |
| GGCATCAACTTGAACAGCAT | ||||
| NF1 | 17q | (CA)n | CAGAGCAAGACCCTGTCT | 170-190 |
| CTCCTAACATTTATTAACCTTA | ||||
| WT1 | 11p3 | (CA)n | AATGAGACTTACTGGGTGAGG | 120-155 |
| TTACACAGTAATTTCAAGCAACGG |
| Name* . | Chromosomal Location . | Repeat Motif . | Primer Sequence 5′-3′ . | Size (bp) . |
|---|---|---|---|---|
| D2S367 | 2p21-22 | (CA)n | Supplied by Perkin Elmer | 300-330 |
| D2S288 | 2p21-22 | (CA)n | AGGGCCTTGCTCTGGATT | 266-298 |
| GGCCAGTGATTGTTCCCC | ||||
| D2S391 | 2p21-22 | (CA)n | Supplied by Perkin Elmer | 125-155 |
| D2S2294 | 2p21-22 | (CA)n | GGCCCATGACTGTAATGC | 211-243 |
| CTGAATGGACACACTGGTTAAG | ||||
| TNF | 6p21.3 | (GT)n | GCCTCTAGATTTCATCCAGCCACA | 95-123 |
| CCTCTCTCCCCTGCAACACACA | ||||
| D6S264 | 6q26 | (CA)n | AGCTGACTTTATGCTGTTCCT | 102-130 |
| TTTTCCATGCCCTTCTATCA | ||||
| APC | 5q | (CA)n | CCCAATTGTATAGATTTAGAAGTC | 155-192 |
| ATCAGAGTATCAGAATTTCT | ||||
| Rb1 | 13q14 | CTTT(T) 4-5 bp repeat | CTCCTCCCTACTTACTTGT | 255-310 |
| AATTAACAAGGTGTGGTGGTACAC | ||||
| P53 | 17p | (CA)n | ACTGCCACTCCTTGCCCCATTC | 105-135 |
| AGGGATACTATTCAGCCCGAGGTG | ||||
| D7S636 | 7q36 | (CA)n | GGAGTGACTGGGCAGGAA | 115-170 |
| AGCTTGTGTGGGGTTTCA | ||||
| LPL | 8p22 | (CA)n(GT)n | CAGTGGGTTATTTGTGGGATA | 105-140 |
| TAGAGCACACTATCCAGGTGA | ||||
| DCC | 18q21 | (TA)n | TCCCTCTAGAAATTGTGTG | 106-160 |
| TGACTTTATCTCATTGGAG | ||||
| D5S107 | 5q12 | (CA)9AA(CA)19(GT)7 | GATCCACTTTAACCCAAATAC | 115-155 |
| GGCATCAACTTGAACAGCAT | ||||
| NF1 | 17q | (CA)n | CAGAGCAAGACCCTGTCT | 170-190 |
| CTCCTAACATTTATTAACCTTA | ||||
| WT1 | 11p3 | (CA)n | AATGAGACTTACTGGGTGAGG | 120-155 |
| TTACACAGTAATTTCAAGCAACGG |
D2S367, D2S288, D2S391, and D2S2944 for LOH analysis; others for MSI analysis.
Statistical methods.
Analyses of the relationship between the deletion of MSH2 protein and mutation of the p53 gene as well as the relationship between the deletion of MSH2 protein and adverse risk cytogenetics were determined by the χ2 test with Yate’s correction.
RESULTS
Expression of the MSH2 gene.
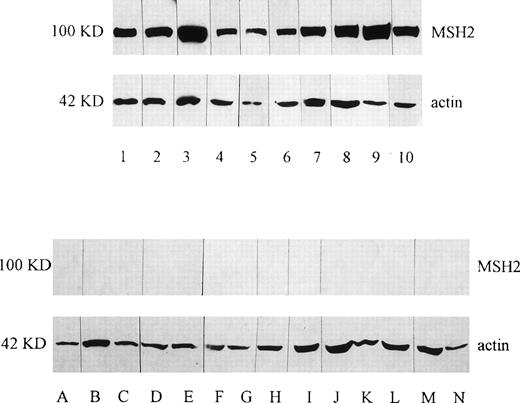
To evaluate the expression of the MSH2 gene, we performed both Western blot analysis using the anti-MSH2 antibody (Ab-1) and RT-PCR of mRNA of the whole of exon 13 and part of exons 12 and 14. Lack of detection of MSH2 protein using this Ab was seen in 14 of 43 cases (32.6%), suggesting either absent or abnormal expression of this protein. As shown in Fig 1, a control antibody to actin was always positive in these cases, thus ensuring the amount and integrity of the sample protein. The clinical characteristics of the group of patients with apparent loss of MSH2 expression are shown in Table 3. The median age was 64 years (range, 17 to 80 years), 9 (64%) were greater than 60 years and 2 patients had therapy-related acute leukemia after treatment for Hodgkin’s disease and breast cancer, respectively. The 29 patients with normal MSH2 expression had a median age of 59 years (range, 24 to 84 years) and 12 (40%) were greater than 60 years of age. Of the 14 patients with deficient MSH2 expression, 5 achieved a complete remission, of whom 4 have relapsed, and only 1 patient (17 years of age) remains in complete remission 6 years after allogeneic bone marrow transplantation.
Western blotting analysis of MSH2 protein in leukemic cells. Thirty micrograms of protein was loaded per lane and blotted with either anti-MSH2 antibody or anti-actin antibody as a control. In the upper panel, the expression of MSH2 protein was shown in the patients from left to right: 1 (JH), 2 (MD), 3 (WA), 4 (CA), 5 (DC), 6 (SR), 7 (PN), 8 (DG), 9 (MC), and 10 (PS). In the lower panel, no MSH2 protein was shown in the patients from left to right A (EH), B (EW), C (KP), D (JG), E (MJ), F (MP), G (GMT), H (MT), I (NS), J (RG), K (SG), L (AH),M(AS), and N (BG). The expression of actin as a control was shown in all of these patients.
Western blotting analysis of MSH2 protein in leukemic cells. Thirty micrograms of protein was loaded per lane and blotted with either anti-MSH2 antibody or anti-actin antibody as a control. In the upper panel, the expression of MSH2 protein was shown in the patients from left to right: 1 (JH), 2 (MD), 3 (WA), 4 (CA), 5 (DC), 6 (SR), 7 (PN), 8 (DG), 9 (MC), and 10 (PS). In the lower panel, no MSH2 protein was shown in the patients from left to right A (EH), B (EW), C (KP), D (JG), E (MJ), F (MP), G (GMT), H (MT), I (NS), J (RG), K (SG), L (AH),M(AS), and N (BG). The expression of actin as a control was shown in all of these patients.
Clinical Characteristics of Patients With Defective MSH2 Protein Expression
| Name . | Diagnosis . | Sex/Age . | Cytogenetics . | p53 Status . |
|---|---|---|---|---|
| 1. AH | M2 | M/65 | 46 XY, t(8;21)(q22;q22) | Wild-type |
| 2. AS | M4 | F/58 | 45 XX, −3, −12, −17, −18, −19, −22, 5q−, 7q−, +t(17q;?), +t(19;?), +i(11q), +2mar | Mutant p53 at codon 248 cgg → tgg (Arg → Trp) |
| 3. BG | M2 | F/69 | 46 XX | Wild-type |
| 4. EH | M6 | M/68 | 41-47 XY, −5, +6, der(7)t(7;?), −8, +8, +9, −13, −14, −14, +t(14;14)(p1;q1), −15, add(16)(q2), −17, −18, −22, −22, +r(?), +mar1, +mar2, +mar3x2(cp10) | Mutant p53 at codon 175 cgc → ggc (Arg → Gly) |
| 5. EW | t-ALL (L3) | M/52 | 46 XY, −4, t(8;14)(q24;q32), −13, −14, +mar1, +mar2, +mar3 | Mutant p53 at codon 249 agg → tgg (Arg → Trp) |
| 6. KP | M2 | M/35 | 42 XY, −5, −7, −8, −13, −13, −14, −15, −17, −18, −19, −21, +der5, +der7, 11p+, 14p+, 18q+, 19q+, i(21q), +mar1, +mar2, +mar3, +r | Wild-type |
| 7. JG | M4 | F/64 | 45 XX, −5, −16, −21, −y(?), +14q, −22q | Mutant p53 at codon 273 cgt → cat (Arg → His) |
| 8. MJ | t-AML (M4) | F/70 | 46, XX | Wild-type |
| 9. MP | CML → AML | M/43 | 46, XY, t(9;22)(q34;q11) | Wild-type |
| 10. MRT | M1 | M/60 | Not available | Wild-type |
| 11. MT | M4 | M/17 | 47 XY, +8 | Wild-type |
| 12. NS | M5 | F/80 | 45-49 X, −X, dup(1)(q10q3), del(1)(q?2), del(2)(q?3), +del(2)(q?3), add(6)(p2), add(12)(q24), −13, −16, +?del(19)(?q1), −20, +3mar(op8) | Mutant p53 at codon 175 cgc → cac (Arg → His) |
| 13. RG | M4 | M/71 | 46 XY | Wild-type |
| 14. SG | M5 | M/77 | 46 XY, inv(6)(p11q23)/8p+ | Wild-type |
| Name . | Diagnosis . | Sex/Age . | Cytogenetics . | p53 Status . |
|---|---|---|---|---|
| 1. AH | M2 | M/65 | 46 XY, t(8;21)(q22;q22) | Wild-type |
| 2. AS | M4 | F/58 | 45 XX, −3, −12, −17, −18, −19, −22, 5q−, 7q−, +t(17q;?), +t(19;?), +i(11q), +2mar | Mutant p53 at codon 248 cgg → tgg (Arg → Trp) |
| 3. BG | M2 | F/69 | 46 XX | Wild-type |
| 4. EH | M6 | M/68 | 41-47 XY, −5, +6, der(7)t(7;?), −8, +8, +9, −13, −14, −14, +t(14;14)(p1;q1), −15, add(16)(q2), −17, −18, −22, −22, +r(?), +mar1, +mar2, +mar3x2(cp10) | Mutant p53 at codon 175 cgc → ggc (Arg → Gly) |
| 5. EW | t-ALL (L3) | M/52 | 46 XY, −4, t(8;14)(q24;q32), −13, −14, +mar1, +mar2, +mar3 | Mutant p53 at codon 249 agg → tgg (Arg → Trp) |
| 6. KP | M2 | M/35 | 42 XY, −5, −7, −8, −13, −13, −14, −15, −17, −18, −19, −21, +der5, +der7, 11p+, 14p+, 18q+, 19q+, i(21q), +mar1, +mar2, +mar3, +r | Wild-type |
| 7. JG | M4 | F/64 | 45 XX, −5, −16, −21, −y(?), +14q, −22q | Mutant p53 at codon 273 cgt → cat (Arg → His) |
| 8. MJ | t-AML (M4) | F/70 | 46, XX | Wild-type |
| 9. MP | CML → AML | M/43 | 46, XY, t(9;22)(q34;q11) | Wild-type |
| 10. MRT | M1 | M/60 | Not available | Wild-type |
| 11. MT | M4 | M/17 | 47 XY, +8 | Wild-type |
| 12. NS | M5 | F/80 | 45-49 X, −X, dup(1)(q10q3), del(1)(q?2), del(2)(q?3), +del(2)(q?3), add(6)(p2), add(12)(q24), −13, −16, +?del(19)(?q1), −20, +3mar(op8) | Mutant p53 at codon 175 cgc → cac (Arg → His) |
| 13. RG | M4 | M/71 | 46 XY | Wild-type |
| 14. SG | M5 | M/77 | 46 XY, inv(6)(p11q23)/8p+ | Wild-type |
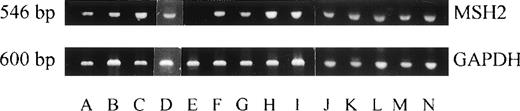
Expression of MSH2 mRNA was studied by RT-PCR of exons 12 to 14 in all 14 cases with apparent loss of MSH2 protein expression. Only 1 patient (MJ) who had t-AML had no detectable MSH2 mRNA (Fig 2).
Expression of MSH2 mRNA was investigated by RT-PCR in cases with abnormal MSH2 protein expression. A specific 546-bp band for MSH2 was detectable in A (EH), B (EW), C (KP), D (JG), F (MP), G (GMT), H (MT), I (NS), J (RG), K (SG), L (AH),M(AS), and N (BG). No MSH2 expression was found in E (MJ) (upper panel). A specific 600-bp band for GAPDH as a control was expressed in all of these samples.
Expression of MSH2 mRNA was investigated by RT-PCR in cases with abnormal MSH2 protein expression. A specific 546-bp band for MSH2 was detectable in A (EH), B (EW), C (KP), D (JG), F (MP), G (GMT), H (MT), I (NS), J (RG), K (SG), L (AH),M(AS), and N (BG). No MSH2 expression was found in E (MJ) (upper panel). A specific 600-bp band for GAPDH as a control was expressed in all of these samples.
Analysis of the MSH2 gene and screening for LOH at the MSH2 locus.
To investigate the possible reason for abnormal expression of MSH2 protein in the 13 patients in whom MSH2 mRNA was detectable, we looked for evidence of LOH at the MSH2 locus. LOH coupled with a germline mutation in the remaining allele is a common mechanism causing inactivation of tumor-suppressor genes and resulting in tumorigenesis. We used 4 polymorphic markers to screen leukemic DNA and T-lymphocyte DNA from patients with normal and abnormal MSH2 protein expression. Control T cells were used to confirm that the patient was informative for the polymorphic marker, but these were unfortunately only available in 11 of the 14 cases with abnormal MSH2 expression and in 14 of 29 cases with normal MSH2 expression. DNA from leukemic cells and T cells was amplified with fluorescently labeled microsatellite markers D2S367, D2S288, D2S391, and D2S2294, and the PCR products were electrophoresed and analyzed on the ABI 377 Sequencer. LOH at the MSH2 locus was detected in 5 of 11 samples with abnormal MSH2 protein expression (Table 4) and 0 of 14 samples with normal MSH2 expression. Examples of LOH are shown in Fig 3. One patient (EW with therapy-related leukemia) had LOH with all 4 markers, 1 patient had LOH with 2 markers, and 3 patients had LOH with 1 marker only. Mutations in the coding region of exon 13 have been described in lymphoblastic lymphomas.30 We therefore sequenced the RT-PCR products of exons 12 to 14 in the 13 cases, and none was found to have a mutation. Also, a mutation has been described in −6 intronic splice acceptor site of exon 13. We also sequenced the PCR product for exon 13 in all cases, and no mutations were found.
LOH Results of Patients With MSH2 Protein Abnormalities
| Patients . | D2S367 . | D2S288 . | D2S391 . | D2S2294 . |
|---|---|---|---|---|
| 1. AH | 2 | 2 | LOH | LOH |
| 3. BG | 2 | 1 | 2 | 2 |
| 4. EH | 2 | 2 | 1 | 1 |
| 5. EW | LOH | LOH | LOH | LOH |
| 6. KP | 2 | 1 | 2 | 1 |
| 7. JG | LOH | 1 | 2 | 1 |
| 9. MP | 2 | 1 | 1 | 2 |
| 10. MRT | 2 | LOH | 1 | 0 |
| 11. MT | 2 | 1 | 1 | 1 |
| 13. RG | 2 | LOH | 1 | 2 |
| 14. SG | 2 | 1 | 2 | 2 |
| Patients . | D2S367 . | D2S288 . | D2S391 . | D2S2294 . |
|---|---|---|---|---|
| 1. AH | 2 | 2 | LOH | LOH |
| 3. BG | 2 | 1 | 2 | 2 |
| 4. EH | 2 | 2 | 1 | 1 |
| 5. EW | LOH | LOH | LOH | LOH |
| 6. KP | 2 | 1 | 2 | 1 |
| 7. JG | LOH | 1 | 2 | 1 |
| 9. MP | 2 | 1 | 1 | 2 |
| 10. MRT | 2 | LOH | 1 | 0 |
| 11. MT | 2 | 1 | 1 | 1 |
| 13. RG | 2 | LOH | 1 | 2 |
| 14. SG | 2 | 1 | 2 | 2 |
Abbreviations: 1, homozygous or uninformative; 2, constitutionally heterozygous with retention of both alleles in leukemic DNA; LOH, constitutionally heterozygous with loss of one allele in leukemic DNA; 0, constitutionally heterozygous with loss of both alleles in leukemic DNA.
Detection of LOH at 4 markers located in 2p22-21. The upper one was normal T cells (N). The lower one was leukemic cells (L). LOH was shown in all of 4 markers.
Detection of LOH at 4 markers located in 2p22-21. The upper one was normal T cells (N). The lower one was leukemic cells (L). LOH was shown in all of 4 markers.
The relationship between MSH2 expression, FAB type, and karyotypic changes.
Data on the FAB type and karyotype were collected for the patients studied. The data relating to the 14 patients with defective MSH2 expression are given in Table 3. All FAB types were represented among these patients except for FAB M3, and no relationship was demonstrated between FAB type and MSH2 expression. Karyotypic data were classified according to the criteria defined in the MRC AML 12 trial.32 The difference in frequency of adverse risk cytogenetics between MSH2-positive and MSH2-negative patients was not statistically significant (χ2 = .68, P = .4), and no correlation was observed between any cytogenetic risk group and MSH2 status.
The relationship between MSH2 expression and p53 mutations.
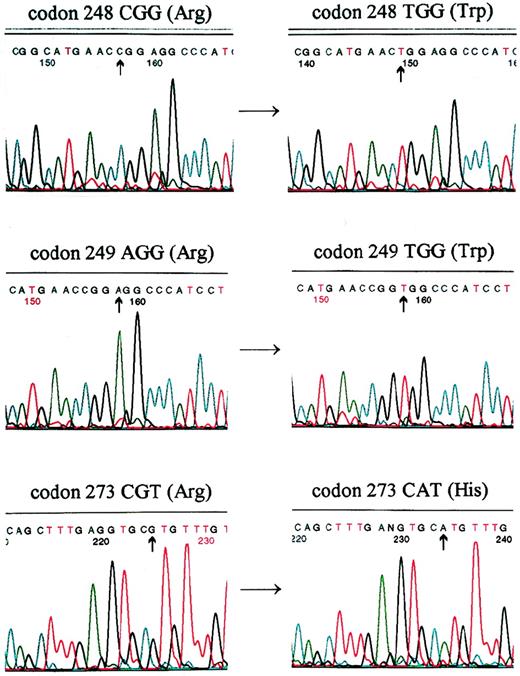
Mutations of the p53 gene in these 43 cases were investigated by direct sequencing of full-length p53 cDNA. Mutations of p53 were found in 5 of 14 (35.7%) cases with abnormal MSH2 expression. All were missense mutations found in exon 5 (codon 175 in 2 cases), exon 7 (codon 248 and 249), and exon 8 (codon 273; see Table 3 and Fig 4). However, mutation of the p53 gene was only found in 1 (missense in codon 273 cgt→tgt, Arg→Cys) of the 29 (3.4%) cases that expressed MSH2 protein. The difference in the frequency of p53 mutations between the MSH2-positive and -negative cells was statistically significant (χ2 = 5.720, P < .02).
Mutations of p53 were detected by direct sequencing in patients: AS (top), EW (middle), and JG (bottom). A cgg→tgg transition occurred in codon 248, resulting in Arg→Trp substitution in patient AS. A agg→tgg transversion occurred in codon 249, resulting in a Arg→Trp substitution in patient EW. A cgt→cat transition occurred in codon 273, resulting in a Arg→His substitution in patient JG.
Mutations of p53 were detected by direct sequencing in patients: AS (top), EW (middle), and JG (bottom). A cgg→tgg transition occurred in codon 248, resulting in Arg→Trp substitution in patient AS. A agg→tgg transversion occurred in codon 249, resulting in a Arg→Trp substitution in patient EW. A cgt→cat transition occurred in codon 273, resulting in a Arg→His substitution in patient JG.
Leukemic cells with abnormal MSH2 expression frequently exhibit MSI.
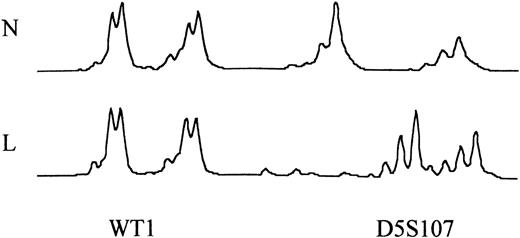
To confirm functional loss of MSH2, the cases with abnormal MSH2 protein expression were analyzed for MSI at 11 different polymorphic loci covering several known tumor-suppressor genes (Table 2). However, DNA from control T lymphocytes was only available from 12 of 14 cases, and in 1 further case control (T-cell) DNA failed to amplify leaving 11 cases for analysis. As shown in Fig 5 and Table 5, which delineates the results for each patient, 7 of 11 had MSI, of whom 6 had MSI at multiple loci and 1 had MSI at one of 5 loci at which DNA amplified. Of the 3 patients tested with a known p53 mutation, all had MSI at multiple loci, including the p53 locus. Likewise, both the cases (EW and MJ) with therapy-related acute leukemia (only 1 of whom had a p53 mutation) had MSI.
Microsatellite analysis comparing normal (N) T-cell and leukemic (L) DNA at the WT1 locus in patient MRT and the D5S107 locus in patient EW. In patient MRT, there was no evidence of MSI. In patient EW, MSI was seen as a shift in the size of the PCR product.
Microsatellite analysis comparing normal (N) T-cell and leukemic (L) DNA at the WT1 locus in patient MRT and the D5S107 locus in patient EW. In patient MRT, there was no evidence of MSI. In patient EW, MSI was seen as a shift in the size of the PCR product.
MSI Results of Patients With MSH2 Protein Deletion
| . | TNF . | D6S264 . | p53 . | APC . | Rb1 . | LPL . | D7S636 . | DCC . | D5S107 . | NF1 . | WT1 . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. AH | + | + | + | − | − | − | − | + | + | + | + |
| 3. BG | NA | + | NA | + | + | NA | NA | NA | NA | NA | NA |
| 4. EH | − | − | + | − | − | + | + | − | − | − | − |
| 5. EW | + | + | + | + | + | + | − | − | + | + | − |
| 6. KP | − | − | − | − | − | − | − | − | − | − | − |
| 7. JG | − | + | + | + | + | + | + | + | + | + | + |
| 8. MJ | − | + | NA | NA | NA | − | NA | NA | − | NA | − |
| 9. MP | − | − | − | − | − | − | − | − | − | − | − |
| 10. MRT | − | − | − | − | − | − | − | − | − | − | − |
| 13. RG | − | + | + | + | − | − | − | + | + | + | + |
| 14. SG | − | − | − | − | − | − | − | − | − | − | − |
| . | TNF . | D6S264 . | p53 . | APC . | Rb1 . | LPL . | D7S636 . | DCC . | D5S107 . | NF1 . | WT1 . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. AH | + | + | + | − | − | − | − | + | + | + | + |
| 3. BG | NA | + | NA | + | + | NA | NA | NA | NA | NA | NA |
| 4. EH | − | − | + | − | − | + | + | − | − | − | − |
| 5. EW | + | + | + | + | + | + | − | − | + | + | − |
| 6. KP | − | − | − | − | − | − | − | − | − | − | − |
| 7. JG | − | + | + | + | + | + | + | + | + | + | + |
| 8. MJ | − | + | NA | NA | NA | − | NA | NA | − | NA | − |
| 9. MP | − | − | − | − | − | − | − | − | − | − | − |
| 10. MRT | − | − | − | − | − | − | − | − | − | − | − |
| 13. RG | − | + | + | + | − | − | − | + | + | + | + |
| 14. SG | − | − | − | − | − | − | − | − | − | − | − |
Abbreviations: +, microsatellite instability; −, no microsatellite instability; NA, DNA would not amplify.
DISCUSSION
MSH2 mRNA and protein levels do not change appreciably throughout the cell cycle in normal human lung fibroblasts.33 Marra et al34 showed that a basic level of MSH2 protein expression is necessary for resting and differentiated cells, although increased MSH2 protein expression is required when DNA replication is activated before mitosis. We examined the expression of the MSH2 gene in 43 leukemic samples by Western blotting. Using the anti-MSH2 antibody (Ab-1), we could not detect MSH2 protein in 14 of 43 samples (32.6%), suggesting absent expression or expression of an aberrant MSH2 protein. We also examined for expression of MSH2 mRNA by RT-PCR of exons 12 to 14 in the 14 cases in which we could not detect MSH2 protein. One patient (MJ) was found to have mRNA deletion. This patient had t-AML after previous chemotherapy for breast and ovarian cancer.
One possible mechanism causing absent or aberrant MSH2 expression is LOH at the MSH2 locus associated with an inactivating mutation in the remaining allele. Detection of LOH has been a technique widely used to look for somatic deletion of tumor-suppressor genes. The MSH2 gene is a large gene comprising 16 exons and 73 kb; therefore, we decided to use 4 polymorphic microsatellite markers linked to 2p22-21 to analyze for LOH of the MSH2 gene. In our study, LOH at the MSH2 locus was identified in 5 of 11 patients in whom both leukemic and T-cell DNA could be studied. No control DNA was available from the remaining 3 cases, but the leukemic DNA was studied using the 4 markers. One of these cases (MJ) who was also known to have absent MSH2 mRNA expression showed only a single allele (or apparent LOH) at each of the 4 markers. These findings strongly suggest the presence of LOH of MSH2 in this case; also, it would be unusual to be uninformative at all 4 loci. A pathogenetic role for LOH in loss of MSH2 expression was further supported by the absence of LOH in the 14 cases with normal MSH2 expression that were studied.
To look for MSH2 mutations, the RT-PCR products of exons 12 to 14 of MSH2 were sequenced in the 13 cases in which mRNA was expressed and were all found to be normal. Genomic DNA for MSH2 exon 13 was also sequenced in all 14 cases, because T to C mutations in the −6 position of a splice acceptor site have been detected in human lymphoblastic lymphoma,30 and no mutations were detected. Recently, Bubb et al35 reported that the majority of MSH2 mutations detected in sporadic colorectal tumors were reductions in length of intronic mononucleotide tracts, and only 1 of 24 (4%) mutations was exonic. These intronic mutations were postulated to compromise transcriptional efficiency of MSH2 possibly by a mechanism involving exon skipping and may have arisen as the result of a primary defect in another mismatch repair gene.35 It has been recognized that multiple mutations in tumor-suppressor genes may accumulate as a consequence of loss of expression of DNA mismatch repair genes.36 We have therefore looked for p53 gene mutations in these 43 cases by direct sequencing of full-length p53 cDNA. Mutations of p53 were found in 6 of the 43 cases. Most importantly, 5 of 14 (35.7%) cases with abnormal MSH2 protein expression had p53 mutations, compared with only 1 of 29 (3.4%) cases expressing normal MSH2 protein. These results indicate a significant relationship between p53 mutation and abnormal MSH2 expression (χ2 = 5.720, P < .02) in acute leukemia. Previously, p53 mutations have been reported to occur frequently in t-AML compared with de novo AML.29 Of the 2 patients in our study with therapy-related leukemia, 1 (EW) had a p53 mutation as well as abnormal expression of MSH2 and allelic loss at 2p21, whereas the second patient with t-AML (MJ) did not have a mutant p53.
Defects in the MSH2 gene play a role in the development in the HNPCC.37 These tumors are characterized by the presence of multiple replication errors classically manifest by MSI. These patients have a genetic predisposition to acquire mutations, known as a mutator phenotype. We found a high incidence of MSI affecting 7 of 11 cases with abnormal expression of MSH2. The 3 patients with known p53 mutations all had widespread MSI, including MSI at the p53 locus, a similar finding to that reported by Ben-Yehuda et al29 in t-AML. Likewise, 3 other cases exhibited widespread MSI affecting more than 30% of loci studied, a feature that is reported to be typical of defects in DNA mismatch repair genes.38 Only 1 case had MSI at a single locus, and in this case DNA failed to amplify at 6 of the 11 loci studied. Of the 7 cases with MSI, 2 had therapy-related leukemia, whereas the remaining 5 were greater than 60 years of age. Previous studies in acute leukemia have generally reported a low incidence of MSI,39,40 although it is of interest that both the cases of MSI reported by Tasaka et al40 were in elderly patients. Recently, MSI has been reported in two leukemic cell lines, both of which had defective expression of MSH2 and one of which had a point mutation in exon 13 of the MSH2 gene.31 These cell lines also exhibited bax frameshift mutations and resistance to apoptosis induced by low serum or ionomycin.31 In the present study, 4 of the 11 cases with abnormal MSH2 expression did not exhibit MSI. The reasons for this are not clear but may relate to methodological problems in separating pure populations of blasts and control T cells from frozen samples.
Others have drawn attention to the similarities between t-AML and AML in the elderly, including the high incidence of poor-risk cytogenetics, particularly of chromosome 5 and/or 7, deletions of 5q, and complex chromosomal abnormalities.41 Likewise, both groups of patients have an adverse response to conventional chemotherapy and a higher incidence of expression of the MDR1 gene.29,41 Patients with t-AML have a high incidence of both MSI and p53 mutations, and it has been suggested that this may be the consequence of an inherited defect in a DNA mismatch repair gene leading to accelerated DNA instability in other oncogenes or tumor-suppressor genes occurring as a consequence of treatment of the primary malignancy.29 Our finding of MSI in some elderly AML patients would suggest that a similar mechanism may be operative in these cases in which an environmental mutagen rather than cytotoxic drug therapy may lead to cumulative DNA damage due to defective mismatch repair, and we are currently screening a wider series of elderly AML patients for MSI. Furthermore, our results imply that abnormalities of the MSH2 gene may be one cause of defective mismatch repair occurring in a proportion of patients with elderly AML as well as therapy-related leukemia.
Supported by a grant from Leukaemia Research Fund.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Prof N.H. Russell, MD, Department of Haematology, Nottingham City Hospital, Hucknall Road, Nottingham NG5 1PB, UK.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal