Abstract
Cyclin A1 differs from other cyclins in its highly restricted expression pattern. Besides its expression during spermatogenesis, cyclin A1 is also expressed in hematopoietic progenitor cells and in acute myeloid leukemia. We investigated mechanisms that might contribute to cyclin A1 expression in hematopoietic cells. Comparison of cyclin A1 and cyclin A promoter activity in adherent and myeloid leukemia cell lines showed that the cyclin A1 promoter is preferentially active in myeloid cell lines. This preferential activity was present in a small, 335-bp cyclin A1 promoter fragment that contained several potential c-myb binding sites. Coexpression of a c-myb expression vector with the cyclin A1 promoter constructs significantly increased the reporter activity in adherent CV-1 as well as in myeloid U937 cells. Gel-shift assays demonstrated that c-myb could bind to the cyclin A1 promoter at a binding site located near the transcription start site. Site-directed mutagenesis of this site decreased promoter transactivation by 50% in both KCL22 cells that express high levels of c-myb and in CV-1 cells that were transfected with c-myb. In addition, transfection of primary human embryonic fibroblasts with a c-myb expression vector led to induction of the endogenous cyclin A1 gene. Taken together, c-myb can directly transactivate the promoter of cyclin A1, and c-myb might be involved in the high-level expression of cyclin A1 observed in acute myeloid leukemia. These findings suggest that c-myb induces hematopoiesis-specific mechanisms of cell cycle regulation.
EXPRESSION OF THE human cyclin A1 gene is restricted to very few tissues.1 Under physiological conditions, the highest concentrations of cyclin A1 are found in testis and lower levels are found in hematopoietic tissue.1 Very high levels can also be detected in several leukemic cell lines and in leukemic blast cells from the majority of patients with acute myeloid leukemia.2 Cyclin A1 is thought to play an important role in the first meiotic cell division in testis and impaired spermatogenesis occurs in mice with cyclin A1 deletion.3The relevance of cyclin A1 for the cell cycle in somatic cells is unclear.4 However, we have recently demonstrated that cyclin A1 associates with the retinoblastoma gene product (Rb) and E2F-1 in leukemia cells in vivo and the interaction can change functional properties of the involved proteins.5 The reason for the high-level expression in leukemic blasts is as yet unknown. In addition, the mechanisms that define the limited expression pattern in vivo have not been elucidated.
The myb family of transcription factors regulates tissue-specific gene expression in the hematopoietic system, as well as in the testis. The v-myb oncogene causes myeloblastosis in chickens.6 The human counterpart, c-myb, was cloned and has been demonstrated to play an essential role in hematopoiesis.7-9 Acute myeloid leukemia is frequently associated with high levels of c-myb expression, and it has been suggested that c-myb might play a role in the pathogenesis of leukemia.10 However, the target genes for c-myb in normal hematopoiesis and in leukemic cells are not yet clearly defined. In addition, why v-myb and deletional mutants of c-myb are oncogenic in hematopoietic cells but not in other tissues is unknown. During hematopoiesis, c-myb is expressed during the stages of differentiation associated with high cellular proliferation, and it may act at least partially by driving expression of genes involved in regulation of the cell cycle. Binding sites for c-myb have been demonstrated in several genes involved in hematopoiesis and cellular proliferation (eg, Brandt et al11 and Ku et al12), although it is unclear which ones are actual target genes for c-myb in vivo.13 The cell cycle-dependent phosphorylation of c-myb further links the protein to the cell cycle machinery.14 Nevertheless, target genes of c-myb that are myelo-specific and are linked to the cell cycle and proliferation remain unknown. Recently, we cloned the promoter of the cyclin A1 gene to elucidate the mechanisms of expression of this gene.15We show here that the cyclin A1 promoter is preferentially active in myeloid leukemia cell lines and is transactivated by c-myb. In addition, forced expression of c-myb in human embryonic fibroblasts induces the endogenous cyclin A1 gene. The high-level expression of cyclin A1 in acute myeloid leukemia might be caused by c-myb, and these findings suggest a specific involvement of c-myb in the cell cycle of hematopoietic cells.
MATERIALS AND METHODS
Constructs and plasmids.
The cloning of the cyclin A1 promoter and the generation of the cyclin A1 promoter-luciferase constructs have been described previously.15 For generating the cyclin A promoter–luciferase construct, a 452-bp fragment of the cyclin A promoter was cloned into the Sac I and Xho I sites of PGL3-Basic after Pfu-PCR amplification of KG-1 genomic DNA and digestion of the product with Sac I and Xho I at the internal sites of the cyclin A promoter. The PGL3-promoter plasmid containing the enhancerless early SV40 promoter upstream of the firefly luciferase gene, the CMV-β-galactosidase, and the pRL-SV40 plasmid were purchased from Promega (Madison, WI). The 2 c-myb expression vectors that were used have been described in detail elsewhere.16 17
Cell culture and transfection.
HeLa, CV-1, and PC3 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) containing 100 U/mL penicillin and 100 μg/mL streptomycin. U937 and KCL22 cells were grown in RPMI containing 10% FCS as well as penicillin and streptomycin. Primary human embryonic fibroblasts were obtained by amniocentesis and chorion villi biopsies for diagnostic reasons and used for experimentation after conclusion of the diagnostic procedures. Human embryonic fibroblasts were grown in RPMI medium supplemented with 20% FCS, penicillin, and streptomycin. For HeLa transfection, 5 × 105 cells were seeded into 60-mm plates 16 hours before transfection. Transfection was performed using lipofectAMINE (GIBCO, Life Technology, Grand Island, NY) according to the manufacturer's protocol. Two micrograms of luciferase reporter plasmid was transfected together with 300 ng of a CMV-β-gal expression vector that was used for standardization. Cells were harvested and assayed for luciferase and β-galactosidase activity after 48 hours. PC3 and CV-1 cells were transfected using Superfect following the protocol of the manufacturer. For PC3 cells, a total of 3.5 μg of DNA was transfected, with 2.5 μg being reporter plasmid and 1 μg being CMV-β-gal. In the c-myb coexpression experiments in CV-1 cells, 0.5 μg of reporter and 1 μg of CMV-β-gal expression vector were used along with the indicated amount of c-myb expression plasmid supplemented with empty expression vector to yield 6.5 μg of total DNA. For analyses of c-myb transactivation effects on the mutant cyclin A1 promoter construct in CV-1 cells, we used the Dual Luciferase Assay system (Promega). A Renilla luciferase plasmid driven by a SV40 promoter (200 ng) was cotransfected with the indicated amounts of luciferase reporter and expression constructs. The myeloid cell lines were transfected by electroporation using a square wave electroporator (BTX-820). A total of 15 μg of DNA (10 μg luciferase reporter and 5 μg of CMV-β-gal expression vector) was added to 8 × 106 cells in a total of 400 μL of DMEM containing 10% FCS in a 4-mm cuvette. One pulse of 340 V was applied for 30 milliseconds on low-voltage setting. After electroporation, the cuvettes were placed on ice for 10 minutes followed by the addition of 3 mL of DMEM containing 10% FCS. Cells were harvested 16 hours after electroporation. In the c-myb cotransfection experiments, a total of 5 μg of plasmid consisting of either the c-myb expression vector, an empty expression vector, or a combination of both was added to the DNA mixture. DNA for transfection experiments was quantified by densitometry after linearization, gel electrophoresis, and ethidium bromide staining. All experiments were performed in duplicate and were independently performed at least 3 times. Data of luciferase assays are shown as the mean ± SEM of 3 independent experiments.
Human embryonic fibroblasts (1 × 106) were seeded into 100-mm cell culture dishes 1 day before transfection. Cells were transfected with 2 μg of either the c-myb expression vector or control vector supplemented with 200 ng of an enhanced green fluorescent protein (EGFP)-expressing plasmid. The transfection was performed using Effectene (Qiagen, Valencia, CA) according to the recommendations of the manufacturer. After 48 hours, cells were harvested by trypsination and transfected, EGFP-expressing cells were enriched by fluorescent-activated cells sorting (FACS). As a control, nontransfected (EGFP-negative) cells were sorted as well.
Reverse transcriptase-polymerase chain reaction (RT-PCR) and Southern blotting.
RNA was prepared from the sorted human embryonic fibroblast populations (∼1 × 105 cells each) using Trizol (GIBCO, Life Technology). The RNA samples were reverse transcribed using Superscript II (GIBCO, Life Technology) and random hexamers following the recommendations of the manufacturer. The cyclin A1 PCR was performed for 28 cycles using primers 5′-CTCCTGTCTGGTGGGAGGA and 5′-CTGATCCAGAATAACACCTGA (382-bp fragment) and β-actin (218-bp fragment) was amplified for 20 cycles using the primer pair: 5′-TACATGGCT GGGGTGTTGAA and 5′-AAGAGAGGCATCCTCACCCT. PCR products were run on a 1.5% agarose gel and blotted on a positively charged nylon membrane. After cross-linking, the blots were hybridized with internal oligonucleotides for cyclin A1 (5′-AGAGTGGAGTTGTGCTGGCT) and β-actin (5′-ATCGAGCACGGCATCGTCAC). Both were labeled with digoxigenin that was nonradioactively detected using digoxigenin antibodies coupled to alkaline phosphatase (Boehringer Mannheim, Mannheim, Germany). A subsequent chemiluminescence reaction (CDP-Star; Tropix, Bedford, MA) was visualized on a film.
Northern blotting.
Expression of cyclin A1 in human tumor cell lines was analyzed by Northern blotting as previously described using 10 μg of total RNA and hybridization with a 32P-random labeled cyclin A1 cDNA probe.1
Electrophoretic mobility shift assays.
Nuclear extracts from Cos-7 cells transfected with either c-myb or empty expression vector were prepared as described.18 For gel retardation experiments, 2 ng of 32P-labeled double-stranded oligonucleotide containing a c-myb consensus binding site (5′-GGGATGGCAGTTGGTGACTC) or the presumed myb binding sites myb-1 (5′-CACTTGCCAGTTGTTCCGGAC), myb-2 (5′-GGCCACCTCTTAACCGCGATCCTCC), or myb-3 (5′-CGGCCCTGCCCAACCCTG CCCCGC) were incubated for 20 minutes on ice with 5 μg of Cos-7 nuclear extract. The final reaction contained the following: 10 mmol/L Tris-HCl, pH 7.5, 5% glycerol, 1 mmol/L MgCl2, 0.5 mmol/L EDTA, 0.5 mmol/L dithiothreitol (DTT), 100 mmol/L NaCl, and 0.4 μg poly (dI-dC)•poly(dI-dC). For competition experiments, 100 ng of double-stranded oligonucleotide containing either the oligonucleotide used for gel retardation (see above) or a nonspecific oligonucleotide was preincubated for 15 minutes at room temperature with the nuclear extracts before the addition of the labeled oligonucleotide. For antibody experiments, 2 μg of monoclonal antibody against c-myb (Upstate Biotechnology, Lake Placid, NY) or an isotype control antibody was added for 30 minutes on ice before reactions were loaded on a 0.5× TBE/6% nondenaturing polyacrylamide gel and run for 2 hours at 10 V/cm. Gels were dried and autoradiographed.
Site-directed mutagenesis.
Site-directed mutagenesis was performed according to the method from Deng and Nickoloff19 with modifications as described.15 All oligonucleotides used in these experiments were 5′ phosphorylated. The following oligonucleotides were used (mutated bases underlined): myb site 1, GCACTTGCCAGAACTTCCGGACACA; myb site 2, GCCACCTCTTCTACGCGATCCTCC; and ets site 1, AACCGCGATCCGAAAGTGCACTT GC.
RESULTS
Cyclin A1 expression and promoter activity in cell lines.
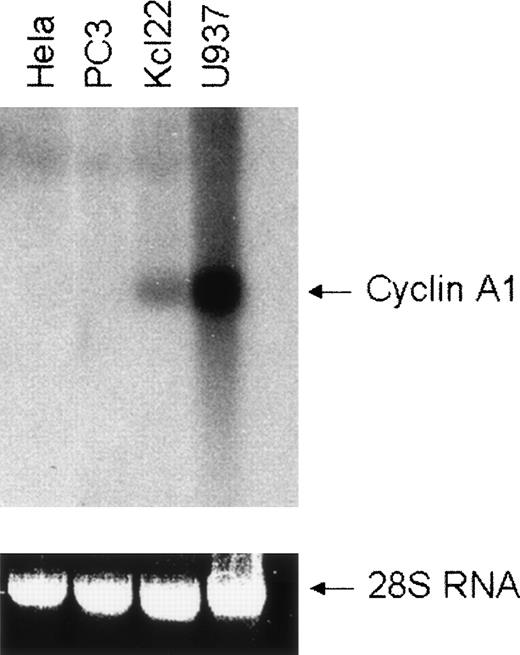
The human cyclin A1 gene is expressed in a highly restricted number of tissues in vivo and cyclin A1 expression in cell lines markedly differs between myeloid and nonmyeloid cell lines.1 To analyze further the differences in expression, we chose 4 human cell lines that differed in the degree of cyclin A1 expression. Two were derived from myeloid cells (U937 and KCL22) and 2 others were derived from solid carcinomas (PC3 from prostate cancer and HeLa from cervical carcinoma). Expression of cyclin A1 was analyzed by Northern blotting, demonstrating that cyclin A1 expression was found in the hematopoietic U937 and KCL22 cell lines, but expression was undetectable in the solid tumor-derived PC3 and HeLa cells (Fig 1).
Expression of cyclin A1 in different cell lines. Expression of human cyclin A1 mRNA in human tumor cell lines was analyzed by Northern blotting of 10 μg of total RNA and hybridization with a 32P-labeled cyclin A1-specific probe. The levels of 28S RNA show that equal amounts of RNA were loaded. HeLa (cervical carcinoma) and PC3 (prostate cancer) cell lines were derived from solid tumors, and U937 and Kcl22 were established from leukemic blasts.
Expression of cyclin A1 in different cell lines. Expression of human cyclin A1 mRNA in human tumor cell lines was analyzed by Northern blotting of 10 μg of total RNA and hybridization with a 32P-labeled cyclin A1-specific probe. The levels of 28S RNA show that equal amounts of RNA were loaded. HeLa (cervical carcinoma) and PC3 (prostate cancer) cell lines were derived from solid tumors, and U937 and Kcl22 were established from leukemic blasts.
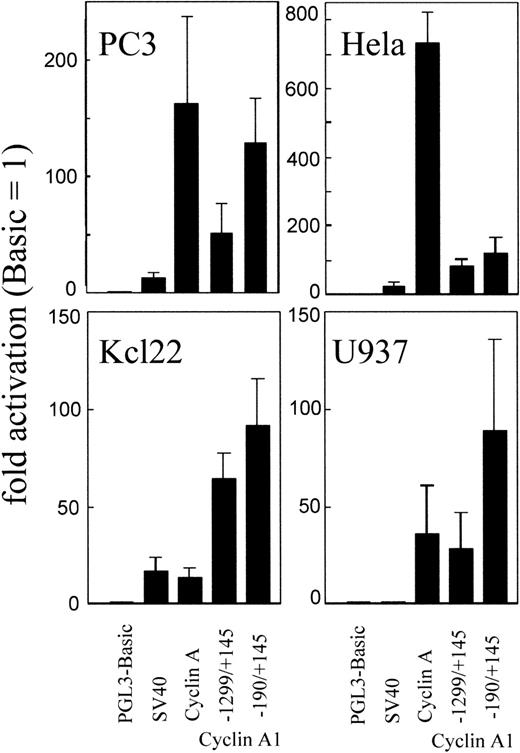
To analyze whether differences in RNA levels could be related to promoter activity, we transiently transfected the cyclin A1 promoter into several myeloid and adherent cell lines (Fig 2). Both cyclin A1 promoter luciferase constructs ranging from −1299 to +145 and from −190 to +145 showed activity in all 4 cell lines (Fig 2). The reporter activity of the shorter (335 bp) promoter fragment was always higher than the activity of the longer fragment. In addition, the activity of the cyclin A1 promoter was higher than that of the SV40 promoter (without enhancer) in all 4 cell lines.
Reporter gene expression of different promoters in cell lines from various tissues. Two fragments (1,444 and 335 bp) of the cyclin A1 promoter, a 452-bp fragment of the cyclin A promoter, and the enhancerless SV40 promoter were transfected into 4 cell lines derived from myeloid cells (U937 and KCL22) or from solid carcinomas (PC3 and HeLa). The cyclin A1 promoter is active in each cell line, and its activity is inversely related to that of the cyclin A promoter. All experiments were performed in duplicates and independently performed at least 3 times.
Reporter gene expression of different promoters in cell lines from various tissues. Two fragments (1,444 and 335 bp) of the cyclin A1 promoter, a 452-bp fragment of the cyclin A promoter, and the enhancerless SV40 promoter were transfected into 4 cell lines derived from myeloid cells (U937 and KCL22) or from solid carcinomas (PC3 and HeLa). The cyclin A1 promoter is active in each cell line, and its activity is inversely related to that of the cyclin A promoter. All experiments were performed in duplicates and independently performed at least 3 times.
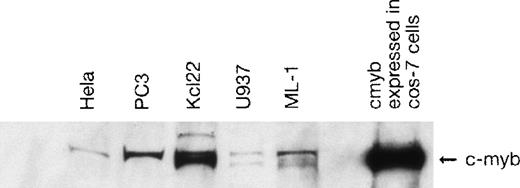
The cyclin A promoter is tightly cell cycle regulated and is assumed to be transactivated in all cycling mammalian cells. Activity of the cyclin A promoter was detectable in all 4 cell lines, but the degree of activity was inversely correlated with the cyclin A1 promoter activity. Cyclin A promoter activity was higher in PC3 and HeLa cells and was lower in the myeloid cell lines as compared with the cyclin A1 promoter activity. Preferential activity of the cyclin A1 promoter in myeloid cells (compared with the cyclin A promoter) was evident for both promoter constructs. The inverse relationship between cyclin A and cyclin A1 was also present at the RNA level in samples from patients with acute myeloid leukemia.2 However, activity of the cyclin A1 promoter by transient transfection was not limited to the myeloid cell lines, but was also present in PC3 and HeLa cells. The tissues from which these cell lines were derived expressed very low levels of cyclin A1. An explanation could be that transcription factors expressed in the cell lines, but not expressed in the normal tissue, lead to aberrant promoter activity. One transcription factor expressed in a wide variety of cell lines is c-myb. Western blot analysis demonstrated expression of c-myb in all 4 cell lines as well as in ML-1, another myeloid cell line that expresses high levels of cyclin A1 (Fig 3).1 The nonmyeloid cell lines appeared to have only a high molecular weight form, whereas the myeloid lines had both a high and a low molecular weight form. This may reflect a phosphorylated and a nonphosphorylated myb protein.
Expression of c-myb in nuclear extracts of cell lines from various tissues. Protein (10 μg) from nuclear extract of the indicated cell lines was loaded in SDS sample buffer on a 7.5% Tris-HCl gel. After blotting, c-myb was detected using a monoclonal antibody (Upstate Biotechnology). Nuclear extract from Cos-7 cells transfected with a c-myb expression vector served as a positive control. A single band with a molecular weight of approximately 80 kD was detected for the nonmyeloid cell lines (PC3 and HeLa), whereas double bands were seen for the myeloid cell lines (U937 and KCL 22). This might indicate differences in the phosphorylation status or the presence of different splice variants.
Expression of c-myb in nuclear extracts of cell lines from various tissues. Protein (10 μg) from nuclear extract of the indicated cell lines was loaded in SDS sample buffer on a 7.5% Tris-HCl gel. After blotting, c-myb was detected using a monoclonal antibody (Upstate Biotechnology). Nuclear extract from Cos-7 cells transfected with a c-myb expression vector served as a positive control. A single band with a molecular weight of approximately 80 kD was detected for the nonmyeloid cell lines (PC3 and HeLa), whereas double bands were seen for the myeloid cell lines (U937 and KCL 22). This might indicate differences in the phosphorylation status or the presence of different splice variants.
Analysis of the cyclin A1 promoter sequence shows potential binding sites for c-myb within the 335-bp fragment (Fig 4). The finding of c-myb expression in all 4 cell lines and the potential myb binding sites in the cyclin A1 promoter led to the hypothesis that c-myb could be involved in transactivation of the cyclin A1 promoter.
Potential myb binding sites in the cyclin A1 promoter. The 3 potential myb binding sites are shown along with the important Sp1 binding sites and the transcription start sites (marked in bold).
Potential myb binding sites in the cyclin A1 promoter. The 3 potential myb binding sites are shown along with the important Sp1 binding sites and the transcription start sites (marked in bold).
c-myb transactivates and binds to the cyclin A1 promoter.
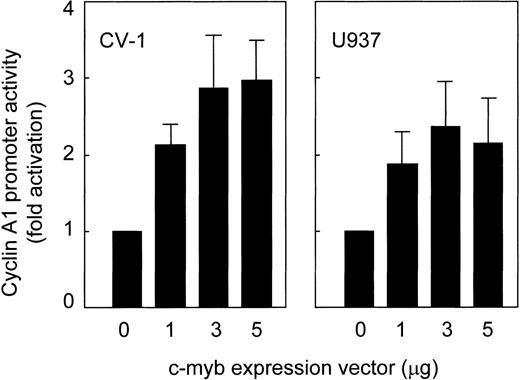
To analyze promoter transactivation by c-myb, a c-myb expression vector was transfected along with the 335-bp cyclin A1 promoter construct into CV-1 cells that do not express c-myb. A dose-dependent increase in cyclin A1 promoter activity occurred (Fig5). No increase in activity was observed when c-myb was cotransfected with the empty reporter plasmid (data not shown). The same experiments were repeated using U937 myeloid cells, which express rather low levels of c-myb. Again, c-myb clearly transactivated the promoter, with maximal activity occurring when 3 μg of c-myb were cotransfected (Fig5). These findings indicate that the cyclin A1 promoter can be transactivated by c-myb in adherent as well as in myeloid cell lines.
Transactivation of the cyclin A1 promoter by c-myb. Different amounts of c-myb were coexpressed with a cyclin A1 promoter construct (335-bp fragment). Empty vector was used to reach the same total amount of DNA in all experiments. The mean and standard error for 3 independent experiments are shown.
Transactivation of the cyclin A1 promoter by c-myb. Different amounts of c-myb were coexpressed with a cyclin A1 promoter construct (335-bp fragment). Empty vector was used to reach the same total amount of DNA in all experiments. The mean and standard error for 3 independent experiments are shown.
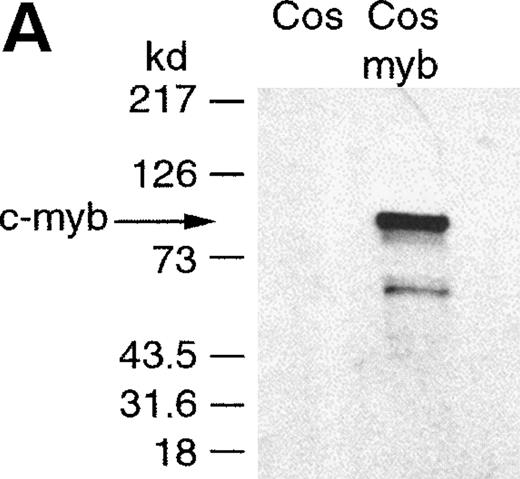
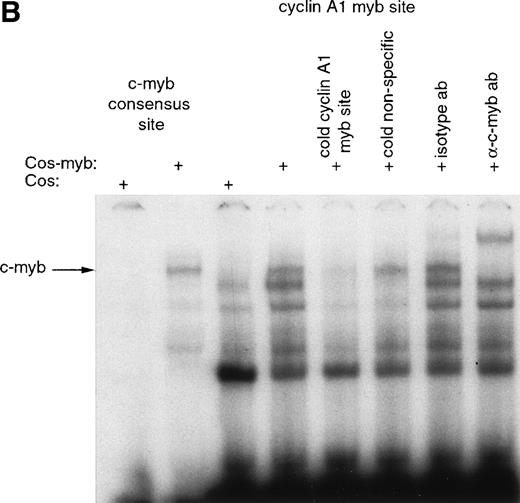
To analyze whether c-myb directly affected the cyclin A1 promoter, we examined whether the c-myb protein bound the predicted myb binding sites in the promoter region. Cos-7 cells were transfected with either empty vector control or a c-myb expression vector. High expression of c-myb was confirmed by Western blotting of nuclear extracts (Fig 6A). Gel-shift experiments were performed using these nuclear extracts and 32P-labeled oligonucleotides constituting a myb consensus site and the different myb-binding sites of the cyclin A1 promoter. The gel-shift binding reaction with c-myb expressing nuclear extract led to the appearance of a new band for the myb consensus site (Fig 6B). A band at a similar position was obtained for the cyclin A1 promoter myb site at +2. Specificity of the binding to the +2 site was confirmed using competitor oligonucleotides and c-myb specific antibody (Fig 6B). In contrast, only weak binding, which could not be confirmed to be c-myb, was seen at the potential myb site at −27, and no specific binding at the site at position −66 could be detected (data not shown).
Binding of c-myb to myb binding sites in the cyclin A1 promoter (EMSA). (A) Expression of c-myb in nuclear extracts of Cos-7 cells either transfected with empty vector (Cos) or with a c-myb expression vector (Cos-myb). Western blot analysis was performed similar to the experiment shown in Fig 3. (B) Binding of the different nuclear extracts to a c-myb consensus binding site and to the cyclin A1 promoter myb binding site at position +2. The nuclear extract containing c-myb protein led to a c-myb containing band at the consensus site as well as at the cyclin A1 promoter myb site. This band was successfully competed away by a 50-fold excess of the nonlabeled oligonucleotide encompassing the cyclin A1 myb site but not by a 50-fold excess of a nonspecific oligonucleotide. Also, the addition of 2 μg of a murine isotype control antibody did not alter the appearance of the band. However, an anti–c-myb antibody led to a supershift of the band.
Binding of c-myb to myb binding sites in the cyclin A1 promoter (EMSA). (A) Expression of c-myb in nuclear extracts of Cos-7 cells either transfected with empty vector (Cos) or with a c-myb expression vector (Cos-myb). Western blot analysis was performed similar to the experiment shown in Fig 3. (B) Binding of the different nuclear extracts to a c-myb consensus binding site and to the cyclin A1 promoter myb binding site at position +2. The nuclear extract containing c-myb protein led to a c-myb containing band at the consensus site as well as at the cyclin A1 promoter myb site. This band was successfully competed away by a 50-fold excess of the nonlabeled oligonucleotide encompassing the cyclin A1 myb site but not by a 50-fold excess of a nonspecific oligonucleotide. Also, the addition of 2 μg of a murine isotype control antibody did not alter the appearance of the band. However, an anti–c-myb antibody led to a supershift of the band.
Mutations in a myb site decrease myb transactivation of the cyclin A1 promoter.
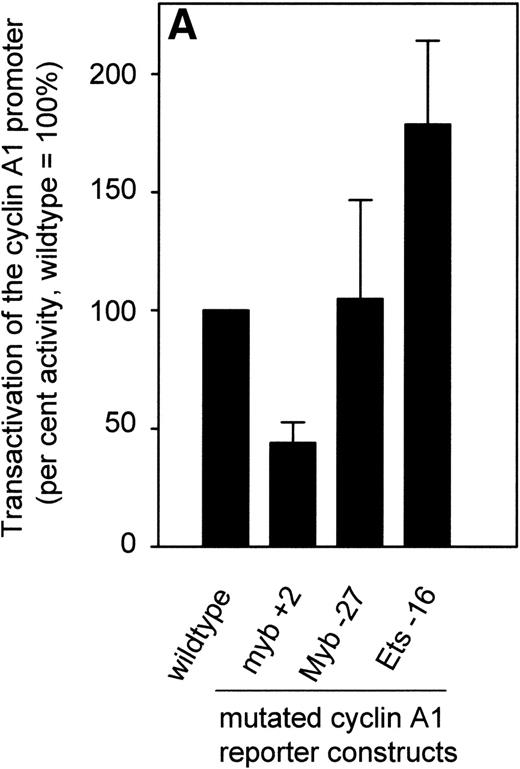
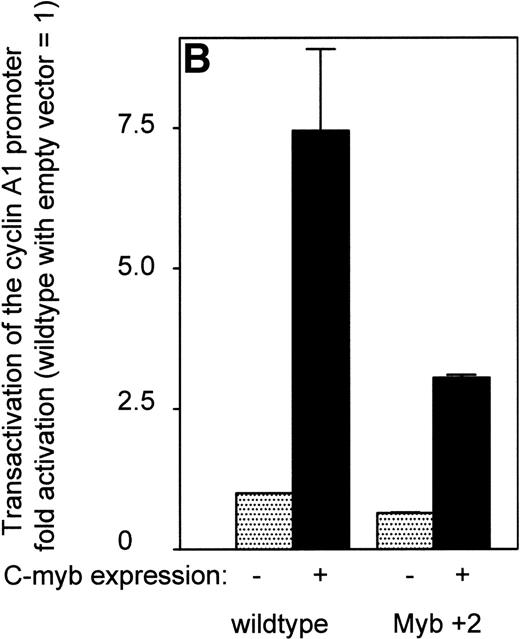
To test whether c-myb activation of the promoter was affected by alteration of the myb binding sites, different sites were mutated and the resulting constructs were transfected in KCL22 cells. These cells showed the highest c-myb expression of all the cell lines (Fig 3). Abrogation of the myb site at +2 clearly diminished promoter activity by 50%, whereas a mutation at either −27 or of the ets site at −15 did not lead to a decrease in promoter activity (Fig 7A). The myb site at +2 is close to the transcriptional start site and the basepairs surrounding the transcriptional start site could function as an Initiator (Inr). To rule out that our observed effects of the mutation at +2 depended on the loss of binding of the basal transcriptional machinery, we transfected the mutated reporter construct together with either the c-myb expression plasmid or an empty vector control into CV-1 cells and compared the results with transfections using the wild-type promoter plasmid (Fig 7B). The mutation at +2 led to a minor reduction in promoter activity when transfected with the empty vector control. However, transactivation of the mutated reporter plasmid by c-myb was reduced by more than 50%, indicating that c-myb can transactivate the cyclin A1 promoter through this site. Other sites or indirect effects may contribute to the cyclin A1 promoter activation, because the mutation at +2 did not abolish the increase in promoter activity entirely.
Effect of point mutations on the activity of the cyclin A1 promoter. (A) After introducing mutations into potential transcription factor binding sites of the cyclin A1 promoter (335-bp fragment), luciferase constructs were transfected into KCL 22 cells that express high levels of c-myb (Fig 3). Bars represent the mean and SEM of at least 3 independent experiments. (B) The wild-type cyclin A1 reporter construct or the myb 1 site mutation of the cyclin A1 promoter was cotransfected with either c-myb or empty vector into CV-1 cells. These experiments were performed using the Dual Luciferase Assay system and the pRL-SV40 vector for standardization.
Effect of point mutations on the activity of the cyclin A1 promoter. (A) After introducing mutations into potential transcription factor binding sites of the cyclin A1 promoter (335-bp fragment), luciferase constructs were transfected into KCL 22 cells that express high levels of c-myb (Fig 3). Bars represent the mean and SEM of at least 3 independent experiments. (B) The wild-type cyclin A1 reporter construct or the myb 1 site mutation of the cyclin A1 promoter was cotransfected with either c-myb or empty vector into CV-1 cells. These experiments were performed using the Dual Luciferase Assay system and the pRL-SV40 vector for standardization.
c-myb can induce cyclin A1 expression in primary human fibroblasts.
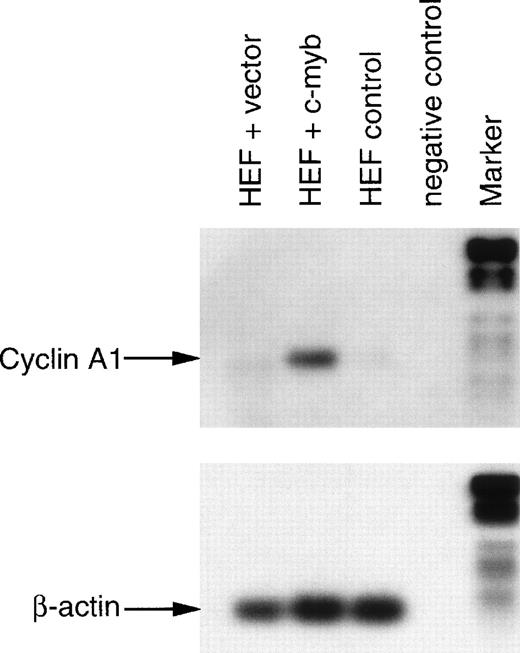
To examine whether c-myb could regulate the endogenous cyclin A1 gene, we transfected primary human embryonic fibroblasts with a c-myb expression vector. An EGFP-expressing plasmid was cotransfected with the c-myb expression vector at a ratio of 1:10, respectively. Forty-eight hours later, the EGFP-expressing cells (∼3% to 5%) were enriched using flow cytometric cell sorting. RNA was extracted and reverse transcribed before PCR was performed to analyze cyclin A1 expression in the different cell populations. As a negative control, we also sorted and analyzed the nontransfected cell population (Fig 8). As a second control, we used the RNA derived from sorted human embryonic fibroblasts that were transfected with an empty control vector instead of the c-myb expression vector. No expression of cyclin A1 was detected in both of the control cell populations. However, strong expression of cyclin A1 was observed in the c-myb–transfected cell population, indicating that the exogenous c-myb expression had induced cyclin A1 expression (Fig8).
c-myb induces endogenous cyclin A1 expression in human embryonic fibroblasts (HEF). Primary human embryonic fibroblasts were transfected, and after 48 hours, they were sorted by flow cytometry using EGFP expression as a marker of transfection (see Materials and Methods). HEF were transfected either with empty vector alone (HEF + vector) or with a c-myb expression vector (HEF + c-myb). Nontransfected cells of the latter transfection were also sorted and served as an additional control (HEF control). Finally, a sample with ddH2O instead of RNA for the reverse transcription reaction served as a negative control (negative control). After the PCR reactions for cyclin A1 (28 cycles, 382 bp) and β-actin (20 cycles, 210 bp), products were run on agarose gels, blotted, and hybridized with digoxigenin-labeled internal oligonucleotides. Detection was performed using antidigoxigenin antibody and a chemiluminescence reaction. The digoxigenin-labeled marker VI (Boehringer Mannheim) was run in parallel: 154, 220/234, 298, 394, 453, 517, 653, and 1,033 bp. The 154-bp band of the marker is too weak to be seen on the β-actin blot.
c-myb induces endogenous cyclin A1 expression in human embryonic fibroblasts (HEF). Primary human embryonic fibroblasts were transfected, and after 48 hours, they were sorted by flow cytometry using EGFP expression as a marker of transfection (see Materials and Methods). HEF were transfected either with empty vector alone (HEF + vector) or with a c-myb expression vector (HEF + c-myb). Nontransfected cells of the latter transfection were also sorted and served as an additional control (HEF control). Finally, a sample with ddH2O instead of RNA for the reverse transcription reaction served as a negative control (negative control). After the PCR reactions for cyclin A1 (28 cycles, 382 bp) and β-actin (20 cycles, 210 bp), products were run on agarose gels, blotted, and hybridized with digoxigenin-labeled internal oligonucleotides. Detection was performed using antidigoxigenin antibody and a chemiluminescence reaction. The digoxigenin-labeled marker VI (Boehringer Mannheim) was run in parallel: 154, 220/234, 298, 394, 453, 517, 653, and 1,033 bp. The 154-bp band of the marker is too weak to be seen on the β-actin blot.
DISCUSSION
The cyclin A1 gene differs from other human cyclin genes in several ways. Cyclin A1 is not related to cellular proliferation and division in general.1,4 But it is robustly expressed in testis and moderately expressed in bone marrow, 2 tissues with an extraordinarily high proliferative potential. Also, high-level expression is found in myeloid leukemia cell lines and in blasts of patients with acute myeloid leukemia.2 Cyclin A1 interacts with RB family members and E2F in vivo.5 The reason for the prominent expression of cyclin A1 in myeloid leukemia cells is unclear; the cyclin A1 gene is not amplified in myeloid leukemia cell lines.1
Recently, we cloned the promoter of the cyclin A1 gene and started to analyze its promoter region.15 The cyclin A1 promoter is TATA-less and starts transcription from a major site around 150 bp upstream of the initiating ATG. A 335-bp cyclin A1 promoter fragment (−190 to +145) showed the highest activity of all constructs upon transient transfection into HeLa cells.15 This fragment contains several GC boxes (Sp1 sites), and promoter activity critically depends on 4 of these sites. The GC boxes are periodically repressed during the G1 phase of the cell cycle and play a major role in the cell cycle regulation of promoter activity.15 After establishing the elements that are relevant for the basic activity of the promoter, we analyzed mechanisms that could provide insight into the high cyclin A1 expression levels observed in acute myeloid leukemia. Transient transfections of the cyclin A1 promoter constructs demonstrated promoter activity in each of the tested cell lines. Also, the construct with the shorter cyclin A1 fragment (−190 to +145) showed stronger activity than the longer fragment in all the cell lines. Interestingly, we detected an inverse correlation for the degree of promoter activity between the cyclin A1 and the cyclin A promoter constructs in adherent and myeloid cell lines. The cyclin A1 promoter was particularly active in myeloid cells, whereas cyclin A promoter activity was strong in the nonmyeloid cell lines. In addition, we have previously reported that RNA levels between cyclin A and cyclin A1 were inversely correlated in samples from leukemia patients.2Also, compared with the SV40 early promoter, the activity of the cyclin A1 promoter constructs was especially high in the 2 myeloid cell lines tested. This indicates that the cyclin A1 promoter is preferentially active in myeloid cells. Nevertheless, the finding that the promoter was active in all the cell lines was initially surprising given the highly restricted expression pattern of cyclin A1 RNA in most nonmyeloid cell types in vivo. Several genes, including transcription factors that are tissue specific in the healthy organism, have been shown to be consistently unregulated in a variety of cell lines. This has long been known for c-myb, and Western blotting confirmed that c-myb is expressed in all 4 human cell lines under investigation. This and the presumed presence of several c-myb binding sites near the transcriptional start site of cyclin A1 led us to analyze the role of c-myb in transactivation of the cyclin A1 promoter.
Our data provide evidence that c-myb can transactivate the cyclin A1 promoter. The transactivation by c-myb leads to a strong increase in cyclin A1 promoter activity in CV-1 cells that do not express c-myb. In U937 cells, the transactivation of the cyclin A1 promoter by extrinsic c-myb expression was approximately 3-fold compared with expression of an empty expression vector. One of the reasons for the weaker transactivation capacity of c-myb in U937 cells might be that U937 already expresses c-myb. Also, the full transactivation potential of c-myb is usually reached in synergy with other transcription factors such as c-ets or NFM.20 A potential c-ets site is present at position −13 of the cyclin A1 promoter, but cotransfection of c-myb with ets-2 did not lead to an increase in promoter transactivation (data not shown), and mutations introduced into the presumed ets binding site did not decrease cyclin A1 promoter activity in KCL22 cells. In addition to transactivating the cyclin A1 promoter in transient transfections, expression of c-myb in primary human embryonic fibroblasts induced endogenous cyclin A1 mRNA, suggesting that c-myb can regulate cyclin A1 expression in vivo.
The proto-oncoprotein c-myb plays an essential role in the proliferation of hematopoietic cells. Knockout mice deficient of c-myb die at day 14.5 of embryogenesis because of the failure of liver hematopoiesis.21 A large body of evidence confirms that c-myb has a crucial role in hematopoiesis, and its expression is high in the proliferating fraction of normal bone marrow cells.7It is also thought to play a role in the pathogenesis of acute myeloid leukemia.22 Some of the genes induced by c-myb appear to be rather specific for hematopoietic cells, whereas others are involved in cell cycle regulation in general, providing a direct link between c-myb expression and cell cycle progression. Cyclin A1 clearly falls into both categories by its expression in myeloid cells and because it is a member of the cell cycle machinery. The finding that c-myb transactivates the cyclin A1 promoter could provide a link to tissue-specific regulation of cell cycle events. In the G1 phase, D-type cyclins integrate signals from cell surface signaling and activated nuclear hormone receptors and drive the cell towards the G1/S boundary.23 The D-type cyclins show tissue specificity and their involvement in tissue specific tumors such as high expression of cyclin D1 in some breast cancers shows the different roles that they might have in distinct tissues.24 Much less is known about tissue-specific influences on cell cycle progression beyond the G1/S boundary. Our finding that c-myb directly transactivates the cyclin A1 promoter links the expression of myeloid-specific genes (eg, c-myb) to tissue-specific cell cycle events that are not related to G1 cyclins. This might imply that tissue-specific mechanisms play a role in cell cycle regulation not only in the G0/G1 phase, but also in the S and G2/M phases.
Transactivation by c-myb could explain why the cyclin A1 promoter constructs are active in a variety of cell lines. However, it does not explain the very low levels of cyclin A1 RNA found in some of these cell lines (eg, PC3 and Hela). The Cyclin A1 promoter is highly GC rich and shows a CpG island that reaches up to 70 bp upstream of the transcription start site. We have started to analyze the potential role of methylation in modulating cyclin A1 expression, and preliminary data show that the cyclin A1 promoter is methylated in nonexpressing adherent cell lines such as PC3 and HeLa (Müller et al, manuscript submitted). Methylation and chromatin remodeling are likely to influence cyclin A1 gene expression in vivo, and studies have shown that the binding of c-myb to its binding sites is prevented by CpG methylation.25
Taken together, we have found that the cyclin A1 promoter is preferentially active in myeloid leukemia cell lines. The finding that c-myb transactivates this promoter through a specific binding site provides an important link between c-myb and cell cycle regulation in hematopoietic cells. The exact role of cyclin A1 in the hematopoietic cell cycle and in the pathogenesis of acute myeloid leukemia awaits further analysis.
ACKNOWLEDGMENT
The authors are grateful to Dr Rhona Schreck for providing the human embryonic fibroblasts.
Supported by grants from the National Institutes of Health (NIH); by US Defense Grants; by the Parker Hughes, C. and H. Koeffler Funds; and by the Gladys Lichtenstein Trust. C.M. is recipient of a fellowship of the German Research foundation (DFG). G.I. is supported by the Howard Hughes Institute Undergraduate Program. H.P.K. holds the Mark Goodson Chair in Oncology Research and is a member of the Jonson Cancer Center. T.P.B.'s work is supported by NIH Grant No. GM55985.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Carsten Müller, MD, ICP-Labor, Department of Hematology/Oncology, University of Münster, Domagkstr. 3, 48149 Münster, Germany; e-mail:muellerc@uni-muenster.de.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal