Abstract
A new platelet-specific alloantigen, termed Sita, was identified in a severe case of neonatal alloimmune thrombocytopenia. The Sita alloantigen is of low frequency (1/400) in the German population. Immunochemical studies demonstrated that the Sita epitopes reside on platelet glycoprotein (GP) Ia. Nucleotide sequence analysis of GPIa cDNA derived from Sita-positive platelets showed C2531→T2531 point mutation, resulting in Thr799Met dimorphism. Analysis of genomic DNA from 22 Sita-negative normal individuals showed that the Thr799 is encoded by ACG2532 (90.9%) or ACA2532 (9.1%). To establish a DNA typing technique, we elucidated the organization of the GPIa gene adjacent to the polymorphic bases. The introns (421 bp and 1.2 kb) encompass a 142-bp exon with the 2 polymorphic bases 2531 and 2532. Polymerase chain reaction-restriction fragment length polymorphism analysis on DNA derived from 100 donors using the restriction enzyme MaeIII showed that the Met799 form of GPIa is restricted to Sita (+) phenotype. Analysis of stable Chinese hamster ovary transfectants expressing allele-specific recombinant forms of GPIa showed that anti-Sita exclusively reacted with the Glu505Met799, but not with the Glu505Thr799 and the Lys505Thr799 isoforms. In contrast, anti-Bra (HPA-5b) only recognized the Lys505Thr799 form, whereas anti-Brb(HPA-5a) reacted with both Glu505Thr799 and Glu505Met799 isoforms. These results demonstrated that the Met799 is responsible for formation of the Sita alloantigenic determinants, whereas amino acid 505 (Lys or Glu) specifically controls the expression of Bra and Brb epitopes, respectively. Platelet aggregation responses of Sita (+) individuals were diminished in response to collagen, indicating that the Thr799Met mutation affects the function of the GPIa/IIa complex.
IMMUNIZATION AGAINST human platelet antigens (HPA) is the central event in the pathophysiology of neonatal alloimmune thrombocytopenia (NAIT) and in posttransfusion purpura (PTP) and is also of importance in some patients refractory to platelet transfusions.1
Currently, 5 HPA systems, HPA-1 (PlA, Zw), -2 (Ko, Sib), -3 (Bak, Lek), -4 (Pen, Yuk), and -5 (Br, Zav), are officially recognized. In addition, a number of low frequency alloantigens, HPA-6bW (Caa, Tua), -7bW (Moa), -8bW (Sra), -9bW (Maxa), -10bW (Laa), Groa, and Iya, have also been reported. Meanwhile, the alloantigenic determinants of all of these HPAs could be localized on platelet membrane glycoprotein (GP) Ia, GPIbα, GPIbβ, GPIIb, and GPIIIa. All of these have been found to result from point mutations in the encoding genes, which lead to single amino acid substitutions.2-13
GPIIb/IIIa, also known as the integrin αIIbβ3,14 which serves as fibrinogen receptor on platelets, carries the majority of these HPAs.
Another integrin, α2β1 (GPIa/IIa), which mediates platelet adhesion to both fibrillar and nonfibrillar collagen,15,16 is known to bear the alloantigenic determinant of the HPA-5.17 Recently, we could demonstrate that the HPA-5a, -5b phenotypes are associated with a single base G1648A substitution of the GPIa gene leading to an aminoacid Glu505Lys dimorphism.6Alloimmunization against HPA-5 has been associated with NAIT,18 PTP,19 platelet transfusion refractoriness,20 and thrombocytopenia due to passively transferred platelet antibodies after transfusion of blood products21 or bone marrow transplantation.22
In addition, the important functional role of GPIa in primary hemostasis is stressed by the observation of congenital and acquired bleeding tendency in patients with deficient GPIa and with autoantibodies. In vitro studies showed impaired reactivity of platelets with collagen in all these cases.23-26
Current evidence suggests that adhesion and activation of platelets by collagen are mediated through distinct receptors. Activation of platelets by collagen is believed to occur via a 2-site, 2-step model in which GPIa/IIa provides an initial interaction (adhesion) that brings a second site in the collagen molecule into the activitory receptor GPVI.27
Recently, Kunicki et al28 were able to identify clusters of linked silent polymorphisms within the coding sequence of the α2 gene that which are responsible for the variation in α2β1 receptor density on the platelet surface. Three allelic differences could now be identified by 4 dimorphisms at positions 807, 837, 873, and 1648. Allele 1 (T807/T837/A873/G1648) is associated with increased levels of α2β1, whereas allele 2 (C807/T837/G873/G1648) and allele 3 (C807/C837/G873/A1648) are associated with lower levels of α2β1.29 Carriers of the allele 1 express high levels of GPIa/IIa, whereas individuals who carry alleles 2 and 3 exhibit lower expression of the platelet integrin. Furthermore, Kritzig et al29 could demonstrate that the density of GPIa/IIa on the platelet surface correlates with the rate of platelet attachment in whole blood to type I collagen. A clinical impact of these findings was demonstrated by the association of the GPIa C807T gene polymorphism with nonfatal myocardial infarction and stroke in younger individuals.30 31
In this report, we describe another case of alloimmunization against a new genetic variant of GPIa that was responsible for a case of severe NAIT, and we describe the biochemical, molecular, biological, and functional properties of platelets carrying this antigen.
CASE REPORT
A healthy 19-year-old white mother (Sit) gave birth to her first child after a normal pregnancy. The male child (2,150 g) was delivered at term with an Apgar score of 8. At birth, a severe thrombocytopenia was observed (31 × 109/L) and the platelet count decreased continually below measurable value. Clinical and laboratory examinations showed no other abnormalities. Petechiae were observed at the age of 4 days and a tentative diagnosis of NAIT was made. Because maternal platelets were not available, the infant was transfused with platelets from a healthy donor prepared by platelet apheresis with good increment.The platelet count increased to 158 × 109/L. In addition, the child was treated with high doses of intravenous IgG from day 3 to 5 of life and with prednisolone from day 2 to 10 of life. After a moderate decrease of the platelet count to 67 × 109/L, the child was discharged with a normal platelet count on day 18. Ultrasound examination showed no signs of cerebral hemorrhage.
MATERIALS AND METHODS
Blood samples.
Blood samples of the mother, the father, and the child were referred to us shortly after delivery because of suspected NAIT. Platelets were isolated from EDTA-anticoagulated blood by differential centrifugation and stored at 4°C in isotonic saline containing 0.1% NaN3. Normal donor platelets for the characterization of platelet antibodies were selected from a large number of donors with known human platelet alloantigens (HPA 1-5) and ABO blood group antigens.
Phenotyping.
Phenotyping of human platelets was performed using the glycoprotein-specific immunoassay (monoclonal antibody-specific immobilization of platelet antigens [MAIPA]), as previously described.32
Antibodies.
Antibodies against Bra and Brb alloantigens were obtained from mothers of children with NAIT and from polytransfused patients, respectively.17 Monoclonal antibody (MoAb) Gi5 and MoAb Gi9 specific for GPIIb/IIIa complex (CD41) and GPIa (CD49b), respectively, were produced and characterized in our laboratory.6 MoAb SAM1 directed against the GPIc (CD49e) subunit and MoAb B1G6 specific for β2 microglobulin of HLA class I molecule were purchased from Immunotech (Hamburg, Germany). MoAb FMC 25 against the GPIX (CD42a) subunit of GPIb/IX/V complex was a generous gift of Dr H. Zola (Adelaide, Australia).
Immunoprecipitation.
Platelets and Chinese hamster ovary (CHO) stable transfectants were surface-labeled with 5 mmol/L NHS-LC-Biotin (Paesel, Frankfurt, Germany) and were immunoprecipitated as previously described.9
Isolation of platelet RNA and leukocyte DNA.
Total platelet RNA was isolated from 20 mL EDTA anticoagulated blood of phenotyped donors by a modification of the guanidium isothiocyanate method as previously described.4 Platelet RNA was resuspended in 100 μL diethyl pyrocarbonate (DEPC)-treated H2O and stored at −70°C. Genomic DNA was obtained from peripheral blood leukocytes derived from 3 mL EDTA anticoagulated blood after removing platelet-rich plasma. The DNA was extracted by the salting out procedure as described by Miller et al,33dissolved in 300 μL TE buffer (10 mmol/L Tris, 1 mmol/L EDTA, pH 8.0), and stored at 4°C.
Primers for GPIa-specific amplification.
To amplify the entire coding region of GPIa by polymerase chain reaction (PCR), 8 overlapping sets of primers were constructed based on the published cDNA sequence of GPIa.34 For cDNA amplification of a region encompassing nucleotides 2304-2858, forward primer P48 (2212-GAAAGGTGCC TGCAGAAG-2229), reverse primer P40 (2858-TCAGCATCATACAGGAGAGG-2839), and nested primer P18 (2304-CTCTTTGGATTTGCGTGTGGACATC-2328) were used. In addition, primer pair P61 (2414-GTGGTGAGGATGGACTTTGC-2433), P42 (2572-CAATTCCAGTGTTGTATGCAC-2552) and primer pair P6 (2551-AGTGCATA CAACACTGGAAT-2570), P7 (2699-TTTAAAGCAGGGTAGCCTAC-2680) were used to amplify GPIa gene from genomic DNA.
PCR amplification of cDNA.
One hundred microliters of platelet RNA were heated to 68°C for 10 minutes and quickly cooled on ice water. The first-strand cDNA was synthesized using 10 μmol/L oligo dT, 40 U RNAse inhibitor (Boehringer Mannheim, Mannheim, Germany), 2 mmol/L of each dNTP (Pharmacia, Freiburg, Germany), 500 U Moloney's murine leukemia virus (MMuLV) reverse transcriptase, and 5× enzyme buffer (GIBCO BRL, Eggenstein, Germany) for 40 minutes at 45°C in a total volume of 30 μL and was stopped by chilling to 0°C. Aliquots of 20 μL of cDNA were preheated to 90°C for 5 minutes and were chilled to 0°C. After digestion with 2 U of RNAseH enzyme (GIBCO BRL) at 37°C for 20 minutes, cDNA was stored at −20°C. Five microliters of cDNA were diluted with 10× PCR buffer, 0.3 μmol/L of each primer (P48, P40), 175 μmol/L dNTP, and 2.5 U Taq Gold polymerase (Perkin Elmer, Norwalk, CT) in a total volume of 50 μL. Amplification was performed on DNA thermal cycler 480 (Applied Biosystem, Weiterstadt, Germany) for 20 cycles. Each cycle consisted of denaturation at 93°C for 1 minute, annealing at 53°C for 1 minute, and extension at 72°C for 2 minutes. In the final cycle, the samples were kept at a temperature of 72°C for 10 minutes and then chilled to 4°C. Two-microliter aliquots of PCR products were reamplified for 30 cycles using nested primer P18 and reverse primer P40 under identical PCR conditions. PCR products were analyzed on 1.6% SeaKemGTG agarose gel (Biozym, Hamel, Germany) containing ethidium bromide.
PCR amplification of genomic DNA.
Two hundred nanograms of genomic DNA were amplified using primer pairs P61, P42 and P6, P7 as described above. Thirty-two cycles of denaturation at 94°C for 1 minute, annealing at 58°C for 1 minute, and extension at 72°C for 2 minutes were performed.
Sequence analysis of PCR products.
Amplified DNA was purified on SeaKemGTG gel (Biozym) by QIAquick (Qiagen, Düsseldorf, Germany). Purified DNA was trimmed using Klenow DNA polymerase (Biolabs, Schwalbach, Germany) and were subcloned into the EcoRV cloning site of the plasmid vector pGEM-5Zf (Promega Biotech, Madison, WI) and then transformed into the DH5α high efficiency competent Escherichia coli (GIBCO BRL). Plasmid DNA was prepared for nucleotide sequencing analysis by QIAprep (Qiagen). The inserts from 8 positive clones were sequenced using Taq-FS Dye-Terminator Cycle Sequencing Kit according to the manufacturer's protocol and were analyzed on ABI PRISM Genetic Analyzer 310 (Applied Biosystem). SP6 and T7 primers that hybridize to vector sequence and flank the cloned insert were used for sequencing.
Direct sequencing of PCR fragment.
The DNA fragment of interest was gel purified from 50 μL PCR reaction and directly sequenced as described above. PCR primers were used as sequencing primers.
Genotyping of Sita and HPA-5 alloantigens by restriction fragment length polymorphism (RFLP).
For genotyping of Sita alloantigen, 3 to 5 μg genomic DNA was amplified using forward primer P61 (see above) and an intronic reverse primer P83 5′-TACCGGTAGGGAGAATGATGC-2602-3 under the following PCR conditions: 30 cycles at 94°C for 1 minute, 58°C for 1 minute, and 72°C for 2 minutes, followed by final extension at 72°C for 10 minutes. Aliquots of 3 μL of PCR products were digested in a thermal cycler with 2 U Mae III endonuclease (Boehringer Mannheim) for 6 hours at 55°C, respectively. Restriction fragments were analyzed on 1.6% agarose gel using Tris borate buffer system (Biozyme). Genotyping for HPA-5 was performed by RFLP using Mnl I endonuclease as previously described.35
Genotyping of GPIa C807T dimorphism by sequence-specific PCR (PCR-SSP).
Genotyping of the C807T dimorphism was performed using PCR-SSP as recently described.30
Generation of allele-specific GPIa cDNA constructs.
A full-length human GPIa cDNA (A1648AG/AC2531A) in the mammalian expression vector pMPSV,36which encodes Lys505Thr799 GPIa isoform, was kindly provided by Dr E. Klein (Department of Dermatology, University of Würzburg, Würzburg, Germany). An allele-specific recombinant form G1648AG/AC2531A encoding the Glu505Thr799 GPIa isoform was produced by cartridge mutagenesis. A 1,435-bp cDNA fragment spanning nucleotides 1486 to 2921 of platelet GPIa mRNA derived from a Brbhomozygous donor was amplified by PCR.6 After digestion with Bgl II and BstEII endonucleases (Biolabs), the 1,374-bp fragments containing the polymorphic base G1648were then shuttled into the pMPSV expression vector containing the GPIa insert (A1648AG/AC2531A), which had been digested with the same enzymes. To exclude contamination of undigested plasmid, the pMPSV expression vector was linearized with Swa I endonuclease (Biolabs) before ligation. After subcloning in E coli, the resulting plasmid constructs were amplified in PCR (bases 1486-1900) and were screened for A1648G dimorphism by RFLP analysis using Mnl I endonuclease. Specific mutations CA→TG at positions 2531 and 2532 were introduced into the Lys505Thr799 and Glu505Thr799 constructs by site-directed mutagenesis using QuickChange Mutagenesis Kit (Strategene, Heidelberg, Germany).
For PCR amplification, 2-nucleotides (underlined) mismatched forward primer 5′-GTTAACATTTTCAGTAATGCTGAAAAATAAAAGGG-3′ and reverse primer 5′-CCCTTTTATTTTTCAGCATTACTGAAAATGTTAACC-3′ from base 2513 to 2548 of GPIa cDNA were constructed. After denaturation for 30 seconds at 95°C, aliquots of 20 ng plasmid were amplified for 12 cycles (denaturation for 30 seconds at 95°C, annealing for 60 seconds at 55°C, and extension for 17 minutes at 68°C). PCR products were digested with Dpn I endonuclease for 1 hour at 37°C and transformed into DH5α high efficiency competent E coli. Plasmid DNA from positive clones were amplified by PCR using primers P18 and P40. PCR products were subjected to RFLP analysis with Mae III as described above. For subsequent transfection studies, all GPIa allele-specific constructs were validated by nucleotide sequence analysis.
Stable expression of allele-specific constructs in CHO cells.
CHO (American Type Tissue Collection, Rockville, MD) cells were grown in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS; Seromed, Berlin, Germany), 1% sodium pyruvate, 1% glutamine, and 1% penicilline/streptomycine (GIBCO BRL; complete medium) and were transfected with allele-specific GPIa constructs by the use of the reagent Lipofectin (GIBCO BRL). In brief, 6 μg of the GPIa construct was mixed with 25 μL Lipofectin in 2 mL OptiMEM Medium (GIBCO BRL) and then added to a subconfluent 10-cm plate of CHO cells for 12 hours. For antibiotic selection, cotransfection with 6 μg RSV neo plasmid (Invitrogen, Leek, Netherlands) containing Neomycin resistent gene was performed. Nine milliliters of complete medium was added and the incubation was continued for 48 hours. After splitting, CHO transfectant was then selected with Genicitin (G418; final concentration, 1 mg/mL; GIBCO BRL) for approximately 2 weeks. Positive clones expressing GPIa/IIa were enriched by adhesion of 3 × 106 stable transfectant onto 3.6-cm petri dishes coated with collagen type I (5 μg/mL of 0.1 mol/L NaHCO3). After subcloning, the surface GPIa/IIa expression was analyzed by flow cytometry (see below). Stable cell lines were grown in complete medium supplemented with 200 μg/mL of G418.
Flow cytometry.
Stable transfectants were harvested with trypsin-EDTA (GIBCO BRL), washed in phosphate-buffered saline (PBS; GIBCO BRL), and fixed with 1% paraformaldehyde for 3 minutes. After three washes, 450 μL (106/μL) of cell suspensions were incubated with 50 μL MoAb of Gi14 (20 μg/mL) or 20 to 30 μL human sera (anti-Bra, anti-Brb, and anti-Sita) for 30 minutes at room temperature. Labeled cells were then washed twice, stained with 500 μL fluorescein isothiocyanate-conjugated rabbit antimouse IgG or antihuman IgG (dilution 1:40; Dako, Hamburg, Germany), and analyzed by flow cytometry (Ortho Diagnostic, Neckargemünd, Germany).
Adhesion of platelets to immobilized collagen.
Adhesion of platelets to immobilized collagen was investigated as recently described.37 Platelets were isolated from ACD-anticoagulated blood by differential centrifugation and washed with AMPL buffer (140 mmol/L NaCl, 5 mmol/L KCl, 1 mmol/L MgCl2, 10 mmol/L glucose, 10 mmol/L sodium pyruvate, 5 mmol/L sodium malate, 10 mmol/L HEPES, and 0.3 mmol/L albumin, pH 7.4) containing 0.02 U/mL apyrase and 5 U/mL hirudin (Paesel & Lorey, Frankfurt, Germany). Aliquots of 1 × 109 washed platelets in PBS (137 mmol/L NaCl, 2 mmol/L KCl, 8 mmol/L Na2HPO4, and 1 mmol/L KH2PO4, pH 7.4) were labeled with 16 μL 75 mmol/L NHS-LC-Biotin (Paesel) at room temperature for 15 minutes. Labeled platelets were washed twice with 5 mL AMPL buffer and resuspended in PBS at a final concentration of 3 × 108/mL. For adhesion, microtiter wells were coated overnight with collagen type I, III, or V (50 μg/mL; Sigma, Dreieich, Germany) or bovine serum albumin (BSA; 10 mg/mL; Sigma), washed 3 times with 200 μL PBS, and blocked with 200 μL 1% BSA in PBS for 1 hour at 37°C. Aliquots of 100 μL of biotinylated platelets were added in triplicate to wells coated either with BSA or collagen and were permitted to adhere at 37°C for 1 hour. To evaluate the functional effect of anti-Sita alloantibodies, platelets were mixed with either 40 μL affinity-purified AB-sera or of anti-Sita for 30 minutes in a 5% CO2atmosphere. Nonadherent cells were removed by gentle absorption onto absorptive pads and by washing of the wells 3 times with 150 μL PBS. Bound platelets were detected by the addition of 100 μL alkaline phosphatase-labeled streptavidin (500 μg/mL, dilution 1:1,500; Dianova) and incubation at 37°C for 1.5 hours. After a further 3 washes with PBS and one wash with diethanolamine buffer, pH 9.8, 100 μL of p-nitro-phenylphosphate (1 mg/mL; Sigma) was added. The reaction was stopped after 5 minutes at room temperature by the addition of 100 μL of 1.0 mol/L NaOH and was read in an enzyme-linked immunosorbent assay (ELISA) plate reader (OD405, Titertex Multiskan; Pharmacia).
Platelet aggregation.
Platelet-rich plasma (PRP) of ACD anticoagulated blood from Sita-phenotyped individuals was ajusted to 3 × 105 platelets/μL by dilution with autologous plasma. To aliquots of 180 μL PRP, 20 μL of various dilutions of ADP (5, 10, and 25 μmol/L; Sigma) or collagen (2.5, 5, and 10 μg/mL; Nycomed, Munich, Germany) were added and the change in the optical density was monitored using APACT aggregometer (Labor Timer, Ahrensburg, Germany) with continuous stirring at 37°C. In some instances, PRP was incubated with anti-Sita alloantibodies as described above. After stimulation with 10 μg/mL collagen, platelet aggregation was recorded as described above.
RESULTS
Serologic identification and family studies.
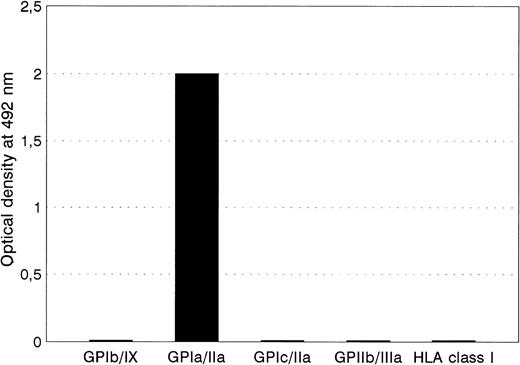
When serum from the mother Sit was tested against autologous, paternal, or donor panel platelets using a platelet adhesion immunofluorescence test, a positive reaction was only observed with paternal platelets (data not shown). An IgG antibody from serum Sita reacted strongly with GPIa/IIa immobilized by MoAb Gi9 from paternal platelets only, but not with epitopes on GPIc/IIa, GPIIb/IIIa, GIb/IX, and HLA class I (Fig 1). Platelets from 11 typed panel donors, including those of Bra and Brbhomozygous individuals, did not react either. These results indicate that the Sita serum recognizes a new low-frequency platelet alloantigen residing on GPIa/IIa complex. Because the Sitaalloantigenic determinants could be immobilized by MoAb directed against GPIa/IIa only, but not by MoAb specific for GPIc/IIa complex, Sita epitopes are probably localized on the GPIa subunit.
The reactivities of anti-Sita with paternal platelets in MAIPA assay using MoAbs FMC25 (anti-GPIb/IX), Gi9 (anti-GPIa/IIa), SAM-1 (anti-GPIc/IIa), Gi5 (anti-GPIIb/IIIa), and B1G6 (anti-β2m) as capture antibodies.
The reactivities of anti-Sita with paternal platelets in MAIPA assay using MoAbs FMC25 (anti-GPIb/IX), Gi9 (anti-GPIa/IIa), SAM-1 (anti-GPIc/IIa), Gi5 (anti-GPIIb/IIIa), and B1G6 (anti-β2m) as capture antibodies.
In a population of 400 unselected donors, GPIa/IIa of 1 individual (Dre) carried the Sita antigen. Figure 2 shows the pedigrees of the family Dre and the index family Sit. Phenotyping analysis of both families demonstrated that Sita antigen is inherited as an autosomal dominant trait. In addition, all Sita (+) individuals were also phenotyped for Bra and Brb(Table 1). Three Brb homozygous Sita (+) individuals (B.II.2, II.4, and III.3) were found (Fig 2B) in family Dre, indicating that Sita alloantigen is inherited with the Brb allele.
The pedigrees of the index family Sit (A) and family Dre (B). Solid symbols represent Sita (+), open symbols represent Sita (−) individuals. The child with NAIT is indicated. The Bra and Brb phenotypes of Sita-phenotyped individuals are shown.
The pedigrees of the index family Sit (A) and family Dre (B). Solid symbols represent Sita (+), open symbols represent Sita (−) individuals. The child with NAIT is indicated. The Bra and Brb phenotypes of Sita-phenotyped individuals are shown.
The 2 cDNA Sequences (Bases 807, 1648, 2531, and 2532) of the Family Sit (A) and Family Dre (B) and Their Br and Sit Phenotypes
| Donor . | Genotypes . | Phenotypes . | ||||
|---|---|---|---|---|---|---|
| 807 . | 1648 . | 2531 . | 2532 . | Brb/Bra . | Sita . | |
| A.I.1 | C/C | G/A | T/C | G/A | +/+ | + |
| A.I.2 | C/T | G/A | C/C | G/A | +/+ | − |
| A.II.1 | C/C | G/G | T/C | G/G | +/− | + |
| B.II.1 | C/C | G/A | T/C | G/A | +/+ | + |
| B.II.2 | C/C | G/G | T/C | G/G | +/− | + |
| B.II.3 | C/T | G/A | C/C | G/A | +/+ | − |
| B.II.4 | C/C | G/G | T/C | G/G | +/− | + |
| B.II.5 | C/T | G/G | C/C | G/G | +/− | − |
| B.II.6 | C/T | G/A | C/C | G/A | +/+ | − |
| B.II.7 | T/T | G/G | C/C | G/G | +/− | − |
| B.III.1 | C/T | G/A | C/C | G/A | +/+ | − |
| B.III.2 | C/C | G/G | C/C | G/G | +/− | − |
| B.III.3 | C/C | G/G | T/C | G/G | +/− | + |
| B.III.4 | C/T | G/A | C/C | G/A | +/+ | − |
| B.III.5 | T/T | G/G | C/C | G/G | +/− | − |
| Donor . | Genotypes . | Phenotypes . | ||||
|---|---|---|---|---|---|---|
| 807 . | 1648 . | 2531 . | 2532 . | Brb/Bra . | Sita . | |
| A.I.1 | C/C | G/A | T/C | G/A | +/+ | + |
| A.I.2 | C/T | G/A | C/C | G/A | +/+ | − |
| A.II.1 | C/C | G/G | T/C | G/G | +/− | + |
| B.II.1 | C/C | G/A | T/C | G/A | +/+ | + |
| B.II.2 | C/C | G/G | T/C | G/G | +/− | + |
| B.II.3 | C/T | G/A | C/C | G/A | +/+ | − |
| B.II.4 | C/C | G/G | T/C | G/G | +/− | + |
| B.II.5 | C/T | G/G | C/C | G/G | +/− | − |
| B.II.6 | C/T | G/A | C/C | G/A | +/+ | − |
| B.II.7 | T/T | G/G | C/C | G/G | +/− | − |
| B.III.1 | C/T | G/A | C/C | G/A | +/+ | − |
| B.III.2 | C/C | G/G | C/C | G/G | +/− | − |
| B.III.3 | C/C | G/G | T/C | G/G | +/− | + |
| B.III.4 | C/T | G/A | C/C | G/A | +/+ | − |
| B.III.5 | T/T | G/G | C/C | G/G | +/− | − |
Immunochemical investigations.
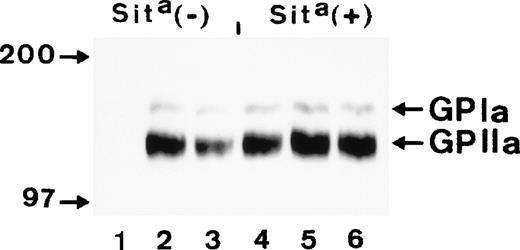
To confirm the localization of Sita alloantigen on GPIa/IIa, immunoprecipitation analysis of biotin-labeled glycoproteins derived from Sita (+) and Sita (−) Bra/Brb heterozygous individuals was performed. As shown in Fig 3, anti-Sita(lanes 1 and 4) only precipitated GPIa/IIa complex from Sita (+) platelets, but not from Sita (−) platelets. In the control experiments, anti-Bra (lanes 2 and 5) and anti-Brb (lanes 3 and 6) precipitated GPIa/IIa complex from both Bra/Brb heterozygous individuals.
Immunoprecipitation analysis of biotin surface-labeled platelets derived from a Sita (−) and a Sita(+) individual with anti-Sita (lanes 1 and 4), anti-Bra (lanes 2 and 5), and anti-Brb (lanes 3 and 6). Immunoprecipitates were separated on 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under nonreduced conditions, transferred onto nitrocellulose membrane, and visualized using streptavidin-horseradish peroxidase and chemiluminescent substrate.
Immunoprecipitation analysis of biotin surface-labeled platelets derived from a Sita (−) and a Sita(+) individual with anti-Sita (lanes 1 and 4), anti-Bra (lanes 2 and 5), and anti-Brb (lanes 3 and 6). Immunoprecipitates were separated on 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under nonreduced conditions, transferred onto nitrocellulose membrane, and visualized using streptavidin-horseradish peroxidase and chemiluminescent substrate.
Genetic analysis.
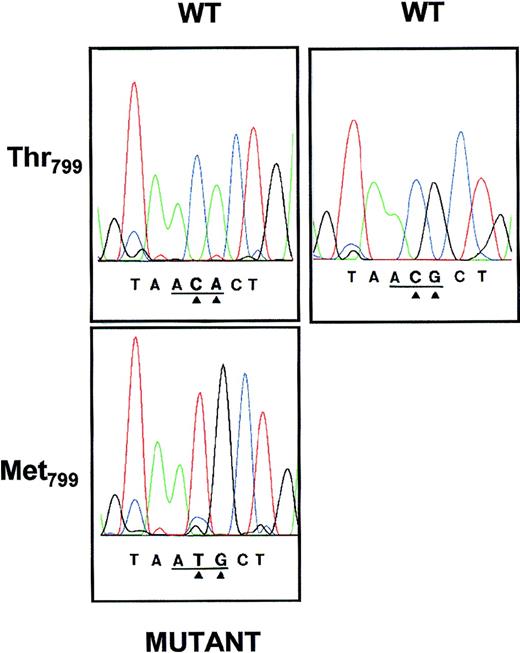
To analyze the nucleotide sequence encoding for GPIa, platelet mRNA was sequentially amplified by reverse transcription using 8 sets of primers, subcloned, and sequenced. Nucleotide sequence analysis of the 554 bp encompassing nucleotides 2304-2858 derived from a Sita (+) individual showed 2-base mutations C2531A2532 →T2531G2532(Fig 4) in 10 of 12 subcloned examined, consistent with the hypothesis that all Sita (+) individuals found to date are heterozygous for this character. These mutations predicted Thr799(AC2531A2532) in the Sita (−) and Met799 (AT2531G2532) in the Sita (+) phenotype. Analysis of the other 7 regions of GPIa mRNA showed no other nonconservative nucleotide differences.
Nucleotide sequence analyses of amplified GPIa cDNA derived from 2 Sita (+) individuals (A.I.1 and A.II.1; Fig 2). PCR product encompassing nucleotides 2304-2854 was subcloned into the plasmid vector pGEM-5Zf and sequenced on both strands using primers corresponding to the SP6 and T7 RNA polymerase promotor sequences. The base exchanges of the wild-type (WT) CA or CG to the mutant TG at positions 2531 and 2532 (arrows) result in a Thr799 (ACA or ACG) → Met799 (ATG) substitution.
Nucleotide sequence analyses of amplified GPIa cDNA derived from 2 Sita (+) individuals (A.I.1 and A.II.1; Fig 2). PCR product encompassing nucleotides 2304-2854 was subcloned into the plasmid vector pGEM-5Zf and sequenced on both strands using primers corresponding to the SP6 and T7 RNA polymerase promotor sequences. The base exchanges of the wild-type (WT) CA or CG to the mutant TG at positions 2531 and 2532 (arrows) result in a Thr799 (ACA or ACG) → Met799 (ATG) substitution.
Genotyping analysis of Sita.
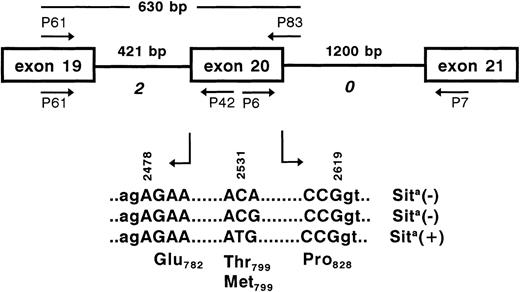
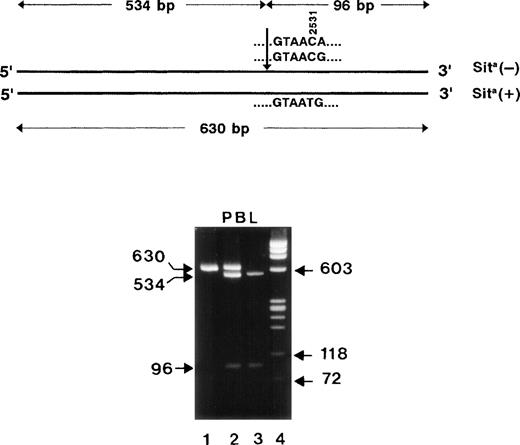
To establish genomic DNA typing, we first elucidated the exon-intron boundaries surrounding the Sit polymorphic base by PCR. When PCR with genomic DNA was performed using exonic primer pairs P61, P42 and P6, P7, fragments of 580 bp and nearly 1,400 bp were obtained. Nucleotide sequence analysis of both PCR products identified 421-bp and 1.2-kb introns (Fig 5) with conserved donor and acceptor splice junctions, which are classified as phase 2 and 0, respectively. These results demonstrate that these 2 introns enclose a 142-bp (bases 2478-2619) exon encoding 47 amino acids. Based on these intron sequences, reverse primer P83 downstream of the 142-bp exon was constructed. After 35 cycles of amplification using primer P61 as forward primer, the expected 630-bp product was obtained from genomic DNA derived from peripheral blood lymphocytes (Fig 6). The PCR product was digested withMae III endonuclease, which cleaves 5′-GTAAC-3′ (wild-type) but not 5′-GTAAT-3′ sequences (mutant). Sita-negative individuals could be clearly distinguished from Sita heterozygous individuals by the absence of the uncut 630-bp band. The results of genotyping by PCR-RFLP of 400 Sita-phenotyped donors were consistent with the serological phenotyping.
PCR strategy for the elucidation of the GPIa gene surrounding the polymorphic base at position 2531 (arrow). Genomic DNA was amplified by PCR using primer pairs P61, P42 and P6, P7. PCR products were sequenced for the determination of exon-intron boundaries. The polymorphic exon 20 (bases 2478-2619) encoding the amino acids Glu782-Pro828 is flanked by 2 introns (421 and 1,200 bp) of phases 2 and 0 (italic). For genotyping analysis of the polymorphic bases 2531 and 2532 (bold), a 630-bp fragment was amplified using primer pair P61, P83.
PCR strategy for the elucidation of the GPIa gene surrounding the polymorphic base at position 2531 (arrow). Genomic DNA was amplified by PCR using primer pairs P61, P42 and P6, P7. PCR products were sequenced for the determination of exon-intron boundaries. The polymorphic exon 20 (bases 2478-2619) encoding the amino acids Glu782-Pro828 is flanked by 2 introns (421 and 1,200 bp) of phases 2 and 0 (italic). For genotyping analysis of the polymorphic bases 2531 and 2532 (bold), a 630-bp fragment was amplified using primer pair P61, P83.
Restriction map of the 630-bp PCR products (top). The arrow indicates the position of the restriction site for MaeIII endonuclease. The length of the restriction fragments is shown above. RFLP analysis of PCR-amplified genomic DNA derived from peripheral blood cells (PBL) of Sita-phenotyped individuals. DNA fragments were analyzed on 1.6% agarose gel stained with ethidium bromide (bottom). The undigested 630-bp PCR product is shown in lane 1. Lanes 2 and 3 represent the analysis of MaeIII-digested PCR product derived from DNA of a Sita (+) heterozygous individual and a Sita (−) individual, respectively. Lane 4, DNA size standards (pBR328 DNA.BglI + pBR328 DNA.HinfI).
Restriction map of the 630-bp PCR products (top). The arrow indicates the position of the restriction site for MaeIII endonuclease. The length of the restriction fragments is shown above. RFLP analysis of PCR-amplified genomic DNA derived from peripheral blood cells (PBL) of Sita-phenotyped individuals. DNA fragments were analyzed on 1.6% agarose gel stained with ethidium bromide (bottom). The undigested 630-bp PCR product is shown in lane 1. Lanes 2 and 3 represent the analysis of MaeIII-digested PCR product derived from DNA of a Sita (+) heterozygous individual and a Sita (−) individual, respectively. Lane 4, DNA size standards (pBR328 DNA.BglI + pBR328 DNA.HinfI).
Amino acid 799 of α2 integrin is encoded by different codons.
Because different triplets theoretically could code the wild-type Thr799 (ACA, ACC, ACG, and ACT), we analyzed the members of the families Sit and Dre (Table 1) by direct nucleotide sequencing. Two different codons, ACA and ACG (Fig 4), both of which encode the wild-type Thr799 form of GPIa, were found among Sita (−) homozygous and Sita (+) heterozygous individuals (Table 1). To determine the distribution of the codon usage for Thr799, we analyzed 22 unselected blood donors by DNA sequencing. Nineteen individuals presented with codons ACG/ACG, 2 with ACG/ACA, and 1 with ACA/ACA. Our results suggest that the codon usage for Thr799 is 90.9% ACG and 9.1% ACA. Other conceivable codons (ACC and ACT) were not found in our population so far. It is of note that the Thr799 ACA allele is uniquely recognized by the restriction enzyme TspRI and therefore represents a potentially useful genetic marker for RFLP (data not shown).
Thus, the mutation C2531T in Sita (+) individuals most probably occurred in the high-frequeny codon ACG (Thr) to ATG (Met) due to a 1-point mutational event.
Linkage association between C2531T dimorphism with A1648G, C807T and A2532G polymorphisms.
To analyze the linkage associations between C2531T with A2532G, A1648G, and C807T polymorphisms, we genotyped all members of the families Sit and Dre for A1648G, C807T, and A2532T dimorphisms by PCR-RFLP, PCR-SSP, and direct nucleotide sequencing analysis, respectively. Their genotypes are summarized in Table 1.
In accordance with our previous phenotyping results by MAIPA assay, we could confirm that the Sita allele segregated together with the Brb allele. Four Sita (+) individuals (A.II.1, B.II.2, B.II.4, and B.III.3) are homozygous for the G1648 allele.
Furthermore, we observed that all Sita (+) individuals were homozygous for the C807, indicating the linkage association between the Sita allele with the C807 allele.
Finally, we found that the G1648 haplotype exclusively segregated with the G2532 allele. All Brbhomozygous family members are homozygous for the G2532allele. In contrast, the A1648 haplotype most probably segregates with the A2532 allele, because all Bra heterozygous individuals were found to be heterozygous G/A at position 2532.
These observations indicate a linkage disequilibrium between polymorphisms at positions 807, 1648, 2531, and 2532. Under the consideration of the linkage associations between the polymorphisms at bases 807/837/873,28 we could now define 4 α2gene alleles by 6 dimorphisms at positions 807/837/873, 1648, 2531, and 2532 (Table 2).
The Frequencies of 4 2 Gene Alleles Defined by 6 Dimorphisms
| Allele . | cDNA Positions of α2 Dimorphisms . | Genfrequency . | |||||
|---|---|---|---|---|---|---|---|
| 807 . | 837 . | 873 . | 1648 . | 2531 . | 2532 . | ||
| 1 | T | T | A | G | C | G | 0.394* |
| 2 | C | T | G | G | C | G | 0.529* |
| 3 | C | C | G | A | C | A | 0.076* |
| 4 | C | T | G | G | T | G | <0.01 |
| Allele . | cDNA Positions of α2 Dimorphisms . | Genfrequency . | |||||
|---|---|---|---|---|---|---|---|
| 807 . | 837 . | 873 . | 1648 . | 2531 . | 2532 . | ||
| 1 | T | T | A | G | C | G | 0.394* |
| 2 | C | T | G | G | C | G | 0.529* |
| 3 | C | C | G | A | C | A | 0.076* |
| 4 | C | T | G | G | T | G | <0.01 |
Kritzig et al.29
Analysis of recombinant GPIa allelic isoforms.
To examine the involvement of amino acid 799 in the formation of the Sita antigenic determinants, we transfected allele-specific expression vectors encoding for the Lys505Thr799, Glu505Thr799, and Glu505Met799 GPIa into CHO cells. Stable transfectants expressing GPIa recombinant proteins were surface-labeled with biotin and analyzed by immunoprecipitation. As shown in Fig 7, anti-Sita (lanes 2) precipitated exclusively the Met799 GPIa isoform, but not both Thr799 isoforms having either glutamic acid or lysine at position 505. The associated band (110 kD) most probably represents endogenous hamster β1 integrin subunit, which is coprecipitated with the human GPIa subunit (Mr 150 kD). In the control experiment, anti-Brb (lanes 1) reacted with Thr799 and Met799 GPIa isoforms, which have the amino acid glutamic acid at position 505. In contrast, anti-Bra (lane 3) only recognized the Lys505GPIa isoform.
Immunoprecipitation analysis of allele-specific recombinant GPIa isoforms. Recombinant forms of GPIa/IIa complex were produced in CHO cells transfected either with Glu505Thr799 (left panel), Glu50Met799 (middle panel), or Lys505Thr799 (right panel) form of GPIa. After surface labeling with biotin, cell lysates were immunoprecipitated with anti-Brb (lanes 1), anti-Sita (lanes 2), and anti-Bra (lanes 3). Immunoprecipitates were analyzed on 7.5% SDS-PAGE under nonreduced conditions, transferred onto nitrocellulose membrane, and visualized using chemiluminescence substrate.
Immunoprecipitation analysis of allele-specific recombinant GPIa isoforms. Recombinant forms of GPIa/IIa complex were produced in CHO cells transfected either with Glu505Thr799 (left panel), Glu50Met799 (middle panel), or Lys505Thr799 (right panel) form of GPIa. After surface labeling with biotin, cell lysates were immunoprecipitated with anti-Brb (lanes 1), anti-Sita (lanes 2), and anti-Bra (lanes 3). Immunoprecipitates were analyzed on 7.5% SDS-PAGE under nonreduced conditions, transferred onto nitrocellulose membrane, and visualized using chemiluminescence substrate.
These findings demonstrate that amino acid Met799 directly controls the expression of the Sita epitopes, whereas residue 505 (lysine or glutamic acid) is responsible for the formation of Bra and Brb alloantigenic determinants, respectively.
Effect of the Sit Thr799Met dimorphism and anti-Sita alloantibodies on platelet function.
To determine the possible effects of Thr799Met mutation on platelet function, we studied platelet ad- hesion and platelet aggregation of Sita-phenotyped individuals.
The results of platelet adhesion to immobilized collagen type I of Sita phenotyped platelets were summarized in Table 3. The platelet adhesion response to high collagen (50 μg/mL) and low collagen concentration (1 μg/mL) of Sita (+) platelets was indistinguishable from Sita (−) platelets.
Adhesion of Platelets to Collagen Type I in the Absence (None) and in the Presence of Affinity-Purified IgG From Normal Human Serum, Anti-Sita, or Murine MoAb Gi9 Specific for GPIa/IIa
| Platelets Phenotypes . | IgG . | Collagen Type I (OD410) . | |
|---|---|---|---|
| 50 μg/mL . | 1 μg/mL . | ||
| Sita (−) | None | 0.79 ± 0.11 | 0.68 ± 0.11 |
| Normal serum | 1.15 ± 0.20 | 0.86 ± 0.20 | |
| Anti-Sita | 1.25 ± 0.15 | 1.01 ± 0.08 | |
| MoAb Gi9 | 0.29 ± 0.04 | 0.20 ± 0.03 | |
| Sita (+) | None | 0.85 ± 0.25 | 0.62 ± 0.18 |
| Normal serum | 1.20 ± 0.15 | 0.94 ± 0.13 | |
| Anti-Sita | 1.32 ± 0.12 | 1.01 ± 0.08 | |
| MoAb Gi9 | 0.28 ± 0.12 | 0.21 ± 0.05 | |
| Platelets Phenotypes . | IgG . | Collagen Type I (OD410) . | |
|---|---|---|---|
| 50 μg/mL . | 1 μg/mL . | ||
| Sita (−) | None | 0.79 ± 0.11 | 0.68 ± 0.11 |
| Normal serum | 1.15 ± 0.20 | 0.86 ± 0.20 | |
| Anti-Sita | 1.25 ± 0.15 | 1.01 ± 0.08 | |
| MoAb Gi9 | 0.29 ± 0.04 | 0.20 ± 0.03 | |
| Sita (+) | None | 0.85 ± 0.25 | 0.62 ± 0.18 |
| Normal serum | 1.20 ± 0.15 | 0.94 ± 0.13 | |
| Anti-Sita | 1.32 ± 0.12 | 1.01 ± 0.08 | |
| MoAb Gi9 | 0.28 ± 0.12 | 0.21 ± 0.05 | |
Biotinylated platelets from Sita (−) individuals or Sita (+) individuals (B.III.2 and 3 in Fig 2) were allowed to adhere to microtiter wells coated with either BSA (10 mg/mL) or collagen type I (50 and 1 μg/mL). Results are represented as the means of optical density (OD410) ± SD for triplicate samples. The OD410 of platelet adhesion to wells coated with BSA was less than 0.16 ± 0.07. The number of donors tested was 2.
Furthermore, we analyzed the effect of anti-Sitaalloantibodies on platelet adhesion. In comparison to the control experiment with MoAb Gi9 specific for a functional epitope of the GPIa/IIa complex, anti-Sita did not inhibit platelet adhesion to type I collagen. Similar results were obtained with type III and V collagens (data not shown).
However, in standard aggregation assay (Fig8), the platelet aggregation response to low collagen concentration (2.5 μg/mL) in the Sita (+) individual (B.III.3, see Fig2) was diminished in comparison to the Sita (−) individual (B.III.2). In contrast, the platelet aggregation of the Sita (+) individual after stimulation with high concentration of collagen (10 μg/mL) was indistinguishable from the Sita (−) individual. The addition of other agonists (ADP and ristocetin) to platelets from Sita (+) individuals resulted in completely normal aggregation response (data not shown).
Platelet aggregation of a Sita (+) donor (B.III.3; curves 1 and 3) and a Sita (−) individual (B.III.2; curves 2 and 4) after stimulation with 2.5 μg/mL collagen (curves 1 and 2) or 10 μg/mL collagen (curves 3 and 4).
Platelet aggregation of a Sita (+) donor (B.III.3; curves 1 and 3) and a Sita (−) individual (B.III.2; curves 2 and 4) after stimulation with 2.5 μg/mL collagen (curves 1 and 2) or 10 μg/mL collagen (curves 3 and 4).
Similar results were observed with 3 other Sita (+) individuals (B.II.1, B.II.2, and B.II.4; data not shown). Because GPIa of both individuals (B.III.3 v B.III.2) only differs at nucleotide 2531 (T or C), but carries identical nucleotide sequences at polymorphic positions 807 (homozygous C) and 1648 (homozygous G), our results suggest that only the mutation Thr→Met at position 799 affects the function of GPIa/IIa as a collagen receptor. The differences in response to collagen thus cannot be explained by possible differences in GPIa expression density related to the base C807T polymorphism.
When platelet aggregation by collagen was evaluated in the presence of purified anti-Sita, no inhibition was found (data not shown).
DISCUSSION
We report here the identification and characterization of a new low-frequency platelet alloantigen, Sita, in whites that was involved in a case of severe NAIT. A study in 400 unrelated individuals identified another Sita (+) individual, showing that this low frequency alloantigen is not restricted to a single family. Studies with immunochemical techniques allowed us to localize the Sita alloantigenic determinant on GPIa. Examination of the nucleotide sequence derived from a Sita (+) father showed a mutation comprising 2 bases CA→TG at positions 2531 and 2532, respectively, in a heterozygous state in the GPIa mRNA transcripts.
An analysis of the exon-intron structure allowed us to define the polymorphic site on an exon with a length of 142 bp. A comparison with other α subunits of integrin showed that this exon corresponds to exon 20 of the αx and αMgenes.38 39 All 3 exons have identical length (142 bp) and are flanked by introns with identical phases (2 and 0).
Interestingly, we also discovered an additional silent polymorphism at base 2532 (G or A) adjacent to the polymorphic base 2531. By direct nucleotide sequencing of genomic DNA derived from Sita(−) individuals, we could define 2 different codons, ACG and ACA, that encode the wild-type Thr799 form of GPIa.
Because the Sita alloantigen is a low-frequency antigen, it seems to represent a recent mutational event. According to the current observations that most polymorphisms responsible for platelet alloantigens are point mutations of the wild-type allele,40the Sita alloantigen most probably occurred due to the single base mutation C→T at position 2531 of the high-frequency wild-type allele ACG (90.9%) rather than the low-frequency ACA allele (9.1%).
Recently, Kritzig et al29 could define 3 allelic differences by 4 dimorphisms at positions 807, 837, 873, and 1648. Our data indicate that the A2532 allele may be linked to the C807A1648 allele, whereas the G2532allele can be associated either with the C807G1648 or T807G1648allele. These observations are in accordance with the frequencies of C807A1648 and A2532 genotypes in our population (10% v 9.1%). However, analysis of A1648 and G1648 genotyped individuals among larger populations is necessary to establish this association. Interestingly, the GPIa cDNA derived from fibroblast cell library also represented the rare C807A1648A2532allelic form.34
The mutation C2531T results in a substitution of the polar uncharged amino acid threonine into a hydrophobic nonpolar residue methionine at position 799 on mature GPIa polypeptide. Stable expression of recombinant allele-specific GPIa in CHO cells led to the confirmation that the single amino acid substitution Thr799Met was sufficient to induce Sita epitope formation recognized by the Sita antibody present in the mother's serum. As expected from our previous observations,6 we could demonstrate in this study that the residue 505 (Lys or Glu) specifically controls the formation of Bra and Brb alloantigenic determinants, respectively.
Functional studies showed no influence of the Sit-polymorphism on platelet adhesion to immobilized collagen. In contrast, after stimulation with a low concentration of collagen, the platelet aggregation response of Sita (+) platelets was reduced as compared with the response of platelets from Sita (−) individuals.
Until now, the region comprising the amino acid 799 has so far not been implicated in the adhesion to collagen, which was mapped to the I-domain of the GPIa subunit.41,42 Recently, several lines of evidence suggest that activation of platelets occurs via a 2-site, 2-step model in which the initial interaction occurs via the GPIa/IIa, allowing binding to GPVI leading to activation.27 It may be speculated that the mutation Thr→Met at position 799 affects the activation of platelets, possibly through interaction between GPIa/IIa and GPVI.
Although the aggregation response to collagen in Sita (+) carriers was diminished, none of these individuals had signs of an altered hemostasis.
In NAIT, alloantibodies against the Bra epitopes on GPIa often induces moderate thrombocytopenia in affected children.18 It is interesting to note that NAIT due to anti-Sita was associated with severe thrombocytopenia. It is conceivable that the additional functional defect of GPIa/IIa complex resulted in the pronounced clinical course of this child. An additional influence of anti-Sita alloantibodies on platelet adhesion and aggregation by collagen could not be observed.
In conclusion, we have identified that a single point mutation Thr799Met adjacent to a hot spot in the GPIa gene is responsible for the formation of a new low-frequency platelet alloantigen, which we termed Sita. In contrast to other known platelet alloantigens, this point mutation appears to affect platelet function.
ACKNOWLEDGMENT
The authors thank Dr H. Schachinger, who kindly referred this NAIT case to us. We are grateful to Micaela Boehringer and Monika Kummel for their technical assistance. Our gratitude is also extended to the families concerned for their cooperation in this study.
Supported by Grant DFG Sa480/2-1. This work is part of the PhD theses of J.A. and U.J.H.S.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Sentot Santoso, PhD, Institute for Clinical Immunology and Transfusion Medicine, Justus Liebig University Giessen, Langhansstr. 7, D-35392 Giessen, Germany; e-mail:Sentot.Santoso@immunologie.med.uni-giessen.de.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal