Expression of the von Willebrand factor (vWF) gene is restricted to the endothelial and megakaryocyte lineages. Within the endothelium, expression of vWF varies between different vascular beds. We have previously shown that the human vWF promoter spanning a region between −2182 (relative to the start site of transcription) and the end of the first intron contains information for environmentally responsive, vascular bed-specific expression in the heart, skeletal muscle, and brain. In the present study, we cloned the mouse vWF (mvWF) promoter and studied its function in cultured endothelial cells and transgenic mice. In transient transfection assays, the mvWF gene was found to be regulated by distinct mechanisms in different endothelial cell subtypes. In independent lines of transgenic mice, an mvWF promoter fragment containing DNA sequences between −2645 and the end of the first intron directed endothelial cell-specific expression in the microvascular beds of the heart, brain, and skeletal muscle as well as the endothelial lining of the aorta. In 1 line of mice, reporter gene activity was also detected in bone marrow megakaryocytes. Taken together, these findings suggest that both the mouse and human vWF promoters are regulated by vascular bed-specific mechanisms.
ALL BLOOD VESSELS AND arteries are lined with a continuous monolayer of endothelial cells. One feature of the endothelium is its rich diversity of regional and organ-specific phenotypes.1-3 This high degree of heterogeneity is observed at morphological, structural, and functional levels as well as in antigen composition and response to growth factors. Endothelial diversity arises from the physiological adaptation of individual cells to the surrounding environment. Indeed, the endothelium may be viewed as a consortium of small enterprises of cells located within blood vessels of different tissues. Although united in certain common functions, each enterprise is uniquely adapted to meet the specific demands of the local environment. The molecular basis of vascular diversity is poorly understood; little is known about the mechanisms that underlie the adaptive response of the endothelium to the microenvironment. One approach to this problem is to study the mechanisms underlying endothelial cell-specific gene regulation.
von Willebrand factor (vWF) is a multimeric glycoprotein that mediates adhesion of platelets to the underlying endothelium and serves as a carrier for the coagulation factor VIII.4,5 Expression of the vWF gene is restricted to endothelial and megakaryocytic lineages. Previous investigations have demonstrated the existence of regional variations in vWF protein and mRNA levels within the vascular tree.6-10 The vWF gene is therefore a marker of endothelial cell heterogeneity, and the elucidation of its transcriptional regulatory mechanisms may provide important insights into genetic programs that govern the production of vascular diversity.
In recent studies of transgenic mice,11-13 the human vWF promoter was found to direct expression to discrete regions of the vascular tree. For example, a 733-bp sequence of the human vWF gene contained information for expression in blood vessels of the brain,11 whereas a larger promoter fragment containing 2,182 bp 5′ flanking sequence, the first exon, and the first intron also directed LacZ expression to the microvascular endothelial cell lining of skeletal muscle and heart.12Expression in the microvascular bed of the heart was shown to be regulated by a cardiomyocyte-dependent signaling pathway.13
These results suggested that the vWF gene is regulated in a modular fashion. According to this model, differential expression of vWF is mediated not by the relative activity of a single transcriptional pathway, but rather by the sum of distinct vascular bed-specific signaling pathways, each beginning in the extracellular milieu and ending at distinct regions of the promoter. However, an alternative explanation was that the mouse endothelium lacked the capacity to activate the human promoter in an authentic pattern. To exclude the existence of interspecies differences in vWF gene regulation, we have cloned the mouse vWF (mvWF) promoter and tested its function in transgenic mice. Under in vivo conditions, the mouse promoter was found to direct cell type-specific expression in subpopulations of endothelial cells within the heart, skeletal muscle, brain, and aorta. The overlapping expression pattern of the mouse and human transgenes strongly suggest that the vWF gene is indeed regulated by vascular bed-specific mechanisms.
MATERIALS AND METHODS
Library screening and DNA cloning.
A 300-bp polymerase chain reaction (PCR) amplicon, generated from murine genomic DNA with oligonucleotides corresponding to exon 4-5 sequence, was used to screen a mouse ES cell P1 library (Genome Systems Inc, St Louis, MO). Primer sequences used were sense 5′ CTCCGTGTATCTTGGGG and antisense 5′ TAGGTAGAGTCCTTCGG. Three separate P1 clones obtained from the screen were hybridized with a32P-labeled probe spanning the −487 to +246 region of the human vWF promoter under low stringency conditions. One of the 3 clones (P1-7910) that yielded a positive signal was digested withSac I, and the resulting DNA fragments were purified and cloned into pUC19. Colonies containing mvWF promoter sequences were identified by low-stringency hybridization with a 32P-labeled probe spanning the −487 to +246 region of the human vWF gene. Sequence analysis of 1 of the positive clones (mvWF-pUC19-5) showed high homology to the human vWF promoter and contained a region between −110 and +3000 bp relative to the transcriptional start site. Additional radiolabeled DNA probes, generated from the 5′ end of the mvWF-pUC19-5 mouse sequence, were used to screen PstI-digested fragments of P1-7910 cloned into the pBluescript vector (Stratagene, La Jolla, CA). One of these clones (pBlue-PP26) contained mvWF promoter sequence between −2645 and +1092. For restriction mapping, P1-7910 and E129 mouse tail-derived DNA was digested with restriction enzymes, loaded on a 0.8% agarose gel, and transferred to nylon membrane. The membrane was hybridized with32P-labeled DNA probes, corresponding to sequences within mvWF promoter. All sequences were determined using automated sequencing technology on a Model 373A DNA (Perkin-Elmer Corp, Applied Biosystems Division, Foster City, CA).
For transient transfections, various lengths of the mouse promoter were inserted into the pGL2-Basic vector (Promega Corp, Madison, WI). To generate the full-length PIE construct, mvWF-pUC19-5 was digested withSpe I, blunt-ended with Klenow polymerase, and then digested with Sac I. The 1.3-kb fragment containing promoter sequences between −110 and +1257 was ligated into the Sac I and blunted Xho I sites of pGL2-Basic. A PCR-generated fragment corresponding to the 3′ end of the first intron was ligated into the above-described plasmid digested with Nco I andHindIII (blunt-ended with Klenow polymerase). To generate the −110 construct, a PCR-generated fragment was obtained (with the following primers: sense, 5′ TGTGGTTTGTCCAAACTCATCAAT; and antisense, 5′ TGCAATAGCTCCAAGTTGCCA), digested with SacI, and ligated into the Xho I (blunt-ended with Klenow polymerase) and Sac I sites of pGL2-Basic. To generate the −110E construct, a PCR generated fragment was generated (with the following primers: sense, 5′ TGTGGTTTGTCCAAACTCATCAAT; and antisense, 5′ CTTGCCCATACAAACAGGGGC), digested with SacI, and ligated into the Xho I (blunt-ended with Klenow polymerase) and Sac I sites of pGL2-Basic. To generate the P and PE constructs, the 2.8-kb Sac I/Kpn I fragment of pBlue-PP26 was ligated into Sac I/Kpn I sites of −110 and −110E, respectively. To generate the −110IE construct, mvWF-pUC19-5 was digested with Spe I (blunted) andSac I. The mvWF fragment containing DNA sequence between −110 and +1258 was inserted into pGL2-Basic cut with XhoI (blunt-ended with Klenow polymerase) and Sac I. A PCR-generated fragment spanning the 5′ end of the first intron was then inserted into the Nco I-HindIII sites of the plasmid described above. All constructions are verified by dideoxy DNA sequencing.
To create the transgenic construct, mvWFlacZ, aHindIII/Nco I fragment of mvWF-pUC19-5 was inserted into a plasmid containing LacZ cDNA coupled to the SV40 poly(A) tail (gift from Janet Rossant, Mount Sinai Hospital, Toronto, Ontario, Canada). An Nco I-digested PCR-generated fragment corresponding to the 3′ end of the first intron was ligated into the above-described plasmid digested with Nco I and Nru I (blunt-ended with Klenow polymerase). In the final step, a 3.7-kbPst I fragment from pBlue-PP26 containing promoter sequences −2645 to +1092 was ligated into the above-described plasmid.
RNA isolation, primer extension, and reverse transcriptase-PCR (RT-PCR).
Mouse tissue was harvested for total RNA using a guanidinium thiocyanate phenol-chloroform single-step extraction (Stratagene). Primer extension analysis was performed using an antisense oligonucleotide corresponding to the sequence from position +114 to +153 of the mvWF gene (5′ AAGCCATTTTCCTCCTGCGCAACTGCTGGATGGATCTGCTCAG). The oligonucleotide was end-labeled with [γ-32P]ATP using polynucleotide kinase and 5 × 105 counts/min were incubated with 20 μg total RNA. The mixture was denatured at 80°C for 15 minutes and annealed at 42°C in a reaction containing 50 μg total RNA, 50 mmol/L KCl, and 50 mmol/L Tris, pH 8.3, for 3 hours. First-strand synthesis was performed with avian myleoblastosis virus (AMV) reverse transcriptase (Promega Corp) and dNTP. Samples were extracted with phenol chloroform/isoamyl alcohol and precipitated with ethanol, and the reaction products were resolved on a 6% polyacrylamide denaturing gel. A dideoxy sequencing reaction of the mvWF-pUC19-5 template was performed with the same primer and included as size markers. Reactions were repeated from different RNA samples to confirm the results. For RT-PCR, first-strand cDNA synthesis and PCR reactions were performed as previously described.12 The primers used were as follows: (A) 5′ GCCATTGTTTCCCGCTGG; (B) 5′ CCAGGGCTCTGTGGTGGC; (C) 5′ CTTGGAGCTATTGCAGGCAG; (D) CTTGACAGCAGGTCGGCTCA; and (E) 5′ TGCCCATACAAACAGGGGCT.
Tissue culture and transient transfections.
Bovine aortic endothelial cells (BAEC) were generously provided by Robert D. Rosenberg (Massachusetts Institute of Technology, Cambridge, MA). Calf pulmonary endothelial cells (CPAE) were obtained from Vikas Sukhatme (Beth Israel Deaconess Medical Center, Boston, MA). Py-4-1 endothelial cells, a cell line derived from hemangiomas of transgenic mice expressing the polyoma early region gene, were a generous gift from Victoria Bautch (University of North Carolina, Chapel Hill, NC).14 NIH 3T3 cells (CRL-1658) and HEK 293 cells (CRL-1573) were obtained from the American Type Culture Collection (Manassas, VA). All cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies Inc, Gaithersburg, MD) supplemented with 10% heat-inactivated fetal calf serum (FCS; Life Technologies Inc) at 37°C and 5% CO2.
Luciferase reporter plasmids were introduced into cultured cells by lipofectamine-mediated gene transfer (Life Technologies Inc). All transfections were performed in triplicate. Cells were seeded at a density of 25,000 to 50,000 cells per well in 12-well plates. At 60% to 70% confluence, the cells were incubated with liposome:DNA complexes containing 3 μL of lipofectamine and 1 μg of test plasmid and 50 ng of a control plasmid containing the Renillaluciferase reporter gene under the control of a cytomegalovirus (CMV) enhancer/promoter (Promega Corp). The transfection mixture was replaced by regular growth medium 5 hours later. Twenty-four hours later, cells were harvested and lysed according to the dual-luciferase assay system (Promega Corp). Standard firefly andRenilla luciferase activity were serially measured in a luminometer (Lumat LB 9507 model; EG&G Berthold, Bad Wilbad, Germany). Standard luciferase activity was normalized to the activity of both the Renilla luciferase reporter gene and the pGL2-Basic control plasmid.
Generation and analysis of transgenic mice.
The mvWFlacZ construct was digested with Not I andXho I, and the fragment containing the upstream promoter region, the first exon, and intron coupled to LacZ was separated from vector sequences through agarose gel electrophoresis, as previously described.11 Microinjections of fertilized mouse eggs and oviduct transfer techniques were performed as previously described.11 Founder mice were identified by Southern blot analysis and mated with wild-type FVB mice to generate stable lines of transgenic mice.11 Tissues from F1 adult mice were perfused with paraformaldehyde-containing solution, embedded in OCT compound, and quickly frozen on dry ice. Frozen sections of 10 μm were collected in a cryostat, attached to polylysine-coated slides, and incubated in a solution containing 5-bromo-4-chloro-3-indolylβ-D-galactopyranoside (X-Gal) for 24 hours at 4°C as previously described.11
Accession number.
The DNA sequences that are reported in this manuscript have been submitted to GenBank with accession no. AF152417.
RESULTS
Cloning and sequence analysis of the mvWF promoter.
To isolate the 5′-flanking region of the mvWF promoter, a mouse ES cell P1 library was screened with a probe containing mouse exon 4/5 sequence. One of the 3 positive clones (P1-7910) was found to hybridize with the human vWF probe comprising nucleotides −487 to +246. Restriction mapping and Southern blot analysis of the P1 clone and E129 mouse tail-derived genomic DNA showed the organization of the mouse promoter (Fig 1). A 3,000-bp Sac I fragment containing 110 bp 5′ flanking region, the first and second exons, the first full intron, and part of the second intron was subcloned into the pUC19 vector, and a 3,745-bp Pst I insert containing sequences between −2645 and +1100 was subcloned into the pBluescript vector. The resulting plasmids, designated mvWF-pUC19-5 and pBlue-PP26, respectively, were used for further analysis.
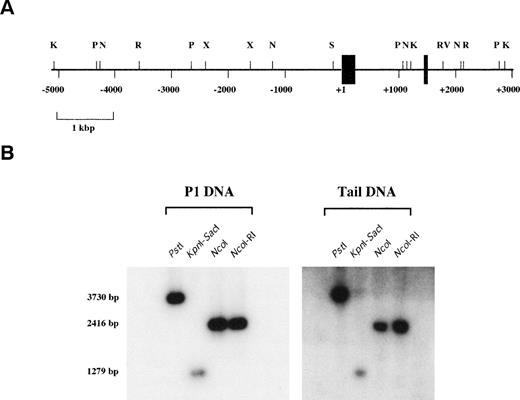
Structure of the mvWF promoter. (A) Restriction map of an 8-kb region of the mvWF gene is shown. Exons I and II are depicted as thick and thin solid boxes, respectively. Introns and upstream promoter sequence are represented by thin lines. Restriction enzymes areKpn I (K), Pst I (P), Nco I (N), EcoRI (R), Xba I (X), Sac I (S), and EcoRV (RV). The numbers shown are relative to the start site of transcription. (B) Restriction fragments of P1 DNA or DNA derived from the tail of E129 mice were resolved on 0.8% agarose gel, transferred to a nylon membrane, and then hybridized with a 32P-labeled probe derived from sequences within the first exon.
Structure of the mvWF promoter. (A) Restriction map of an 8-kb region of the mvWF gene is shown. Exons I and II are depicted as thick and thin solid boxes, respectively. Introns and upstream promoter sequence are represented by thin lines. Restriction enzymes areKpn I (K), Pst I (P), Nco I (N), EcoRI (R), Xba I (X), Sac I (S), and EcoRV (RV). The numbers shown are relative to the start site of transcription. (B) Restriction fragments of P1 DNA or DNA derived from the tail of E129 mice were resolved on 0.8% agarose gel, transferred to a nylon membrane, and then hybridized with a 32P-labeled probe derived from sequences within the first exon.
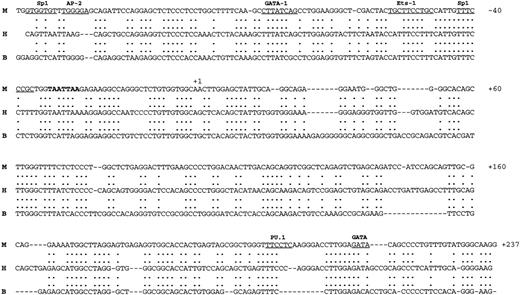
The sequence of the upstream promoter region, the first 2 exons, and the first intron of the mvWF gene was determined. As in the case of the human and bovine vWF genes,15 16 the first exon encodes 5′ untranslated sequences and the second exon contains the ATG translational start site. Exons 1 and 2 are separated by an intron of approximately 1,260 bp. The exon/intron boundaries are highly conserved between mouse and human, and the splice donor and acceptor sites conform with the GT and AT rule. The 5′ flanking region contains a TATA box that is identical to that of the human gene. Overall, the sequence between −140 and the end of the first exon is 70% conserved between mouse and human and 54% conserved between all 3 species (Fig 2). An upstream GATA-binding site at −80 relative to the start site of transcription is conserved in the mouse, human, and bovine promoters, whereas a GATA-binding site at the 3′ end of the first exon is conserved only in the mouse and human promoters. Consensus sequences for SP1- and Ets-binding sites are also present in the upstream promoter region of the mouse gene. In addition, the 5′ flanking region contains a hexamer GAGCTC and a CA repeat between −1835 and −1804.
Nucleotide sequence of the mvWF promoter. Alignment of the 5′ flanking region and first exon of the mouse, human, and bovine vWF genes. The numbers of the nucleotide sequence relative to the start site of transcription are on the right. The transcriptional start site is marked as +1. The TATA box is in bold. Potential consensus sites for DNA-binding proteins in the 5′ flanking region and first exon are underlined. The upper strand represents the mouse sequence (M), the middle strand the human sequence (H), and the lower strand the bovine sequence (B).
Nucleotide sequence of the mvWF promoter. Alignment of the 5′ flanking region and first exon of the mouse, human, and bovine vWF genes. The numbers of the nucleotide sequence relative to the start site of transcription are on the right. The transcriptional start site is marked as +1. The TATA box is in bold. Potential consensus sites for DNA-binding proteins in the 5′ flanking region and first exon are underlined. The upper strand represents the mouse sequence (M), the middle strand the human sequence (H), and the lower strand the bovine sequence (B).
Determination of the transcriptional start site of the mvWF promoter.
The transcriptional start site was mapped by reverse transcriptase primer extension analysis. A radiolabeled antisense primer, corresponding to the sequences between +114 and +153, was used in extension reactions with total RNA from mouse lungs. The major extension product was 153 bp in length and ended at an A residue (Fig 3A). This is identical to the transcriptional start site reported for the human vWF gene. In complementary studies, RT-PCR analyses were performed with mouse lung- and heart-derived cDNA. Primer pairs that included sense primers beginning 3′ of the putative start site and an antisense primer at +153 yielded PCR products of expected size, whereas primer pairs derived from the 5′ flanking region and first exon did not (Fig3B).
Determination of the transcriptional start site of the mvWF gene. (A) In primer extension assays, purified total RNA from mouse lung tissue (lanes 1 and 2) was used as template. Primer extension analysis was performed using a 32P-labeled antisense oligonucleotide spanning the region between +114 and +153 and AMV reverse transcriptase. DNA size markers and a dideoxy sequencing reaction of mvWF-pUC19-5 generated with the same primer are included on the left. The arrow indicates the start site of transcription. (B) In RT-PCR analyses, first-strand cDNA synthesis and PCR reactions were performed with total RNA from mouse lung (lanes 2, 6, 9, and 12) and heart (lanes 3, 7, 10, and 14) tissue. P1-7910 DNA template was included as a positive control (lanes 1, 5, 8, and 11). A 100-bp DNA ladder is shown in lanes 4 and 11. The location of the primers relative to the transcriptional start site (depicted by an asterisk) is shown above. The primer sequences are described in Materials and Methods. PCR-generated fragments from cDNA were obtained only with primer sets D-E (144 bp) and C-E (232 bp).
Determination of the transcriptional start site of the mvWF gene. (A) In primer extension assays, purified total RNA from mouse lung tissue (lanes 1 and 2) was used as template. Primer extension analysis was performed using a 32P-labeled antisense oligonucleotide spanning the region between +114 and +153 and AMV reverse transcriptase. DNA size markers and a dideoxy sequencing reaction of mvWF-pUC19-5 generated with the same primer are included on the left. The arrow indicates the start site of transcription. (B) In RT-PCR analyses, first-strand cDNA synthesis and PCR reactions were performed with total RNA from mouse lung (lanes 2, 6, 9, and 12) and heart (lanes 3, 7, 10, and 14) tissue. P1-7910 DNA template was included as a positive control (lanes 1, 5, 8, and 11). A 100-bp DNA ladder is shown in lanes 4 and 11. The location of the primers relative to the transcriptional start site (depicted by an asterisk) is shown above. The primer sequences are described in Materials and Methods. PCR-generated fragments from cDNA were obtained only with primer sets D-E (144 bp) and C-E (232 bp).
Functional analysis of the murine vWF promoter in transient transfection assays.
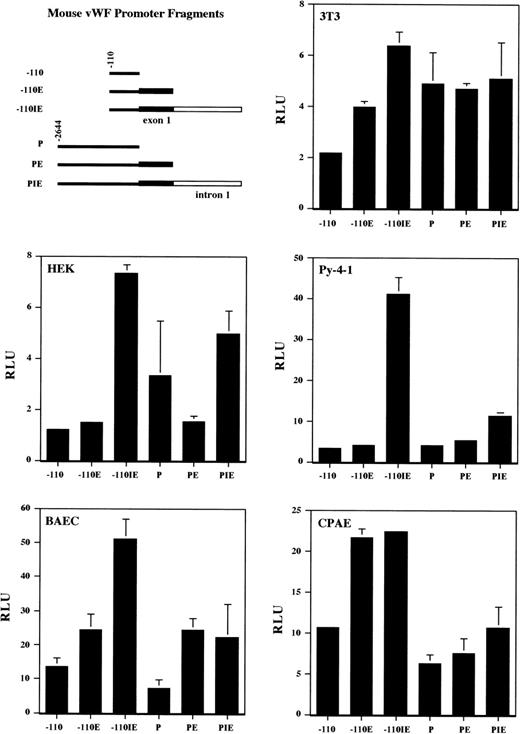
To test the function of the upstream promoter region, the first exon, and the first intron under in vitro conditions, the various promoter fragments (Fig 4) were fused to the luciferase reporter gene in pGL2-Basic, and the resulting constructs were transiently transfected into cultured cells. The plasmid constructs were cotransfected with a Renilla luciferase reporter gene to control for transfection efficiency, and the luciferase activity was further normalized to that of the promoterless pGL2-Basic vector. The vWF promoter was active in NIH 3T3 and HEK 293 cells (Fig 4). The intron had enhancing activity in both cell types, particularly in the context of the −110 upstream promoter region. In general, the various vWF constructs expressed at higher levels in endothelial cells compared with nonendothelial cells. In Py-4-1 endothelial cells, the intron contained significant enhancing activity (9.6-fold), the first exon had no overall effect on expression levels, and the 5′ flanking region demonstrated a strong repressive activity in the context of the full-length promoter (Fig 4). In BAEC, the first intron and first exon both had a moderate enhancing effect (2.1- and 2-fold, respectively), whereas the 5′ flanking sequence had an overall repressing effect (1.9-fold). Finally, in CPAE, the first intron had no effect, the first exon had a moderate enhancing effect (1.8-fold), and the 5′ flanking sequence served to repress activity (1.7-fold).
Promoter activity of the mvWF 5′ flanking region, first exon, and first intron in various cell lines. The mvWF promoter fragments were coupled to the luciferase reporter gene and named according to the scheme shown (upper left). Nonendothelial cells (NIH 3T3 and HEK) and endothelial cells (Py-4-1, CPAE, and BAEC) were transiently transfected with the mvWF luciferase constructs and harvested 24 hours later for luciferase activity. The results show the mean and standard deviation of luciferase light units obtained in triplicate from 1 representative experiment. More than 3 independent experiments were performed with each cell line. Luciferase light units are corrected both for transfection efficiency (as described in Materials and Methods) and for the activity of a promoterless PGL2-Basic vector.
Promoter activity of the mvWF 5′ flanking region, first exon, and first intron in various cell lines. The mvWF promoter fragments were coupled to the luciferase reporter gene and named according to the scheme shown (upper left). Nonendothelial cells (NIH 3T3 and HEK) and endothelial cells (Py-4-1, CPAE, and BAEC) were transiently transfected with the mvWF luciferase constructs and harvested 24 hours later for luciferase activity. The results show the mean and standard deviation of luciferase light units obtained in triplicate from 1 representative experiment. More than 3 independent experiments were performed with each cell line. Luciferase light units are corrected both for transfection efficiency (as described in Materials and Methods) and for the activity of a promoterless PGL2-Basic vector.
Functional analysis of the murine vWF promoter in transgenic mice.
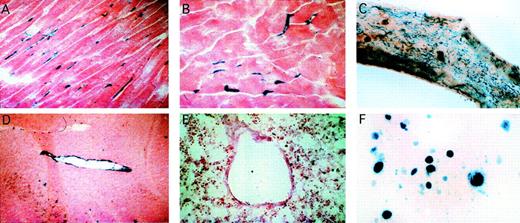
A total of 6 independent lines of transgenic mice were generated with a fragment of the mvWF promoter containing 2,645 bp 5′ flanking sequence, the first exon, and first intron. In 3 of these lines, transgene expression was not detectable in any tissue. In the other 3 lines, the X-Gal reaction product was detected in a subpopulation of endothelial cells of brain, heart, skeletal muscle, and aorta (Fig 5A through E). In the brain, reporter gene activity was detected in small- and medium-sized blood vessels. In the heart and skeletal muscle, expression was limited to the endothelial cell lining of myocardial capillaries. The level of transgene expression, as judged by the degree of LacZ staining, varied slightly between the 3 lines. In 1 of the 3 expressing lines of mice, the X-Gal reaction product was also detected in bone marrow megakaryocytes (Fig 5F).
The mvWF LacZ transgene is expressed in a vascular bed-specific pattern. LacZ staining of 10-μm tissue sections from mvWFlacZ line no. 47 shows reporter gene activity in endothelial cells of the heart (A), skeletal muscle (B), and brain (D). An en face preparation of the thoracic aorta shows diffuseLacZ staining of endothelial cells (C). A bone marrow aspirate shows the X-Gal reaction product in megakaryocytes (F). There is no detectable β-galactosidase activity in other tissues (E shows lung). The blue staining in the lung represents backgound.
The mvWF LacZ transgene is expressed in a vascular bed-specific pattern. LacZ staining of 10-μm tissue sections from mvWFlacZ line no. 47 shows reporter gene activity in endothelial cells of the heart (A), skeletal muscle (B), and brain (D). An en face preparation of the thoracic aorta shows diffuseLacZ staining of endothelial cells (C). A bone marrow aspirate shows the X-Gal reaction product in megakaryocytes (F). There is no detectable β-galactosidase activity in other tissues (E shows lung). The blue staining in the lung represents backgound.
DISCUSSION
The structural organization of the mouse, human, and bovine vWF promoters, including the intron-exon boundaries, the length of the first exon and first intron, and the sequence of the 5′ flanking region, is closely related. Several consensus sequences forcis-acting elements are found in the immediate upstream promoter region and first exon. Two GATA-binding consensus sequences, located at positions −80 and +215 relative to the start site of transcription, are conserved between the mouse and human vWF promoters. The upstream GATA-binding site is also present in the bovine vWF promoter. A palindromic hexamer GAGCTC at position −110 of the mvWF gene is also found within the intronic enhancer of the endothelial cell-specific Tie-2 gene17 and is a putative binding site for bZIP-related transcription factors.18 The poly(dG-dT) · poly(dA-dC) elements that are present in the upstream promoter region of the mvWF gene are also found in the human and bovine vWF promoters15,16 as well in as the Tie-2 intronic enhancer.17
Our understanding of the functional role of promoter elements in mediating endothelial cell-specific expression is largely based on transient transfection assays. For example, the human vWF promoter has been shown to be regulated by a repressor-derepressor mechanism under in vitro conditions. An NF1-binding site between −440 and −487 and an Oct-1-binding site between −133 and −125 were found to repress transcription in cultured endothelial and nonendothelial cell types, an effect that was offset in endothelial cells by a GATA-binding site within the first exon.19-21Binding sites for other transcription factors, including Sp1,22 NFκ-b,23 Ets-1,24 and PEA-3,17 have been implicated in the regulation of endothelial cell-specific genes. Of these, an Ets-1 motif in the upstream promoter region has been shown to play a role in mediating expression of the human vWF gene.25
In transient transfection assays, the mvWF promoter was found to be regulated by different mechanisms in different endothelial cell types. For example, the first exon conferred enhancing activity in bovine aortic and pulmonary artery endothelial cells, but not in mouse-derived Py-4-1 endothelial cells. On the other hand, sequences within the first intron enhanced expression in Py-4-1 endothelial cells and bovine aortic endothelial cells, but not in calf pulmonary endothelial cells. It is tempting to speculate that these differences reflect biologically relevant cell subtype-specific transcriptional control mechanisms that are intrinsic to the cells. However, in keeping with current models of endothelial cell gene regulation, we predict that the discordant mechanisms represent variable degrees of phenotypic drift. According to this notion, the uncoupling of endothelial cells from their natural environment induces an alteration in transcriptional regulatory mechanisms and programmed gene expression. The extent to which the phenotype of the cultured cell resembles its in vivo counterpart is likely to depend on the origin of the cell, the culture conditions, and the passage number. Regardless of the mechanism of differential gene regulation in tissue culture, these findings underscore the need for caution in interpreting the results of in vitro transfection assays.
Recent transgenic studies of the human vWF gene have uncovered a complex model of transcriptional regulation in which expression is governed by vascular bed-specific signaling pathways.11-13This paradigm of gene regulation is extended in the present study. In transgenic mice, a region of the mouse promoter that spans the 2,645 bp 5′ flanking region, the first exon, and first intron was found to direct vascular bed-specific expression in subpopulations of endothelial cells within the heart, skeletal muscle, and brain. By contrast, β-galactosidase activity was consistently absent in organs such as the lung, liver, and spleen. This pattern is unlikely to be explained by differences in the stability of the X-Gal reaction product, because widespread β-galactosidase activity has been demonstrated in the endothelium of other transgenic models.17 26 Indeed, the pattern of expression is similar to that reported for the human transgene and suggests that vascular bed-specific gene regulation is evolutionarily conserved between species.
In contrast to the human vWF transgene, the mouse promoter also contained information for expression in aortic endothelial cells. Moreover, in 1 of the 3 lines of mice, the X-Gal reaction product was detected in megakaryocytes. Although it is difficult to draw conclusions from 1 expressing line, this finding suggests that the mouse promoter contains information for megakaryocyte-specific expression when integrated into an appropriate chromatin environment.
Recent studies of other endothelial cell-specific transgenes have also demonstrated differential expression within the vascular tree. For example, the upstream promoter of the Tie-2 gene was shown to direct expression to distinct endothelial cell subpopulations within transgenic embryos,27 whereas the inclusion of an intronic enhancer region conferred widespread expression in adult endothelium.17 A 5.9-kb fragment of the murine preproendothelin-1 promoter directed differential expression within the endothelium and vascular smooth muscle cells of adult transgenic mice.28 In the latter study, expression levels in endothelial cells varied not only between arteries, veins, and capillaries, but also between vascular beds of different organs.28 In a transgenic analysis of the human endothelial nitric oxide synthase gene, a 1,600-bp region of the promoter was also shown to contain information for vascular bed-specific expression.29 Taken together, these results are consistent with our own observations of differential vWFlacZ expression and provide strong support for the existence of regional differences in the mechanisms of endothelial cell gene regulation.
The potential importance of combinatorial gene regulation as a more general transcriptional control mechanism is supported by studies of other cell lineages. In transgenic mice, the α1(1) collagen gene was shown to possess different cis elements required for expression in fibroblasts of the skin as compared with fibroblasts within the fascia.30 In another investigation,31expression of the CD4 gene in transgenic mice was shown to be governed by distinct regulatory elements in separate T-cell subsets. In a recent study of the muscle-specific SM22α gene,32 a 445-bp region of the promoter directed expression in the vascular smooth cells of arteries as well as cardiac and skeletal myocytes in a temporospatial pattern similar to that of the endogenous gene. However, in contrast to the endogenous gene, transgene expression was absent in venous and visceral smooth muscle cells. The promoter region of yet another muscle-specific gene (MLC-3F) was found to contain distinct DNA regions capable of distinguishing between regulatory programs within the various chambers of the transgenic heart.33 These reports provide additional evidence that transcriptional control mechanisms may differ between subpopulations of cells and reinforce the notion that not all cell types within a given lineage are alike.
In summary, we have shown that the mvWF promoter is highly homologous to the human vWF gene in both structure and function. The results support the notion that the vWF gene is regulated in a modular fashion from one vascular bed to another. The existence of multiple vascular bed-specific signaling pathways would enhance the capacity of endothelial cells to adapt to the needs of the local environment. At the same time, it might also render the endothelium more vulnerable to focal dysfunction and pathology. Indeed, further elucidation of these pathways would provide an initial framework for manipulating and regulating activity in specific subpopulations of endothelial cells in vivo.
ACKNOWLEDGMENT
The authors thank Cecil Denis and Denisa Wagner for providing us with mvWF exon 4/5 DNA sequence. We also thank Nadia Jahroudi and Nick Shworak for their helpful discussions.
Supported by National Institutes of Health Grant No. HL60585-01.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to William C. Aird, MD, Molecular Medicine, RW-663, Beth Israel Deaconess Medical School, Boston, MA 02215; e-mail:waird@caregroup.harvard.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal