To study the effects of major histocompatibility complex (MHC) class II expression on T-cell development, we have investigated T-cell immune reconstitution in two MHC class II–deficiency patients after allogeneic bone marrow transplantation (allo-BMT). Our study showed that the induction of MHC class II antigen expression on BM graft-derived T cells in these allo-BMT recipients was hampered upon T-cell activation. This reduction was most striking in the CD8+ T-cell subset. Furthermore, the peripheral T-cell receptor (TCR) repertoire in these graft-derived MHC class II–expressing CD4+ and in the CD8+ T-cell fractions was found to be restricted on the basis of TCR complementarity determining region 3 (CDR3) size profiles. Interestingly, the T-cell immune response to tetanus toxoid (TT) was found to be comparable to that of the donor. However, when comparing recipient-derived TT-specific T cells with donor-derived T cells, differences were observed in TCR gene segment usage and in the hydropathicity index of the CDR3 regions. Together, these results reveal the impact of an environment lacking endogenous MHC class II on the development of the T-cell immune repertoire after allo-BMT.
IN ORDER TO EVALUATE the effects of an environment devoid of endogenous major histocompatibility complex (MHC) class II antigen expression on T-cell immune reconstitution, we have performed a detailed analysis of the overall peripheral T-cell receptor (TCR) repertoire and the tetanus toxoid (TT) specific TCR repertoire in two MHC class II–deficiency patients after allogeneic bone marrow transplantation (allo-BMT). We have generated CD4+ and CD8+ T-cell lines and studied MHC expression patterns and TCR complementarity determining region 3 (CDR3) size profiles for the analysis of the overall T-cell repertoire. TT-specific T-cell lines were used for the analysis of the antigen-specific T-cell repertoire upon analysis of TCR gene segment usage, and hydropathicity index of the CDR3 region.
MHC class II deficiency, also referred to as bare lymphocyte syndrome (BLS), is a rare immunodeficiency disease, inherited in an autosomal recessive fashion. It is characterized by defective expression of MHC class II antigens on all cell types, in conjunction with a varying degree of MHC class I antigen expression, resulting in severely impaired cellular and humoral immune responses upon antigenic challenge. As a consequence, these patients are extremely susceptible to viral, bacterial, and fungal infections.1-4 The underlying genetic abnormality involves mutations in genes that encode transcription factors controlling MHC class II expression: CIITA (group A)5 and subunits of the RFX complex, RFXANK (group B),6 RFX5 (group C),7 and RFXAP (group D).8,9 Currently, the treatment of choice for this otherwise lethal immunodeficiency is allo-BMT. However, the success rate of engraftment and immunological recovery in BLS patients is lower than in patients with other immunodeficiency syndromes, especially following allo-BMT with BM grafts that are not HLA-identical.10-12
During normal T-cell development, BM-derived precursor T cells home to the thymus, where they are subjected to positive and negative selection processes upon interaction with MHC class I and II molecules expressed on thymic epithelial and dendritic cells of the cortex and medulla, respectively.13-17 These selection processes within the thymic microenvironment result in a peripheral pool of T cells that does not respond to self-peptides but is able to recognize foreign peptides in the context of self-MHC. Within the thymic microenvironment, MHC class II–mediated interactions result mostly in CD4+ T-cell development, whereas MHC class I–mediated interactions result in CD8+ T-cell development.18 Despite a general lack of endogenous MHC class II expression19,20 in the thymic microenvironment, a small number of CD4+ T cells is still found in the peripheral compartments of BLS patients.20,21 These observations imply that alternative ligands, such as MHC class I antigens, may have mediated the development of CD4+CD8+ thymocytes into CD4+ T cells.22 On the other hand, it cannot be excluded that these CD4+ T cells have reached the circulation without any selection. Interestingly, these BLS patient-derived CD4+ T cells show diverse TCRAV and TCRBV gene family usage20,23,24 with inverse TCRAV/TCRBV skewing patterns.20,25 Moreover, the lack of MHC class II expression has been shown to have an effect on the net charge and hydropathic index of the TCR CDR3 region within this subset.25
Theoretically, two mechanisms of T-cell reconstitution are expected after allo-BMT: First, peripheral expansion of graft-derived mature (memory) donor T cells provides the first wave of T cells after allo-BMT.26-30 These cells can be maintained in the periphery for over 10 to 20 years.31,32 The second mechanism involves thymic and/or extra-thymic selection processes and expansion of positively selected donor precursor T cells. The latter process of T-cell selection probably accounts for the more durable reconstitution of the T-cell immune repertoire.33-35 The MHC class II–mediated selection may present some special problems in an MHC class II–deficient environment with respect to the generation of a fully competent T-cell immune repertoire. In particular, positive and negative selection events after allo-BMT may be hampered due to the lack of MHC class II expression in these BLS patients on the thymic epithelial cells of the cortex and medulla, which are not of hematopoietic origin.19 36
MATERIALS AND METHODS
Patients.
Between 1985 and 1995, six unrelated infants suffering from MHC class II–deficiency were treated with an allo-BMT in the Department of Pediatrics at the Leiden University Medical Center. Conditioning of the patients was performed according to protocols of the European Society for Immunodeficiencies (ESID) and the European Group for Bone Marrow Transplantation (EBMT). Only two of the transplanted patients showed successful engraftment with immunological recovery and are alive from 1.5 to 4 years after allo-BMT. These unrelated patients: Patients 1 (unique patient number [UPN] 235; OSE) and 2 (UPN 293; EBA) who received transplants in 1993 and 1995, respectively, with a full BM graft from their healthy HLA-identical sibling donors: Donors 1 [D(UPN235); MSE] and 2 [D(UPN293); CBA] were analyzed in this study. The characteristics of these patients, the transplant-related variables, and the allo-BMT outcome are presented in Table 1. The BM graft consisted of 4.6 × 108 nucleated cells/kg BW in the case of patient 1, and 2.5 × 108 nucleated cells/kg body weight (BW) in the case of patient 2. Acute graft-versus-host disease (GVHD) grade I with involvement of the skin was observed in patient 1 and was lacking in patient 2. Limited chronic GVHD was observed in patient 1 after allo-BMT and was absent in patient 2. Both children received intravenous Igs immediately after allo-BMT, ie, patient 2 for 1.5 months, after which supplementation was discontinued. At the time of this study, patient 1 was still on Ig supplementation. The chimerism patterns after allo-BMT were determined via a polymerase chain reaction (PCR)-based CA repeat analysis in fluorescence-activated cell (FACS)-sorted cell populations, adapted from Van Leeuwen et al.38 For the purpose of this study, both BM donors and recipients received a TT (booster) vaccination at approximately 1 year after allo-BMT, and blood was drawn 4 weeks after the vaccination. The use of this human material has been approved by the Committee on Medical Ethics of the Leiden University Medical Center (Protocol: P254/96).
Clinical Characteristics of the BLS Patients and Allo-BMT Outcome
| Features . | Patients . | |
|---|---|---|
| Patient 1 . | Patient 2 . | |
| CID type defective gene | Type III BLS,* RFX5 (group C) | Type III BLS, RFXANK (group B) |
| Age at BMT | 23 mo | 8 mo |
| Donor | HLA-identical sibling | HLA-identical sibling |
| Age donor | 12 yr | 7 yr |
| HLA† type | A2 A3 B51 B50 Cw6 | A2 A11 B18 B51 Cw5 |
| DR4 DR17 DQ2 DQ8 | DR7 DR11 DQ2 DQ7 | |
| Preparative regimen‡ | Bu20/Cy200 | Bu20/Cy200 |
| T-cell depletion | − | − |
| GVHD prophylaxis1-153 | + | + |
| Chimerism1-155 | Mixed | Mixed |
| T | D/R | D |
| B | R/d | R/D |
| NK | R/d | D |
| MM | R/d | D/r |
| Current status | Alive and well >4 yr | Alive and well >1.5 yr |
| Features . | Patients . | |
|---|---|---|
| Patient 1 . | Patient 2 . | |
| CID type defective gene | Type III BLS,* RFX5 (group C) | Type III BLS, RFXANK (group B) |
| Age at BMT | 23 mo | 8 mo |
| Donor | HLA-identical sibling | HLA-identical sibling |
| Age donor | 12 yr | 7 yr |
| HLA† type | A2 A3 B51 B50 Cw6 | A2 A11 B18 B51 Cw5 |
| DR4 DR17 DQ2 DQ8 | DR7 DR11 DQ2 DQ7 | |
| Preparative regimen‡ | Bu20/Cy200 | Bu20/Cy200 |
| T-cell depletion | − | − |
| GVHD prophylaxis1-153 | + | + |
| Chimerism1-155 | Mixed | Mixed |
| T | D/R | D |
| B | R/d | R/D |
| NK | R/d | D |
| MM | R/d | D/r |
| Current status | Alive and well >4 yr | Alive and well >1.5 yr |
Abbreviations: CID, combined immunodeficiency disease; NK, natural killer cells; MM, myeloid/monocytic cells.
Type III BLS: MHC class II-deficiency, with reduced levels of MHC class I expression. The genetic defect of these patients was determined by heterokaryon fusions or by complementation analysis.37
Patient = Donor, HLA-DP typing of these patients/donors was not performed.
Bu20 = total dose of busulfan in mg/kg BW; Cy200 = total dose of cyclophosphamide in mg/kg BW.
GVHD prophylaxis consisted of cyclosporin A 2 mg/kgBW/d intravenously for 1 to 2 months, 6 mg/kg/OS (OS = oral administration) 2 to 6 months after BMT; methotrexate 10 mg/m2 on days +1, +3, +6.
Chimerism determined via FACS-PCR-CA repeat analysis38approximately 1 year after allo-BMT. The predominant origin of the cell populations is given in capital letters, either donor (D) or recipient (R), whereas a minor population (<10%) is indicated in lowercase letters (d or r). If donor and recipient were present in equal amounts, both populations are given in capital letters (R/D).
Processing of PBMCs.
Approximately 20 mL of heparinized blood was drawn from the BLS patients before allo-BMT and from the donors and allo-BMT recipients 4 weeks after TT booster vaccination. Peripheral blood mononuclear cells (PBMCs) were separated over a Ficoll-Isopaque gradient (LUMC Hospital Pharmacy, Leiden, The Netherlands). Immunophenotypical analysis of PBMCs was performed at regular intervals after allo-BMT as described previously.38 Normalization of a lymphocyte subset was defined as reaching the fifth percentile of age-matched reference values.39,40 Approximately 2 × 106 PBMCs of both donor and recipient was used for FACS-sorting to separate the CD4+ and CD8+T-cell subsets using fluorescein-conjugated anti-CD4 and R-phycoerythrin-conjugated anti-CD8 monoclonal antibodies (MoAbs) (DAKO, Dakopatts, Glostrup, Denmark) and a FACScan (Becton Dickinson, Mountain View, CA). The recovered CD4+ and CD8+T cells were 98% to 99% pure. CD4+ and CD8+single positive T-cell lines were generated by culturing ≈2 × 105 to 5 × 105 sorted cells of each subset in RPMI 1640 medium containing 5% human AB serum supplemented with 2 mmol/L L-glutamine, 100 IU/mL penicillin, 100 μg/mL streptomycin, 20 IU/mL recombinant interleukin-2 (rIL-2), and 1 μg/mL phytohemagglutinin (PHA) for 14 days. The TCR CDR3 size distributions41 in the HLA-DR− and -DR+,high T-cell fractions were investigated after a second FACS-sort using fluorescein-conjugated anti–HLA-DR MoAb (Becton Dickinson) in combination with R-phycoerythrin-conjugated anti-CD4 or anti-CD8 MoAbs (Dakopatts). To avoid contamination with HLA-DR+,dull T cells, only truly HLA-DR−and HLA-DR+,high T cells were collected. The recovered HLA-DR− and HLA-DR+,high T-cell fractions contained at least 5 × 103 T cells and were greater than 98% pure. The CD4+ and CD8+ T-cell subsets were also tested for MHC class I expression using an indirect staining with the anti-MHC class I MoAb W6/32 and fluorescein-conjugated goat anti-mouse MoAb (Becton Dickinson).
Antigen-specific T-cell lines/clones.
For the analysis of the antigen-specific repertoire, TT-specific T-cell lines were generated as described previously.42 Briefly, 3 to 6 × 106 PBMCs were grown in culture medium in the presence of 1.9 limes flocculationis (lf)/mL TT (RIVM, Bilthoven, The Netherlands). After two rounds of stimulation and subsequent TT-specificity testing in a 3H-thymidine incorporation assay, T-cell clones were generated from these lines by limiting dilution.43 The T-cell lines and clones that, after repeated testing, had a stimulation index (SI) > 3, with more than 500 counts per minute, were considered to be specific for TT (SI =3H incorporation of T cells with APC + TT/3H incorporation of T cells with APC only) and used for further molecular analysis. MHC class II–positive B-cell lines derived from the healthy HLA-identical siblings were used as antigen-presenting cells (APC) in these experiments.
RNA extraction, cDNA synthesis, PCR amplification.
Total RNA was extracted from TT-specific T-cell clones, and CD4+DR−/+,high and CD8+DR−/+,high T-cell fractions (104-3 × 106 cells) and converted into cDNA using oligo dT (Promega, Madison, WI). The cDNA was subjected to PCR amplification for the determination of TCR gene segment usage. PCR amplification and primers were as described previously.42,44 45
Spectratyping.
The distribution of the TCR CDR3 sizes was analyzed by PCR.41 For these purposes, a PCR reaction was performed with 5′ TCRBV family specific primers (TCRBV 1-23) and a 3′ TCRBC internal primer labeled with [γ-32P]ATP as described previously.41,42 To avoid a hampered size distribution due to very small numbers of T cells in the starting material, we used cDNA from at least 5 × 103 sorted cells for each spectratype analysis, because we (results not shown) and others46 have determined that ≈2 × 103cells is the lowest number of T cells at which a more or less normal distribution of TCR CDR3 sizes can be observed among sorted PBMC.
PCR fragment purification, DNA sequencing.
The PCR fragments were purified by electrophoresis in a 1% low-melting-point agarose gel. The desired fragment was isolated and purified using Wizard Columns (Promega) and used for direct sequencing47 based on the dideoxy-nucleotide chain termination method.48 The sequencing reactions were done with the T7 sequencing Kit (Pharmacia LKB, Uppsala, Sweden) using 5 to 10 pmol of TCRAC or TCRBC internal primer (listed below), approximately 0.25 pmol of PCR fragment, and [α-33P]dATP (0.5 μCi) (Dupont New England Nuclear Research Products, Boston, MA). DNA sequences were compared with TCR sequences contained in the GenBank using the PCGENE Computer Software Program (Release 6.85; Intelligenetics Inc, Palo Alto, CA). The hydropathic index for amino acids within the CDR3 component of each different TCRBV rearrangement of the TT-specific T-cell clones was calculated using the SOAP program included in the PCGENE computer software program. The resultant figure for the amino acid sequence is given under the name of Grand Average of Hydropathicity (GRAVY). The CDR3 region was defined for the purpose of this analysis as being the number of amino acids between five amino acids before the conserved cysteine at position 92 in the TCRBV element49 and three amino acids after the conserved FGXG motif found in the TCRBJ element.
Oligonucleotide sequencing/spectratyping primers.
The constant region sequencing and spectratyping primers used were: TCRAC internal 5′ GGT ACA CGG CAG GGT CAG GGT TC 3′; and TCRBC internal 5′ TGT GGG AGA TCT CTG CTT CTG 3′.
RESULTS
Standard immunophenotypical analysis and monitoring of T-cell proliferative responses before and at 1 year after allo-BMT.
Standard immunophenotypical analysis of PBMCs (Table 2) showed that both BLS patients had a normal percentage of CD3+ T cells before allo-BMT. In patient 1, a decreased percentage of CD4+ T cells and an increase of CD8+ T cells was observed, whereas in patient 2 normal percentages of both T cell subsets were present. At 1 year after allo-BMT the percentage of CD3+ T cells was low to normal, the percentage of CD4+ T cells was low in both patients, and the percentage of CD8+ T cells was either normal (patient 1) or increased (patient 2) when compared with age-matched healthy controls.39,40 Analysis of CD45RA/RO expression50 showed that in patient 1 before allo-BMT the majority of the CD4+ T cells was of the CD45RO+memory-phenotype, whereas the majority of T cells in patient 2 and the CD8+ T cells of patient 1 was of the CD45RA+naive-phenotype (Table 3). After allo-BMT, a substantial population of the CD4+ (27% to 43%) and of the CD8+ T cells (54% to 85%) was of the CD45RA+ naive-phenotype in both allo-BMT recipients (Table3).
Immunological Recovery: Analysis of Lymphocyte Subsets at Approximately One Year After Allo-BMT
| Individual . | Lymphocytes* (×103/μL) . | % CD3† . | % CD4 . | % CD8 . | % CD16/56 . | % CD20 . |
|---|---|---|---|---|---|---|
| Patient 1 pre-BMT | 1.1 | 65 | 12 | 50 | 29 | 4 |
| Patient 1 post-BMT | 1.3 | 45 | 10 | 29 | 12 | 24 |
| Patient 2 pre-BMT | 2.8 | 50 | 34 | 13 | 6 | 39 |
| Patient 2 post-BMT | 6.2 | 57 | 20 | 37 | 10 | 26 |
| Reference value‡ | ||||||
| 1-2 yr | 3.6 (2.9-5.1) | 63 (52-74) | 41 (27-52) | 21 (14-28) | 8 (4-15) | 27 (15-39) |
| 3-4 yr | 3.7 (2.3-5.4) | 62 (45-76) | 39 (27-47) | 18 (10-29) | 12 (7-17) | 20 (13-39) |
| Donor 1 | 2.1 | 77 | 37 | 25 | 16 | 10 |
| Donor 2 | 3.5 | 65 | 30 | 23 | 14 | 17 |
| Reference value | ||||||
| 5-9 yr | 2.6 (1.7-3.5) | 72 (51-80) | 42 (28-54) | 24 (17-32) | 8 (5-16) | 14 (9-22) |
| 10-14 yr | 2.4 (1.6-3.1) | 68 (49-83) | 39 (32-52) | 21 (13-31) | 13 (5-23) | 15 (9-28) |
| Individual . | Lymphocytes* (×103/μL) . | % CD3† . | % CD4 . | % CD8 . | % CD16/56 . | % CD20 . |
|---|---|---|---|---|---|---|
| Patient 1 pre-BMT | 1.1 | 65 | 12 | 50 | 29 | 4 |
| Patient 1 post-BMT | 1.3 | 45 | 10 | 29 | 12 | 24 |
| Patient 2 pre-BMT | 2.8 | 50 | 34 | 13 | 6 | 39 |
| Patient 2 post-BMT | 6.2 | 57 | 20 | 37 | 10 | 26 |
| Reference value‡ | ||||||
| 1-2 yr | 3.6 (2.9-5.1) | 63 (52-74) | 41 (27-52) | 21 (14-28) | 8 (4-15) | 27 (15-39) |
| 3-4 yr | 3.7 (2.3-5.4) | 62 (45-76) | 39 (27-47) | 18 (10-29) | 12 (7-17) | 20 (13-39) |
| Donor 1 | 2.1 | 77 | 37 | 25 | 16 | 10 |
| Donor 2 | 3.5 | 65 | 30 | 23 | 14 | 17 |
| Reference value | ||||||
| 5-9 yr | 2.6 (1.7-3.5) | 72 (51-80) | 42 (28-54) | 24 (17-32) | 8 (5-16) | 14 (9-22) |
| 10-14 yr | 2.4 (1.6-3.1) | 68 (49-83) | 39 (32-52) | 21 (13-31) | 13 (5-23) | 15 (9-28) |
Absolute lymphocyte numbers are given: value × 103cells/μL.
The percentages were determined after staining with appropriate MoAbs and analysis on a FACSscan.
Immunological Recovery: Analysis of Naive (CD45RA+) and Memory (CD45RO+) T Cells Before and at Approximately One Year After Allo-BMT
| Individual . | % CD4+ CD45RA+ . | % CD4+ CD45RO+ . | % CD8+ CD45RA+ . | % CD8+ CD45RO+ . |
|---|---|---|---|---|
| Patient 1 pre-BMT | 34 | 69 | 81 | 21 |
| Patient 1 post-BMT | 27 | 71 | 85 | 14 |
| Patient 2 pre-BMT | 60 | 34 | 66 | 32 |
| Patient 2 post-BMT | 43 | 65 | 54 | 66 |
| Reference3-150 value | ||||
| 0-11 mo | 81 (66-88) | |||
| 1-6 yr | 71 (66-77) | |||
| Donor 1 | 60 | 35 | 82 | 19 |
| Donor 2 | 48 | 60 | 40 | 63 |
| Reference value | ||||
| 7-17 yr | 61 (55-67) |
| Individual . | % CD4+ CD45RA+ . | % CD4+ CD45RO+ . | % CD8+ CD45RA+ . | % CD8+ CD45RO+ . |
|---|---|---|---|---|
| Patient 1 pre-BMT | 34 | 69 | 81 | 21 |
| Patient 1 post-BMT | 27 | 71 | 85 | 14 |
| Patient 2 pre-BMT | 60 | 34 | 66 | 32 |
| Patient 2 post-BMT | 43 | 65 | 54 | 66 |
| Reference3-150 value | ||||
| 0-11 mo | 81 (66-88) | |||
| 1-6 yr | 71 (66-77) | |||
| Donor 1 | 60 | 35 | 82 | 19 |
| Donor 2 | 48 | 60 | 40 | 63 |
| Reference value | ||||
| 7-17 yr | 61 (55-67) |
The percentage CD45RA+ and CD45RO+ T cells was determined after staining with FITC-labeled anti-CD45RA or anti-CD45RO MoAbs and PE-labeled anti-CD4 or anti-CD8 MoAbs; CD45RA+RO+ T cells were calculated as well.
Age-matched reference values39 are given as median with ranges from 25th to 75th percentiles in parentheses; reference values for CD8+ naive and memory T cells and for CD4+CD45RO+ T cells are not available.
The general T-cell proliferative responses upon repeated vaccination were evaluated before and after allo-BMT in a standard whole-blood culture. This showed a lack of antigen-specific proliferative responses before transplantation (results not shown). After allo-BMT, low to normal proliferative T-cell responses to TT (SI 17 to 19) as well as to diphtheria toxin (SI 3 to 9) were observed in both recipients (results not shown).
MHC expression patterns and chimerism analysis.
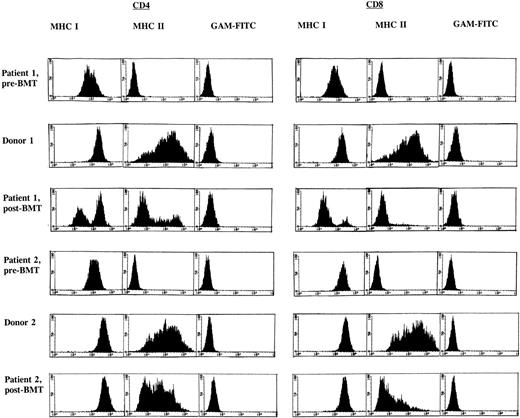
FACS analysis of the PHA/rIL-2–activated CD4+ and CD8+ T-cell subsets in both BLS patients before BMT showed an absence of MHC class II (HLA-DR) expression and dull expression of MHC class I when compared with the donors as shown in Fig 1. A similar analysis after allo-BMT revealed a deficiency in the induction of MHC class II in both recipients, in particular in the CD8+ T-cell subset (Fig1). Both BM donors showed approximately 70% MHC class II–positive cells in both T-cell subsets. In patient 1, after allo-BMT, expression of MHC class II was observed in 24% of the CD4+ T cells, whereas 5% of the CD8+ T-cell subset was MHC class II–positive. Similar observations were made in patient 2, after allo-BMT, with 46% and 15% MHC class II–positive T cells in CD4+ and CD8+ T-cell subsets, respectively. However, in contrast to MHC class II, other activation markers, such as CD25 and CD45RO, were found to be expressed at normal levels in both allo-BMT recipients (results not shown). Because recipient-derived T cells lack the capacity to express MHC class II upon activation, a chimerism analysis was performed to determine the origin of these MHC class II–negative T-cell fractions. These analyses revealed a mixed recipient-donor T-cell chimerism in patient 1 (Table 4). Approximately 40% of the CD4+ T cells and less than 10% of the CD8+ T cells were of donor origin (Table 4), and all other hematopoietic lineages showed a similar distribution (Table 1). Patient 2 displayed a different phenotype, as the majority of the hematopoietic cell lineages were found to be of donor origin (Tables 1 and 4). Investigation of MHC class I expression in these T-cell subsets (Fig 1) supported the observation based on chimerism analysis as described above. Together, these data indicate that after allo-BMT, in particular, donor-derived CD8+ T cells are hampered in the induction of MHC class II expression after T-cell activation.
FACS analysis of CD4+ and CD8+ T-cell subsets before and 1 year after allo-BMT. FACS profiles are given for BLS patients 1 (top) and 2 (bottom) and their respective BM donors, after labeling of PHA/rIL-2–stimulated CD4+ and CD8+ T-cell subsets with MoAb W6/32 directed against MHC class I and subsequent staining with fluorescein (FITC)-conjugated goat anti-mouse MoAb or after a direct staining with an FITC-conjugated MHC class II–directed MoAb (BD -HLA-DR); GAM-FITC was used as a negative control.
FACS analysis of CD4+ and CD8+ T-cell subsets before and 1 year after allo-BMT. FACS profiles are given for BLS patients 1 (top) and 2 (bottom) and their respective BM donors, after labeling of PHA/rIL-2–stimulated CD4+ and CD8+ T-cell subsets with MoAb W6/32 directed against MHC class I and subsequent staining with fluorescein (FITC)-conjugated goat anti-mouse MoAb or after a direct staining with an FITC-conjugated MHC class II–directed MoAb (BD -HLA-DR); GAM-FITC was used as a negative control.
Detailed Chimerism Analysis in Activated CD4+ and CD8+ T-Cell Subsets One Year After Allo-BMT
| Sample . | T-Cell Population4-150 . | Chimerism4-151 . |
|---|---|---|
| Patient 1 | CD4+ HLA-DR− | R/D |
| CD4+ HLA-DR+ | D | |
| CD8+ HLA-DR− | R/d | |
| CD8+ HLA-DR+ | D | |
| Patient 2 | CD4+ HLA-DR− | D |
| CD4+ HLA-DR+ | D | |
| CD8+ HLA-DR− | D | |
| CD8+ HLA-DR+ | D |
| Sample . | T-Cell Population4-150 . | Chimerism4-151 . |
|---|---|---|
| Patient 1 | CD4+ HLA-DR− | R/D |
| CD4+ HLA-DR+ | D | |
| CD8+ HLA-DR− | R/d | |
| CD8+ HLA-DR+ | D | |
| Patient 2 | CD4+ HLA-DR− | D |
| CD4+ HLA-DR+ | D | |
| CD8+ HLA-DR− | D | |
| CD8+ HLA-DR+ | D |
The T-cell populations were FACS-sorted CD4+ and CD8+ T-cell subsets, cultured for 10 to 14 days in the presence of PHA/rIL-2 and subsequently subjected to a second FACS-sort to generate the DR−/DR+,high T-cell fractions, using anti-CD4, anti-CD8, and anti-HLA-DR specific MoAbs.
The chimerism patterns were determined via FACS-PCR-CA repeat analysis.38 The predominant origin of the T-cell population is given in capital letters, either donor (D) or recipient (R), whereas a minor population (<10%) is given in lowercase letters (d or r). If donor and recipient were present in equal amounts, both populations were given in capital letters (D/R). The origin of all other hematopoietic cell lineages is depicted in Table 1.
TCR CDR3 size distribution patterns.
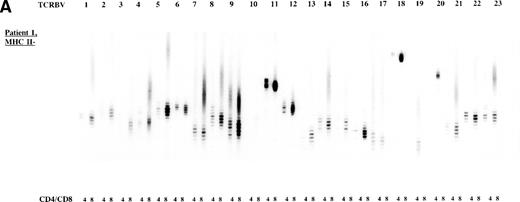
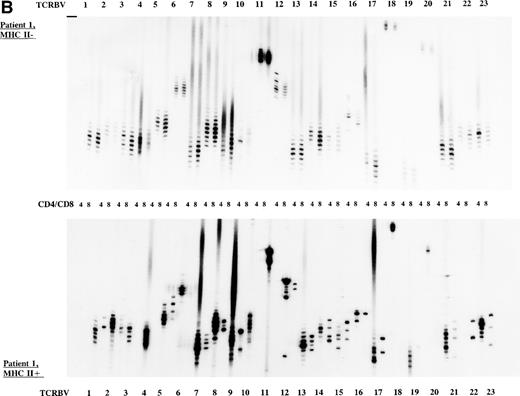
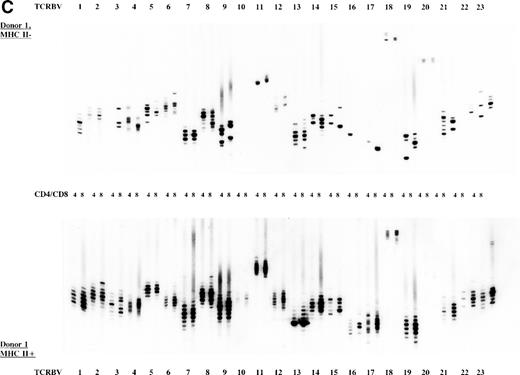
To study the overall TCR reconstitution in the periphery in more detail, we analyzed the TCR β-chain CDR3 size profiles. Both BLS patients before and after allo-BMT showed similar TCR CDR3 size distribution patterns, as demonstrated by the results of patient 1 (Fig 2A and B). Before allo-BMT, a more or less normal distribution of TCR CDR3 sizes was observed in the MHC class II–deficient CD8+T-cell subset, whereas a less complex CDR3 size profile was seen in the MHC class II–deficient CD4+ T-cell subset (eg, TCRBV 1, 2, 3, 4, 13) (Fig 2A). Similar analysis of the donor showed a normal distribution of TCR CDR3 sizes in the MHC class II–positive fractions and skewed size profiles, ie, reduction in the number of bands and/or with different band-intensity in the MHC class II–negative fractions (Fig 2C). However, when comparing the CDR3 size profiles of the patient before allo-BMT with the MHC class II–positive subset of the donor, a less complex CDR3 size pattern could be observed (compare Fig 2A with 2C). Interestingly, after allo-BMT the TCR CDR3 sizes of donor-derived MHC class II–positive fractions in both CD4+ and CD8+ T-cell subsets of the recipient were not normally distributed, whereas those in the MHC class II–negative fractions were (Fig 2B v C). Spectratypes of BLS patient 2 both before and after allo-BMT showed similar patterns when compared with patient 1, ie, a heterogeneous but less complex TCR CDR3 size distribution than the donor before BMT (results not shown). After allo-BMT a normal distribution of TCR CDR3 sizes in the MHC class II–negative T-cell fractions was noted, whereas skewed patterns were noted in the MHC class II–positive fractions (results not shown). Similarly, both donors were comparable, ie, with a normal distribution of TCR CDR3 sizes in the MHC class II–positive T-cell fractions and with skewed patterns in the MHC class II–negative fractions (Fig 2C, results not shown).
TCR CDR3 size distribution (spectratype) analysis of overall peripheral TCRBV repertoires in CD4+ and CD8+ T-cell subsets after sorting for MHC class II expression. (A) Results of the TCR CDR3 size distribution analysis in patient 1 before allo-BMT, ie, MHC class II–negative fractions in CD4+ and CD8+ T-cell subsets. (B) Patient 1 one year after allo-BMT, T-cell subsets divided in MHC class II–expressing (MHC II+) or lacking (MHC II−) T-cell fractions. (C) Donor 1. From left to right, TCRBV families 1-23 are depicted in CD4+ and CD8+ T-cell subsets.
TCR CDR3 size distribution (spectratype) analysis of overall peripheral TCRBV repertoires in CD4+ and CD8+ T-cell subsets after sorting for MHC class II expression. (A) Results of the TCR CDR3 size distribution analysis in patient 1 before allo-BMT, ie, MHC class II–negative fractions in CD4+ and CD8+ T-cell subsets. (B) Patient 1 one year after allo-BMT, T-cell subsets divided in MHC class II–expressing (MHC II+) or lacking (MHC II−) T-cell fractions. (C) Donor 1. From left to right, TCRBV families 1-23 are depicted in CD4+ and CD8+ T-cell subsets.
Generation of TT-specific T-cell lines and clones.
After stimulation of PBMC derived from patients 1 and 2 after allo-BMT with TT, the responding TT-specific T cells showed specificity for TT in a 3H-thymidine incorporation assay with SI ranging from 7 to 30 (Table 5). This was found to be in a similar range when compared with their donors. Mitogenic stimulation of these donor or recipient T-cell lines with PHA showed a similar proliferative capacity (SI 48 to 84) as shown in Table 5. Subsequently, TT-specific T-cell clones were generated from these T-cell lines by limiting dilution. The percentage of TT-specific T-cell clones derived from these lines varied from approximately 10% in donor-recipient couple 1 to approximately 45% in donor-recipient couple 2 (Table 5, right panel), all of which exhibited a similar proliferative capacity (results not shown). Furthermore, all generated TT-specific T-cell clones demonstrated a donor phenotype since they expressed MHC class II upon activation.
Results of Proliferation Assays of the TT-Specific T-Cell Lines Derived From Allo-BMT Recipients and Their Donors, and Overview of the Generation of TT-Specific T-Cell Clones
| Sample . | SI5-150 ± SD . | No. Clones5-152 . | % TT Specific . | No. Identical Clones5-153 . | D/R Type5-155 . | |
|---|---|---|---|---|---|---|
| TT5-151 . | PHA . | |||||
| Donor 1 | 28 ± 9 | 84 ± 42 | 178 | ≈9%5-154 | 4/13 | D |
| Patient 1 | 7 ± 3 | 52 ± 26 | 125 | ≈7%5-154 | 3/10# | D |
| Donor 2 | 18 ± 9 | 50 ± 25 | 76 | ≈38% | 11/28 | D |
| Patient 2 | 30 ± 15 | 48 ± 24 | 43 | ≈56% | 11/22 | D |
| Sample . | SI5-150 ± SD . | No. Clones5-152 . | % TT Specific . | No. Identical Clones5-153 . | D/R Type5-155 . | |
|---|---|---|---|---|---|---|
| TT5-151 . | PHA . | |||||
| Donor 1 | 28 ± 9 | 84 ± 42 | 178 | ≈9%5-154 | 4/13 | D |
| Patient 1 | 7 ± 3 | 52 ± 26 | 125 | ≈7%5-154 | 3/10# | D |
| Donor 2 | 18 ± 9 | 50 ± 25 | 76 | ≈38% | 11/28 | D |
| Patient 2 | 30 ± 15 | 48 ± 24 | 43 | ≈56% | 11/22 | D |
Stimulation indices were calculated: SI = 3H thymidine incorporation of T cells with APC + antigen/3H thymidine incorporation of T cells with APC only ± standard deviation (SD). The background values varied between 100 and 900 cpm, the values of interest varied between 20,000 and 50,000 cpm for PHA and between 2,000 and 10,000 cpm for TT.
In the proliferation assay, TT and PHA were used as antigen.
All minicultures that were derived from 96-well plates with at least 33% negative wells/plate after limiting dilution were considered to be clonal, and used for the proliferation assays and molecular analysis.
Numbers of identical TT-specific T-cell clones (identical TCR sequence, Table 6) divided by total numbers of TT-specific T-cell clones.
Donor (D) or recipient (R) type T-cell clone as determined via FACS analysis of HLA-DR expression, after staining with anti–HLA-DR MoAbs.
% of TT specific clones of donor 1 and patient 1: mean of 3 independent cloning experiments.
#Due to the low number of clones, all clones once specific for TT were included for further molecular analysis.
Molecular characterization of TT-specific T-cell clones.
When comparing TCRBV sequences of TT-specific T-cell clones derived from the two BM donors, a marked overrepresentation of TCRBV5 containing TCRs was observed. This preferential utilization of TCRBV5 was not found after transplantation in the allo-BMT recipients (Table 6). Furthermore, no apparent sharing of TCRBV, TCRBJ, and amino acid composition of the TCR CDR3 region was observed between donor- and recipient-derived sequences (Table 6). When analyzing the DNA sequences of the TT-specific T-cell clones in more detail, no striking differences were observed in the length of the TCR CDR3 region (Table 6) or in the amount of nongermline modification at the sites of recombination between donor and recipient (results not shown). T-cell clones with an identical β-chain CDR3 amino acid motif (QGLAGV) were found in donor-recipient couple 1 (Table 6), but with a different TCRB nucleotide composition and TCR α-chain (results not shown).
DNA Sequence Analysis of TT-Specific T-Cell Clones Derived From MHC Class II–Deficiency Allo-BMT Recipients and Their Donors
| No. of Clones . | TCRBV . | N1-D-N2 . | TCRBJ . | |||
|---|---|---|---|---|---|---|
| Donor 1 | 1 | 1S1 | ASSV | YPRPP | QF | 2S1 |
| 1 | 2S1 | SA | TKVGLGF | TDTQY | 2S3 | |
| 1 | 5S1 | ASSL | VEG | NTEAF | 1S1 | |
| 1 | 5S1 | ASSL | RAFFVD | EQF | 2S1 | |
| 4 | 5S2 | ASS | YGV | SGANVLT | 2S6 | |
| 1 | 5S6 | ASS | QGLAGV | TDTQY | 2S3 | |
| 1 | 5S6 | ASS | QGLAGI | TDTQY | 2S3 | |
| 1 | 5S6 | ASS | RDSK | NTGELF | 2S2 | |
| 1 | 5S6 | ASS | SGLAVSG | DTQY | 2S3 | |
| 1 | 13S3 | AS | TPDSFMA | T | 1S2 | |
| Patient 1 | 1 | 2S1 | SAR | GAGART | PLH | 1S6 |
| 1 | 4S1 | SVE | ERRRR | ETQY | 2S5 | |
| 2 | 4S1 | SV | VSTGAGTAA | EQY | 2S7 | |
| 3 | 5S6 | ASS | QGLAGV | TDTQY | 2S3 | |
| 1 | 10S1 | ASS | PRGGSSLG | TDTQY | 2S3 | |
| 1 | 11S1 | ASS | PMGQIP | EQY | 2S7 | |
| 1 | 13S3 | ASSE | IFVKGADP | TDTQY | 2S3 | |
| Donor 2 | 1 | 1S1 | ASSV | ATGQ | QETQY | 2S5 |
| 1 | 2S1 | SA | TGTGKS | EQY | 2S7 | |
| 1 | 5S1 | ASSL | EVH | NYGYT | 1S2 | |
| 2 | 5S1 | ASSL | AGS | SYNEQF | 2S1 | |
| 2 | 5S1 | ASS | SDGQGR | PQH | 2S5 | |
| 2 | 5S1 | AS | GFTAGVS | ETQY | 2S5 | |
| 1 | 5S6 | ASS | SGLAGVS | DTQY | 2S3 | |
| 1 | 6S5 | ASS | FRREP | SYNEQ | 2S1 | |
| 1 | 8S1/2 | ASSL | GRGG | TDTQY | 2S3 | |
| 1 | 8S1/2 | ASSL | GQAI | GYT | 1S2 | |
| 3 | 8S1/2 | ASS | SSGKH | EQF | 2S1 | |
| 1 | 8S1/2 | AS | RWGA | EAF | 1S1 | |
| 11 | 21S3 | ASS | FTRK | NTEAF | 1S1 | |
| Patient 2 | 1 | 2S1 | SAR | DKNAA | QF | 2S1 |
| 2 | 4S1 | SV | QRLD | TIY | 1S3 | |
| 11 | 6S1 | ASS | QQLGGA | DTQY | 2S3 | |
| 3 | 6S7a | ASS | PALAGGP | YEQY | 2S7 | |
| 2 | 13S3 | AS | GGQGR | GY | 1S2 | |
| 1 | 15S1 | ATS | VGQG | SYEQY | 2S7 | |
| 2 | 22S1 | ASSE | GQAGG | PQH | 1S5 | |
| No. of Clones . | TCRBV . | N1-D-N2 . | TCRBJ . | |||
|---|---|---|---|---|---|---|
| Donor 1 | 1 | 1S1 | ASSV | YPRPP | QF | 2S1 |
| 1 | 2S1 | SA | TKVGLGF | TDTQY | 2S3 | |
| 1 | 5S1 | ASSL | VEG | NTEAF | 1S1 | |
| 1 | 5S1 | ASSL | RAFFVD | EQF | 2S1 | |
| 4 | 5S2 | ASS | YGV | SGANVLT | 2S6 | |
| 1 | 5S6 | ASS | QGLAGV | TDTQY | 2S3 | |
| 1 | 5S6 | ASS | QGLAGI | TDTQY | 2S3 | |
| 1 | 5S6 | ASS | RDSK | NTGELF | 2S2 | |
| 1 | 5S6 | ASS | SGLAVSG | DTQY | 2S3 | |
| 1 | 13S3 | AS | TPDSFMA | T | 1S2 | |
| Patient 1 | 1 | 2S1 | SAR | GAGART | PLH | 1S6 |
| 1 | 4S1 | SVE | ERRRR | ETQY | 2S5 | |
| 2 | 4S1 | SV | VSTGAGTAA | EQY | 2S7 | |
| 3 | 5S6 | ASS | QGLAGV | TDTQY | 2S3 | |
| 1 | 10S1 | ASS | PRGGSSLG | TDTQY | 2S3 | |
| 1 | 11S1 | ASS | PMGQIP | EQY | 2S7 | |
| 1 | 13S3 | ASSE | IFVKGADP | TDTQY | 2S3 | |
| Donor 2 | 1 | 1S1 | ASSV | ATGQ | QETQY | 2S5 |
| 1 | 2S1 | SA | TGTGKS | EQY | 2S7 | |
| 1 | 5S1 | ASSL | EVH | NYGYT | 1S2 | |
| 2 | 5S1 | ASSL | AGS | SYNEQF | 2S1 | |
| 2 | 5S1 | ASS | SDGQGR | PQH | 2S5 | |
| 2 | 5S1 | AS | GFTAGVS | ETQY | 2S5 | |
| 1 | 5S6 | ASS | SGLAGVS | DTQY | 2S3 | |
| 1 | 6S5 | ASS | FRREP | SYNEQ | 2S1 | |
| 1 | 8S1/2 | ASSL | GRGG | TDTQY | 2S3 | |
| 1 | 8S1/2 | ASSL | GQAI | GYT | 1S2 | |
| 3 | 8S1/2 | ASS | SSGKH | EQF | 2S1 | |
| 1 | 8S1/2 | AS | RWGA | EAF | 1S1 | |
| 11 | 21S3 | ASS | FTRK | NTEAF | 1S1 | |
| Patient 2 | 1 | 2S1 | SAR | DKNAA | QF | 2S1 |
| 2 | 4S1 | SV | QRLD | TIY | 1S3 | |
| 11 | 6S1 | ASS | QQLGGA | DTQY | 2S3 | |
| 3 | 6S7a | ASS | PALAGGP | YEQY | 2S7 | |
| 2 | 13S3 | AS | GGQGR | GY | 1S2 | |
| 1 | 15S1 | ATS | VGQG | SYEQY | 2S7 | |
| 2 | 22S1 | ASSE | GQAGG | PQH | 1S5 | |
Summary of the complete results of the DNA sequencing. Listed are the TCRBV/BD/BJ usage of the TT-specific T-cell clones, as well as the CDR3 region of the β chain. Nomenclature according to Arden et al51 (TCRBV) and Toyonaga et al52 (TCRBJ). When TCRBD was determined, the amino acids encoded by the D gene segment are given in bold type. The underlined CDR3 amino acids denote the boundaries of the CDR3 region on the TCRBV and TCRBJ side, respectively.
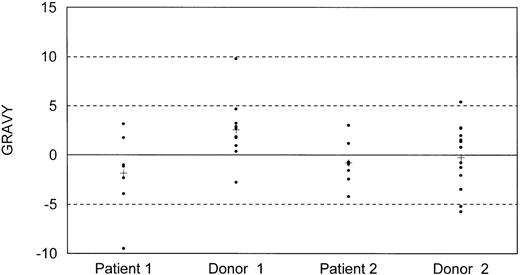
As a next step we analyzed the structural properties of the obtained TCR CDR3 region sequences of the TT-specific T-cell clones derived from both recipients and their donors. The use of certain amino acids in recipient-derived TCR CDR3 sequences resulted in an overall alteration of the grand average of hydropathicity (GRAVY) in both recipients (Fig 3). These GRAVY values were skewed toward a more hydrophillic CDR3 profile when compared with values of their donors. The mean GRAVY values for different TCR CDR3 sequences in patient 1, donor 1, patient 2, and donor 2 were −1.9, 2.5, −0.80, and −0.26, respectively, with only the difference between patient-donor couple 1 achieving statistical significance (P = .026). This difference was not due to the overrepresentation of TCRBV5 in the donors, because elimination of TCRBV5+ CDR3 sequences from the analysis still resulted in a significant difference (mean GRAVY patient 1: −2.06, donor 1: 2.91). Similar differences in GRAVY values between donors and recipients were not observed in allo-BMT recipients treated for leukemia (results not shown).
Distribution of hydropathicity index (GRAVY) values of TCR CDR3 sequences of TT-specific T-cell clones derived from BLS allo-BMT recipients 1 and 2 after allo-BMT, and their corresponding donors. A hydropathic index of −5 is considered to be neutral, values above −5 are considered to be hydrophobic, and values below −5 are considered to be hydrophillic; each dot represents a single CDR3 region. (+) Mean value of all different CDR3s of one individual.
Distribution of hydropathicity index (GRAVY) values of TCR CDR3 sequences of TT-specific T-cell clones derived from BLS allo-BMT recipients 1 and 2 after allo-BMT, and their corresponding donors. A hydropathic index of −5 is considered to be neutral, values above −5 are considered to be hydrophobic, and values below −5 are considered to be hydrophillic; each dot represents a single CDR3 region. (+) Mean value of all different CDR3s of one individual.
DISCUSSION
The aim of this study was to investigate the T-cell immune reconstitution after allo-BMT in an environment lacking endogenous MHC class II. For these purposes, we have analyzed the functional and phenotypical properties of the overall peripheral TCR repertoires, as well as of the TT-specific TCR repertoires in two MHC class II–deficiency patients.
Chimerism analysis showed different patterns of engraftment in the two patients despite a similar conditioning regimen given before transplantation (Table 1). In patient 1, hematopoietic cells were primarily of recipient origin with the exception of the T-cell lineage, which was of mixed recipient/donor origin, whereas in patient 2 all cell lineages were primarily of donor origin. The mixed T-cell chimerism that was observed in patient 1, despite the myeloablative chemotherapy given before allo-BMT, has also been observed in BMT recipients treated for leukemia who received myeloablative chemotherapy and additional total body irradiation before transplantation.53
T-cell FACS analysis revealed an impairment in MHC class II expression after activation in particular within the allo-BMT recipient-derived CD8+ T-cell subsets (Fig 1), irrespective of the T-cell chimerism pattern observed after allo-BMT (Table 4). In contrast, CD25 and CD45RO were expressed normally by both T-cell subsets, showing that the expression of these activation-associated markers is not hampered. This suggests that the donor-derived precursor and/or memory CD8+ T cells and, to a lesser degree, CD4+ T cells, have lost the capacity to express MHC class II upon T-cell activation after allo-BMT. The exact mechanism causing the aberrant MHC class II expression remains to be investigated.
TCR CDR3 size distribution analysis of the CD4+ and CD8+ T-cell subsets derived from both patients before allo-BMT revealed a heterogeneic TCR repertoire which was less diverse than that of the donors (Fig 2A/C, results not shown). After allo-BMT, the MHC class II–positive (donor-derived) fractions of the patients displayed a skewed TCR CDR3 size distribution pattern (Fig 2B). This is in contrast to the normal size distribution patterns that were observed in the MHC class II–positive fractions of the BM donors (Fig 2C). These skewed size profiles were not due to small numbers of cells in the analysis because sufficient T cells were used for each spectratype to ensure a normal distribution. Moreover, it is not likely that limited numbers of donor T cells or GVHD have contributed to these skewed patterns because the BM grafts were not T-cell depleted and because no or only limited GVHD was observed after allo-BMT. Therefore, these data argue for a qualitative incomplete T-cell immune reconstitution after allo-BMT in an MHC class II–negative environment.
The proliferative capacities of recipient- and donor-derived T-cell lines and clones with either mitogen or tetanus toxoid were comparable (Table 5). The TT-specific T-cell clones of both recipients were of donor-origin. These T cells presumably represent recently selected precursor T cells that have gained the capacity to respond to TT, since direct evidence for peripheral reconstitution with memory-type donor T cells was not obtained. TT-specific T-cell clones with completely identical TCR α and β chains were not found when comparing recipients with donors. Moreover, the presence of donor-derived APC (Table 1) may allow for efficient interactions between donor-derived T cells and APC, and could, therefore, enable adequate immune responses to other pathogens. This is supported by the observation that both recipients not only display proliferative responses to TT but also to diphtheria toxin.
The differences observed in the structure of the αβ TCR of the TT-specific T-cell clones such as differences in TCRBV/TCRBJ gene segments or in the amino acid composition of the CDR3 region (Table 6) most probably result from recipient-mediated selection of graft-derived precursor T cells either in the thymus or extra-thymically. This is supported by the observation that at least part of the CD4+T cells was of the CD45RA+ naive phenotype50(Table 3), presumably representing recent thymic emigrants. However, recognition of different TT epitopes may have contributed to the observed differences as well, because fine-specificity of these clones to previously established immuno-dominant epitopes of TT54,55 could not be determined. Prolonged survival of these recently selected and/or mature donor T cells may be mediated by graft-derived MHC class II–positive monocytes and dendritic cells that migrate to the periphery.56 57
Of note was the observation that the TCR CDR3 amino acid composition of the above mentioned TT-specific T-cell clones derived from patient 1 and to a lesser extent from patient 2 displayed an overall alteration of the hydropathicity index when compared with their donors (Fig 3). These skewed profiles appear to be characteristic of BLS patients, because no GRAVY differences were observed in allo-BMT recipients treated for leukemia. The altered amino acid composition of the CDR3 region in the two analyzed BLS recipients after allo-BMT agrees with GRAVY differences observed in TCRBV6 rearrangements prior to BMT.25 This alteration of the hydropathicity index is probably the result of T-cell selection in an MHC class II–negative environment and may have functional implications with regard to the affinity and avidity of the interaction between the TCR and the peptide/MHC complex.58
The questions still remain of how and where the peripheral T cells have been selected in BLS patients that are devoid of endogenous MHC class II. During normal T-cell development, BM-derived T-cell precursors undergo positive and negative selection processes in the thymus upon interaction with MHC class I and II molecules expressed on thymic epithelial and dendritic cells.13-17 In mouse models with different levels of MHC class II expression, it has been shown that TCR/MHC class II interactions are required for the initial stages of positive selection, but not during terminal differentiation.59 Introduction of BM-derived MHC-positive cells in MHC-negative hosts/thymus cultures resulted in inefficient positive selection,60,61 which could be restored by the introduction of MHC-positive thymic epithelial cells or fibroblasts in an MHC-negative thymus.62,63 Furthermore, it has been shown that expression of MHC class II in the thymic cortex combined with lack of MHC class II expression in the medulla gives rise to autoreactivity due to positive selection of T cells in the absence of negative selection.36,64 It is important to note that thymi of BLS patients with a defect in RFX5 are devoid of MHC class II expression.19,20,65 This is in contrast to CIITA−/− and RFX5−/−knock-out mice which have residual MHC class II expression in the thymus, on mature dendritic cells and activated B cells.66-68 In BLS recipients after allo-BMT, both positive and negative selection would be hampered by the absence of endogenous MHC class II expression on the thymic epithelial cells of the cortex and medulla,19,20 although alternative negative selection may occur on graft-derived MHC class II–expressing dendritic cells and monocytes57 that have migrated to the thymic medulla. Therefore, thymic selection processes in the absence of endogenous MHC class II expression result in the observed altered functional and phenotypical properties of T cells after allo-BMT.
ACKNOWLEDGMENT
We thank Drs S.J.P Gobin, R.R.P. de Vries, I.I.N. Doxiadis, G.M.Th. Schreuder (Leiden University Medical Center), and J.T. Kurnick (Massachusetts General Hospital and Harvard Medical School, Boston, MA) for critically reading the manuscript, L. Wilson (Leiden University Medical Center) for assisting with FACS analysis, and A. van de Marel and M. van der Keur (Leiden University Medical Center) for FACS-sorting and analysis.
Supported in part by the J.A. Cohen Institute for Radiopathology and Radiation Protection (IRS).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Peter J. van den Elsen, PhD, Department of Immunohematology and Blood Bank, Leiden University Medical Center, Bldg 1, E3-Q, Albinusdreef 2, PO Box 9600, 2300 RC Leiden, The Netherlands; e-mail: pvdelsen@euronet.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal