An isochromosome of the long arm of chromosome 17, i(17q), is the most frequent genetic abnormality observed during the disease progression of Philadelphia chromosome–positive chronic myeloid leukemia (CML), and has been described as the sole anomaly in various other hematologic malignancies. The i(17q) hence plays a presumably important pathogenetic role both in leukemia development and progression. This notwithstanding, the molecular consequences of this abnormality have not been investigated in detail. We have analyzed 21 hematologic malignancies (8 CML in blast crisis, 8 myelodysplastic syndromes [MDS], 2 acute myeloid leukemias, 2 chronic lymphocytic leukemias, and 1 acute lymphoblastic leukemia) with i(17q) by fluorescence in situ hybridization (FISH). Using a yeast artificial chromosome (YAC) contig, derived from the short arm of chromosome 17, all cases were shown to have a breakpoint in 17p. In 12 cases, the breaks occurred within the Smith-Magenis Syndrome (SMS) common deletion region in 17p11, a gene-rich region which is genetically unstable. In 10 of these 12 cases, we were able to further map the breakpoints to specific markers localized within a single YAC clone. Six other cases showed breakpoints located proximally to the SMS common deletion region, but still within 17p11, and yet another case had a breakpoint distal to this region. Furthermore, using chromosome 17 centromere-specific probes, it could be shown that the majority of the i(17q) chromosomes (11 of 15 investigated cases) were dicentric, ie, they contained two centromeres, strongly suggesting that i(17q) is formed through an intrachromosomal recombination event, and also implicating that the i(17q), in a formal sense, should be designated idic(17)(p11). Because i(17q) formation results in loss of 17p material, potentially uncovering the effect of a tumor suppressor on the remaining 17p, the occurrence of TP53 mutations was studied in 17 cases by sequencing the entire coding region. In 16 cases, noTP53 mutations were found, whereas one MDS displayed a homozygous deletion of TP53. Thus, our data suggest that there is no association between i(17q) and coding TP53 mutations, and that another tumor suppressor gene(s), located in proximity of the SMS common deletion region, or in a more distal location, is of pathogenetic importance in i(17q)-associated leukemia.
ISOCHROMOSOME 17q [i(17q)] is the most common isochromosome in hematologic malignancies and has been described both as a primary and a secondary aberration. i(17q) is a frequent secondary chromosomal aberration in the accelerated phase or blast crisis of chronic myeloid leukemia (CML), indicating that this abnormality plays an important role in the disease progression.1,2 The i(17q) is also found in 1.4% to 2.4% of acute myeloid leukemias (AML), chronic myeloproliferative disorders (CMD), myelodysplastic syndromes (MDS), acute lymphoblastic leukemias (ALL), and chronic lymphoproliferative disorders with clonal chromosome aberrations.3
Isochromosome formation is generally assumed to be the result of a break or misdivision of the centromere, resulting in two mirror image arms attached to a single centromere. In the case of i(17q) formation, this would lead to loss of the entire short arm and gain of the entire long arm. However, cytogenetic4-6 and recent molecular genetic studies7,8 on constitutional and acquired isochromosomes suggest that the breakpoints, in the few cases studied, occur in the pericentromeric region. Furthermore, primitive neuroectodermal tumors (PNET) often show an i(17q)9,10 and data from loss of heterozygosity (LOH) studies are consistent with a clustering of breakpoints in 17p11 close to the centromere.8 This region coincides with the Smith-Magenis Syndrome (SMS) common deletion region, a genetically unstable gene-rich region frequently deleted in SMS patients.11-13
Whether i(17q) in hematologic malignancies also is the result of a breakpoint within the pericentromeric region of 17p has not been determined, although a few cytogenetic and fluorescence in situ hybridization (FISH) studies, using chromosome 17 centromere-specific probes, have shown that i(17q) in some cases is dicentric.4-6,14 15 These findings indicate that the i(17q) in hematopoietic disorders also could be the result of a breakpoint in 17p, close to the centromere.
Given the frequent occurrence of i(17q) in hematologic malignancies, and the fact that i(17q) sometimes is found as the sole karyotypic abnormality—suggesting a possible role as a primary genetic alteration in leukemogenesis—we investigated 21 cases of hematologic neoplasms characterized by an i(17q) using a yeast artificial chromosome (YAC) contig from the proximal region of chromosome 17 (17p11) and FISH on metaphase chromosomes. Because i(17q) formation results in loss of 17p material, potentially unmasking the effect of a tumor suppressor gene (TSG), we also studied the occurrence of TP53 mutations in 17 cases by sequencing the entire coding region.
MATERIALS AND METHODS
Patients.
Twenty-one patients with hematologic malignancies—8 CML in blast crisis (BC), 8 MDS, 2 AML, 1 ALL, and 2 chronic lymphocytic leukemias (CLL)—with an i(17q) as a sole or a secondary chromosomal aberration, and where material in fixative was available, were selected for FISH and TP53 mutational analyses (Table1). In addition, six patients with CML in accelerated or blastic phase without chromosome 17 abnormalities were studied. All patients had been cytogenetically analyzed, either at the Department of Clinical Genetics, Lund University Hospital (Sweden) or at the Department of Human Genetics, University of Leuven (Belgium).
Clinical and Genetic Features of the 21 Hematologic Malignancies With i(17q)
| Case No. . | Sex/ Age (yr) . | Disease . | Karyotype . | % i(17q) by Interphase FISH . | TP53 Coding Mutation . |
|---|---|---|---|---|---|
| 1 | M/77 | CML BC | 47,XY,+8,t(9;22)(q34;q11),i(17)(q10)[5] | 91 | No* |
| 2 | F/61 | CLL | 46,XX,i(17)(q10)[11] | 90 | No |
| 3 | M/78 | CLL | 46,XY,i(17)(q10)[9]/46,XY[6] | 69 | No† |
| 4 | M/73 | MDS | 46,XY,i(17)(q10)[10] | 86 | No‡ |
| 5 | M/74 | MDS | 46,XY,i(17)(q10)[7]/46,XY[3] | 75 | No |
| 6 | F/74 | AML | 46,XX,del(6)(q14q22),i(17)(q10)[9] | 95 | No |
| 7 | M/60 | MDS | 47,XY,+13,i(17)(q10)[8]/46,XY[7] | 82 | No |
| 8 | F/3 | ALL | 53-55,XX,+5,+6,+8,+9,+10,+13,i(17)(q10),+18,+19,+21,+mar[19]/46,XX[5] | ND | No |
| 9 | M/57 | MDS | 45,XY,der(13;14)(q10;q10)c[1]/45,idem,i(17)(q10)[24] | ND | ND |
| 10 | M/70 | MDS | 46,XY,i(17)(q10)[23]/46,XY[2] | 82 | No |
| 11 | M/76 | MDS | 46,XY,i(17)(q10)[25] | 77 | No |
| 121-153 | M/16 | CML BC | 46,XY,t(9;22)(q34;q11)[3]/49,idem,+8,+14,i(17)(q10),+19[5] | 95 | No |
| 13 | F/50 | CML BC | 47,XX,+8,t(9;22)(q34;q11),i(17)(q10)[25] | 89 | No1-155 |
| 14 | F/45 | CML BC | 46,XX,t(9;22)(q34;q11),i(17)(q10)[23]/47,idem,+mar[2] | 93 | No |
| 15 | F/28 | CML BC | 46,XX,t(9;22)(q34;q11)[1]/47,idem,i(17)(q10),+21[4] | 52 | ND |
| 16 | M/74 | CML BC | 46,XY,t(9;22)(q34;q11),i(17)(q10)[1]/ 47,idem,add(4)(p12),+8[7]/48,idem,add(4)(p12), +8,+der(22)t(9;22)[2] | 93 | No |
| 17 | M/53 | AML | 46,XY,i(17)(q10)[9]/46,XY[6] | 75 | No |
| 18 | F/30 | CML BC | 46,XX,t(9;22)(q34;q11)[18]/46,idem,i(17)(q10)[3]/ 46,idem, inv(3)(q21q26)[2]/46,idem,inv(3), i(17)(q10)[2] | 55 | No |
| 19 | M/73 | CML BC | 45,X,−Y,t(9;22)(q34;q11),i(17)(q10)[26] | 95 | No1-154 |
| 20 | M/78 | MDS | 46,XY,i(17)(q10)[17]/46,XY[8] | ND | ND |
| 21 | F/82 | MDS | 46,XX,i(17)(q10)[11] | ND | ND |
| Case No. . | Sex/ Age (yr) . | Disease . | Karyotype . | % i(17q) by Interphase FISH . | TP53 Coding Mutation . |
|---|---|---|---|---|---|
| 1 | M/77 | CML BC | 47,XY,+8,t(9;22)(q34;q11),i(17)(q10)[5] | 91 | No* |
| 2 | F/61 | CLL | 46,XX,i(17)(q10)[11] | 90 | No |
| 3 | M/78 | CLL | 46,XY,i(17)(q10)[9]/46,XY[6] | 69 | No† |
| 4 | M/73 | MDS | 46,XY,i(17)(q10)[10] | 86 | No‡ |
| 5 | M/74 | MDS | 46,XY,i(17)(q10)[7]/46,XY[3] | 75 | No |
| 6 | F/74 | AML | 46,XX,del(6)(q14q22),i(17)(q10)[9] | 95 | No |
| 7 | M/60 | MDS | 47,XY,+13,i(17)(q10)[8]/46,XY[7] | 82 | No |
| 8 | F/3 | ALL | 53-55,XX,+5,+6,+8,+9,+10,+13,i(17)(q10),+18,+19,+21,+mar[19]/46,XX[5] | ND | No |
| 9 | M/57 | MDS | 45,XY,der(13;14)(q10;q10)c[1]/45,idem,i(17)(q10)[24] | ND | ND |
| 10 | M/70 | MDS | 46,XY,i(17)(q10)[23]/46,XY[2] | 82 | No |
| 11 | M/76 | MDS | 46,XY,i(17)(q10)[25] | 77 | No |
| 121-153 | M/16 | CML BC | 46,XY,t(9;22)(q34;q11)[3]/49,idem,+8,+14,i(17)(q10),+19[5] | 95 | No |
| 13 | F/50 | CML BC | 47,XX,+8,t(9;22)(q34;q11),i(17)(q10)[25] | 89 | No1-155 |
| 14 | F/45 | CML BC | 46,XX,t(9;22)(q34;q11),i(17)(q10)[23]/47,idem,+mar[2] | 93 | No |
| 15 | F/28 | CML BC | 46,XX,t(9;22)(q34;q11)[1]/47,idem,i(17)(q10),+21[4] | 52 | ND |
| 16 | M/74 | CML BC | 46,XY,t(9;22)(q34;q11),i(17)(q10)[1]/ 47,idem,add(4)(p12),+8[7]/48,idem,add(4)(p12), +8,+der(22)t(9;22)[2] | 93 | No |
| 17 | M/53 | AML | 46,XY,i(17)(q10)[9]/46,XY[6] | 75 | No |
| 18 | F/30 | CML BC | 46,XX,t(9;22)(q34;q11)[18]/46,idem,i(17)(q10)[3]/ 46,idem, inv(3)(q21q26)[2]/46,idem,inv(3), i(17)(q10)[2] | 55 | No |
| 19 | M/73 | CML BC | 45,X,−Y,t(9;22)(q34;q11),i(17)(q10)[26] | 95 | No1-154 |
| 20 | M/78 | MDS | 46,XY,i(17)(q10)[17]/46,XY[8] | ND | ND |
| 21 | F/82 | MDS | 46,XX,i(17)(q10)[11] | ND | ND |
Abbreviations: M, male; F, female; ND, not determined.
A previously described polymorphism in exon 4, Arg72Pro (CGC → CCC), was found (see Results). Exons 2-3 were not analyzed.
Exons 2,3, and 7 were not analyzed.
No PCR products were obtained using primers specific for exons 4-11 of TP53, whereas two microsatellite markers (D17S1322and D17S1326) in 17q, and one (D17S379) in 17p13, yielded fragments of expected sizes (see Results).
Case 12 has previously been reported in ref 48.
Exons 2-5 were not analyzed.
A noncoding polymorphism was found in intron 9 [exon 9 (+12) T → C].
Cytogenetic studies.
Bone marrow and/or peripheral blood cells were cultured and cytogenetically analyzed according to standard procedures. The remaining cell pellets were stored at −20°C in fixative (methanol:acetic acid, 3:1, vol/vol). Description of karyotypes and criteria for clonality followed the recommendation of ISCN (1995).16
Probes and FISH analysis.
The following 10 YACs were used: 845d2, 935a6, 828b9, 951g11, 52b10, 481h11, 912d7, 961f10, 427g11 (TP53), and 436g12 (TP53). The YAC clones were obtained from CEPH (Paris, France) and were selected on the basis of their reported genetic and physical mapping position on chromosome 17 (refs 17 and 18,http://carbon.wi.mit.edu:8000/cgi-bin/contig/phys_map, andhttp://www.cephb.fr/infoclone.html). All clones, except 427g11 and 436g12, are contained within the whole contig 17.3 (WC17.3) constructed at the Whitehead Institute for Genomic Research (MIT, Boston, MA), and have been mapped within or adjacent to the Charcot Marie Tooth (CMT1A) and SMS loci in 17p1117 18 (Fig1). YACs 427g11 and 436g12 contain TP53 (verified by amplifying the entire coding region of TP53). Hybridization of individual YAC clones to normal lymphocyte metaphase cells showed that 845d2 mapped to 17q11, whereas the localization of 427g11 and 436g12 to 17p13, and of the remaining seven YAC clones to 17p11, was confirmed. For identification of the entire chromosome 17, a whole chromosome painting probe (wcp17), obtained by inter-Alu PCR from a somatic cell hybrid containing human chromosome 17 (NA10498; NIGMS Human Genetic Mutant Cell Repository, Camden, NJ), was used. The probe D17Z1 (American Type Culture Collection [ATCC], Manassas, VA ) was used as a chromosome 17 centromere-specific (cen17) probe, and for identification of chromosome arm 17p, a commercially available partial chromosome paint 17 probe (pcp17) was used (ALTechnologies, Arlington, VA).
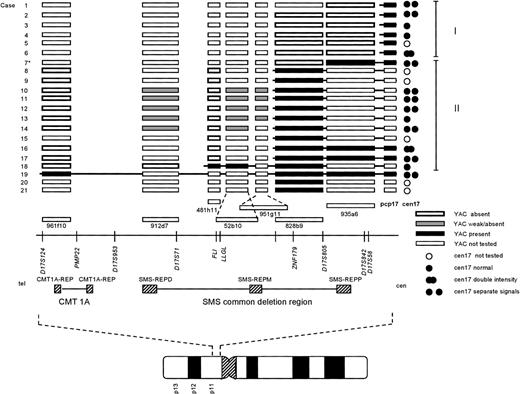
Summary of the FISH results. Each case number (1-21) is indicated to the left. For clarity, the cases were ordered depending on their breakpoint location, allowing further assignment into two different groups (I and II; indicated to the right). The symbols used to designate the result of a FISH experiment are indicated at the bottom right of the figure. The approximate locations of the YAC clones used, in relation to selected markers and genes on the physical map, is based on the results reported by Chen et al, 1997,18 and public available databases (see Materials and Methods). TheCMT1A locus and SMS common deletion region, together with the repetitive sequence elements (CMT1A-REP, and SMS-REP) contained within these regions, are indicated below the physical map. At the bottom, an ideogram of chromosome 17 is depicted, showing the approximate location of the region investigated. *Case 7 showed a split signal using YAC 935a6, with one signal on each q arm (see results and Fig 2C). For abbreviations used, see text.
Summary of the FISH results. Each case number (1-21) is indicated to the left. For clarity, the cases were ordered depending on their breakpoint location, allowing further assignment into two different groups (I and II; indicated to the right). The symbols used to designate the result of a FISH experiment are indicated at the bottom right of the figure. The approximate locations of the YAC clones used, in relation to selected markers and genes on the physical map, is based on the results reported by Chen et al, 1997,18 and public available databases (see Materials and Methods). TheCMT1A locus and SMS common deletion region, together with the repetitive sequence elements (CMT1A-REP, and SMS-REP) contained within these regions, are indicated below the physical map. At the bottom, an ideogram of chromosome 17 is depicted, showing the approximate location of the region investigated. *Case 7 showed a split signal using YAC 935a6, with one signal on each q arm (see results and Fig 2C). For abbreviations used, see text.
YAC probe preparations and FISH analyses were performed essentially as described previously.19 In brief, after amplification of total yeast DNA using Alu primers, probes were generated by labeling with biotin- or digoxigenin-conjugated dUTP (Boehringer Mannheim, Mannheim, Germany), using Amersham’s Mega prime kit (Amersham, UK). Probes were purified on a Sepharose CL-6B column (Pharmacia, Uppsala, Sweden), and 100 ng of each probe was coprecipitated with Cot 1 DNA (Bethesda Research Laboratories [BRL], Gaithersburg, MD) and Sonicated salmon DNA. Cell pellets were resuspended in fresh fixative. After spreading, the slides were kept at 60°C overnight, and subsequently treated in 2X saline sodium citrate (SSC) at 60°C for 1 to 2 hours. Slides were then treated in 10 μg/mL pepsin in 10 mmol/L HCl for 5 to 10 minutes at 37°C, washed in 1X phosphate-buffered saline (PBS), followed by dehydration. The chromosomes were denatured in 2X SSC and 70% formamide at 70°C for varying amounts of time, typically between 15 seconds and 2 minutes. The previously precipitated probes were resuspended in hybridization solution (50% formamide, 50 mmol/L Na2HPO4, 2X SSC, and 10% dextrane sulfate), denatured at 70°C for 10 minutes, prehybridized at 37°C for 1 hour, applied to each slide at a volume of 10 μL, and then covered by a coverslip. Hybridizations were performed overnight in a humidified sealed chamber, and the slides were washed at 70°C in 0.4X SSC for 2 minutes. Biotinylated probes were detected with avidin-Cy3 (Amersham, Amersham Place, UK) and digoxigenin-labeled probes were visualized using one layer of sheep antidigoxigenin-fluorescein isothiocyanate (FITC; Boeringer Mannheim, Mannheim, Germany), at a concentration of 1 and 5 μg/mL, respectively. The hybridization signals were analyzed in a Cytovision Ultra System (Applied Imaging, Sunderland, UK), using a cooled charged coupled device (CCD) camera. Whenever possible, and in the great majority, at least 10 metaphases were analyzed.
Interphase FISH and TP53 mutational analyses.
To evaluate the size of the clones characterized by an i(17q), FISH analysis was performed on interphase cells using a pool of two YACs containing TP53 (427g11 and 436g12) and one YAC localized to 17q11 (845d2). To reduce the false-positive background rate, only nuclei with at least two signals for the 845d2 YAC were scored for the presence of one or two copies of the TP53 locus. At least 100 interphase nuclei were studied in all samples, including two normal controls.
DNA was extracted from remaining fixative or from stored peripheral and/or bone marrow samples using standard methods. Sufficient amounts of DNA were obtained from 17 cases, and 200 ng DNA was used in each polymerase chain reaction (PCR) reaction to amplify the entire coding region of the TP53 gene in seven or eight different fragments. Primer sequences for the TP53 gene are given in Table 2. Primers for the microsatellite markers D17S379, D17S1322, and D17S1326 were obtained from Research Genetics, Inc (Huntsville, AL). PCR was performed in 50-μL reactions with 1.5 mmol/L MgCl2, and 200 μmol/L of each dNTP in PCR buffer II (Perkin Elmer, Branchburg, NJ). Depending on the primer pairs used, the DNA was amplified for 30 cycles at 93°C to 96°C for 30 to 45 seconds, 55°C to 62°C for 30 to 90 seconds, and 72°C for 50 to 90 seconds, followed by a 5- to 10-minute final extension at 72°C. One of the primers of each primer pair contained an M13 sequence (Table 2), allowing direct sequencing of the PCR product using the Dye Primer Cycle Sequencing Ready Reaction −21 M13 kit (Perkin Elmer) on an ABI 373 Sequencer (Applied Biosystems, Foster City, CA) with 4.75% denaturing acrylamide gels.
Sequences of the Primer Pairs Used to Amplify theTP53 Gene
| Primer Pair . | Sequence (5′ → 3′) . |
|---|---|
| Exon 2F | M13- GGT TGT GGT GAA ACA TTG GA |
| Exon 3R | GAT GGG TGA AAA GAG CAG TCA |
| Exon 4F | M13- ACA ACG TTC TGG TAA GGA CAA |
| Exon 4R | ACA CTG ACA GGA AGC CTA AG |
| Exon 5F | M13- AGG AGG TGC TTA CAC ATG TT |
| Exon 6R | CAT CTC ATG GGG TTA TAG GGA |
| Exon 6F | M13- ATG AGC GCT GCT CAG ATA GCG AT |
| Exon 6R | CAT CTC ATG GGG TTA TAG GGA |
| Exon 7F | TCA TCT TGG GCC TGT GTT ATC |
| Exon 7R | M13- CCA ACC ACC CTT GTC CTT T |
| Exon 8F | M13- GAG TAG ATG GAG CCT GGT TT |
| Exon 9R | GGC ATT TTG AGT GTT AGA CTG GA |
| Exon 10F | M13- CGA TGT TGC TTT TGA TCC GTC A |
| Exon 10R | GAG AAT GGA ATC CTA TGG CT |
| Exon 11F | M13- AAT TCC CGT TGT CCC AGC CTT A |
| Exon 11R | TGC TTC TGA CGC ACA CCT ATT |
| Primer Pair . | Sequence (5′ → 3′) . |
|---|---|
| Exon 2F | M13- GGT TGT GGT GAA ACA TTG GA |
| Exon 3R | GAT GGG TGA AAA GAG CAG TCA |
| Exon 4F | M13- ACA ACG TTC TGG TAA GGA CAA |
| Exon 4R | ACA CTG ACA GGA AGC CTA AG |
| Exon 5F | M13- AGG AGG TGC TTA CAC ATG TT |
| Exon 6R | CAT CTC ATG GGG TTA TAG GGA |
| Exon 6F | M13- ATG AGC GCT GCT CAG ATA GCG AT |
| Exon 6R | CAT CTC ATG GGG TTA TAG GGA |
| Exon 7F | TCA TCT TGG GCC TGT GTT ATC |
| Exon 7R | M13- CCA ACC ACC CTT GTC CTT T |
| Exon 8F | M13- GAG TAG ATG GAG CCT GGT TT |
| Exon 9R | GGC ATT TTG AGT GTT AGA CTG GA |
| Exon 10F | M13- CGA TGT TGC TTT TGA TCC GTC A |
| Exon 10R | GAG AAT GGA ATC CTA TGG CT |
| Exon 11F | M13- AAT TCC CGT TGT CCC AGC CTT A |
| Exon 11R | TGC TTC TGA CGC ACA CCT ATT |
M13 indicates that the sequence 5′ TGT AAA ACG ACG GCC AGT 3′ was attached to the 5′ end of the primer, allowing direct sequencing of the PCR product (see Materials and Methods).
RESULTS
Cytogenetic analysis.
The cytogenetic findings are summarized in Table 1. Among the eight CML BC, the i(17q) was the only additional structural aberration in five. Seven of the eight MDS, as well as the two CLL, displayed an i(17q) as the sole acquired cytogenetic aberration. The ALL had several numerical and structural changes in addition to the i(17q), whereas one of the two AML had one additional chromosomal change.
i(17q) is the result of clustered breakpoints in 17p11.
The i(17q) in all 21 cases was shown to be the result of a breakpoint within 17p. The results from the FISH analyses are schematically summarized in Fig 1. Cases 1-18 were ordered and divided into two groups depending on their breakpoint location (I and II; Fig 1). Cases 1-6 (group I) showed no signal on i(17q) using the most proximal YAC (935a6), whereas the pcp17 probe clearly revealed a signal, consistent with the presence of 17p material (Fig 2A and B). Case 7 (included in group II, see below) showed an unexpected pattern of hybridization on i(17q) using YAC 935a6; a split YAC signal with one signal on each q arm was observed (Fig 2C), whereas YAC 828b9 clearly was absent (not shown). This could indicate that an inversion, with the breakpoint localized within the 935a6 YAC, had taken place before the formation of the i(17q). In cases 8-17 (group II), the i(17q) was positive for YAC 828b9 (Fig 2D), but negative for the more telomeric YAC 481h11 (Fig 2E). The two YACs 52b10 and 951g11, located in between 828b9 and 481h11 (Fig 1), showed an inconsistent intra-individual hybridization pattern in cases 8-17 (not shown). In some metaphases, a weak signal was present on i(17q), whereas some metaphases lacked a signal. These YACs, as well as YAC 912d7, contain parts of repetitive sequence elements (SMS-REP) which are present in the SMS common deletion region (Fig 1). Thus, the most likely explanation for the weak hybridization signals observed when using these YACs is a weak crosshybridization to the most proximally located SMS-REP (SMS-REPP). The breakpoint in case 18 (group II) was located between YAC 481h11 and 912d7 (not shown). Only case 19 had a breakpoint telomeric to the SMS common deletion region, because YAC 961f10 was contained within the i(17q) (Fig 2F). In cases 20 and 21, the YAC 828b9 was present on i(17q), but we were unable to further map the breakpoint in these two cases because of a lack of material.
Examples of FISH analyses of i(17q). YAC and pcp17 probes appear as red signals (Cy3), whereas wcp17 and cen17 probes appear as green signals (FITC). When red (Cy3) and green (FITC) signals are located adjacent to each other a yellow signal is obtained. Chromosomes were counterstained using DAPI, yielding a blue color. (A) Case 1 hybridized with a combination of YAC 935a6 and wcp17 showing the absence of a YAC signal on i(17q) and presence of a signal on the normal chromosome 17. (B) Case 1 hybridized with a combination of pcp17 and cen17 showing two separate centromere 17 signals on the i(17q), with 17p material present in between. (C) Case 7 hybridized with a combination of YAC 935a6 and wcp17 showing a split YAC signal on i(17q), with one signal on each q arm. (D) Case 11 hybridized with a combination of YAC 828b9 and wcp17 showing the presence of a YAC signal on i(17q). (E) Case 11 hybridized with a combination of YAC 481h11 and wcp17 showing the absence of YAC signal on i(17q) and presence of a signal on the normal chromosome 17. (F) Case 19 hybridized with a combination of YAC 961f10 and wcp17 showing the presence of YAC signal on i(17q).
Examples of FISH analyses of i(17q). YAC and pcp17 probes appear as red signals (Cy3), whereas wcp17 and cen17 probes appear as green signals (FITC). When red (Cy3) and green (FITC) signals are located adjacent to each other a yellow signal is obtained. Chromosomes were counterstained using DAPI, yielding a blue color. (A) Case 1 hybridized with a combination of YAC 935a6 and wcp17 showing the absence of a YAC signal on i(17q) and presence of a signal on the normal chromosome 17. (B) Case 1 hybridized with a combination of pcp17 and cen17 showing two separate centromere 17 signals on the i(17q), with 17p material present in between. (C) Case 7 hybridized with a combination of YAC 935a6 and wcp17 showing a split YAC signal on i(17q), with one signal on each q arm. (D) Case 11 hybridized with a combination of YAC 828b9 and wcp17 showing the presence of a YAC signal on i(17q). (E) Case 11 hybridized with a combination of YAC 481h11 and wcp17 showing the absence of YAC signal on i(17q) and presence of a signal on the normal chromosome 17. (F) Case 19 hybridized with a combination of YAC 961f10 and wcp17 showing the presence of YAC signal on i(17q).
The majority of the i(17q) contain two centromeres.
A total of 15 cases could conclusively be analyzed for the presence of one or two chromosome 17 centromeres on the i(17q) (Fig 1). In nine cases, two separate cen17 signals, with 17p material present in between, were obtained when using the cen17/pcp17 combination (see, eg, Fig 2B). In cases 6 and 16, no separation of the two cen17 signals was observed, but the signal intensity was roughly two times stronger on the i(17q) than on the normal chromosome 17, consistent with the presence of two centromeres on i(17q). In four cases (cases 3, 4, 13, and 18), no separation of the two cen17 signals was observed, nor was the cen17 signal on the i(17q) stronger than on the normal chromosome 17. This finding may be due to a combination of suboptimal hybridization efficiency and presence of condensed chromosomes, not giving rise to stronger or separate cen17 signals. An alternative explanation could be that the mechanism, by which these i(17q) were formed, is different from the other i(17q). Nevertheless, the great majority of i(17q) (11 of 15 cases) were clearly dicentric.
FISH analysis of CML BC without structural changes of chromosome 17.
Given the clustering of breakpoints in 17p11 in a region, which due to the presence of repetitive sequences has been shown to be genetically unstable and deleted in patients with SMS,17 18 we investigated whether CML BC without structural changes of chromosome 17 may harbor submicroscopic deletions within this region. Six cases were analyzed using YAC 481h11, but no clearly absent or diminished signals were observed.
i(17q) is not associated with mutations in the TP53 gene.
No coding TP53 mutations were identified in any of the 17 cases investigated (Table 1). In 13 cases, the entire coding region of theTP53 gene was sequenced. Because of a lack of material, exons 2 and 3 were not sequenced in case 1, and for the same reason no data were obtained from exons 2, 3, and 7 and from exons 2-5 in cases 3 and 13, respectively. A previously described polymorphism in exon 4 (http://www.iarc.fr/p53/poly.htm), Arg72Pro (CGC → CCC), was found in case 1, whereas case 19 showed a noncoding polymorphism in intron 9 [exon 9 (+12) T → C]. In case 4, no PCR products were obtained when using the primer pairs specific for exons 4-11, whereas amplification of two microsatellite markers located in 17q (D17S1322 and D17S1326), and one in 17p13 (D17S379), yielded fragments of expected sizes (data not shown). These results are consistent with a homozygous deletion of theTP53 gene.
To rule out the possibility that the low occurrence of TP53mutations was the result of a high admixture of normal cells, we performed interphase FISH to estimate the percentage of cells containing an i(17q). The size of the clones was shown to be relatively large (range, 52% to 95%, mean 82%; Table 1), making it highly unlikely that the lack of identified TP53 mutations was due to a large contamination of normal cells. Two normal peripheral blood samples were investigated to determine the false-positive background rate. A total of 349 and 154 interphase nuclei were analyzed in each sample, revealing a background rate of 7% and 6%, respectively.
DISCUSSION
In the present study, we show that i(17q) is the result of a breakpoint in the pericentromeric region of the short arm of chromosome 17 and that most breakpoints occur proximally to, or within, a previously delineated region, the SMS common deletion region in 17p11.11-13 Using a YAC contig from this region and FISH on metaphase chromosomes, we mapped the breakpoints in 19 of 21 hematologic malignancies characterized by an i(17q). In 12 cases, the breaks occurred within the SMS common deletion region and the great majority of the breaks (10 of 12 cases) were localized within or adjacent to YAC 828b9 (Fig 1). Six cases showed breakpoints located proximally to the SMS common deletion region, but still within 17p11, and one case had a breakpoint distal to the SMS common deletion region.
We were also able to show that the majority of the i(17q) chromosomes (11 of 15 investigated cases) contained two centomeres, lending further support to previous results from a limited number of cases using conventional C-banding techniques4-6 and centromere 17–specific FISH probes.14,15 Hence, i(17q) should formally be designated idic(17)(p11).16 It has been proposed that i(17q) may arise through a break in the short arm followed by joining of the two chromatids containing the centromeres.6 It is also possible that i(17q) could occur through a nonreciprocal translocation between the two chromosomes 17, with breakpoints in 17p11 in each homolog. This would require a gain of a chromosome 17 before the translocation event because one normal chromosome 17 is present in most instances. In four cases, the i(17q) did not appear to have two centromeres. Although we believe that the reason for this is methodological, we cannot exclude the possibility that these i(17q) were formed through a different mechanism, eg, a nonreciprocal translocation with breakpoints in 17p11 and 17q11 of each homolog. Again, however, this would require a nondisjunctional event leading to a gain of a chromosome 17 before the translocation. Therefore, we believe that i(17q) arises through an intrachromosomal recombination event. Interestingly, it has been shown that the SMS common deletion region, the region to which most of our breakpoints mapped, contains three copies of SMS-REPs, and that a recombination event between the distal (SMS-REPD) and proximal repeat (SMS-REPP) most likely is responsible for the interstitial deletions observed in SMS patients.18 It is possible that the same mechanism is responsible for the chromosomal disruption and subsequent formation of an i(17q) in hematologic malignancies.
Another tumor type that also frequently shows an i(17q) is PNET. Using LOH and interphase FISH studies, Scheurlen et al8 mapped the breakpoints in nine cases that had been found to have monoallelic deletions encompassing nearly the complete short arm of chromosome 17, suggesting the presence of an i(17q). All nine tumors had breakpoints proximal to, or within, the SMS common deletion region, and in five tumors they were able to determine the chromosomal breakpoint between two adjacent microsatellite markers. The most frequent breakpoint was found between the markers D17S71 and D17S805 (the latter marker is present in YAC 828b9; Fig 1). Thus, it seems as if the breakpoints in hematologic malignancies are quite similar to the ones observed in PNET. Also, we did not detect any clear-cut differences between the localization of the chromosomal breakpoints and the type of hematologic malignancy in our series. This suggests that 17p harbors a “promiscuous” gene which, if structurally rearranged or otherwise deregulated, is pathogenetically important in a wide variety of neoplasms.
The i(17q) formation leads to a loss of genetic material located distally to the breakpoints described above, whereas material located proximally is duplicated. It is presently not known if the loss or the gain, or perhaps a combination of the two, is the pathogenetically important event. It is also unknown if the observed clustering of breakpoints within the SMS common deletion region merely is a result of its genetic instability, and that any gene(s) located distally to the breakpoint on the short arm of chromosome 17 could be a candidate TSG, or if gene(s) located in the breakpoint region could become deleted or altered in their expression or structure due to the chromosomal disruption. Although speculative, one of our cases (case 7; Fig 2C) may suggest that the latter mechanism indeed is possible. Using YAC 935a6, we observed a split YAC signal on i(17q) with one signal on each q arm, indicating that an inversion most likely had taken place before the isochromosome formation. This inversion could delete or alter the structure of critical gene(s) within the SMS common deletion region.
Several genes and ESTs have been mapped to the SMS common deletion region,17,18,20 among them the human homolog (LLGL1)21 of the drosophila TSG D-lgl and a possible transcription factor, ZNF179,21,22 that could be of pathogenetic relevance. In all but two of our cases (18 and 19), the breakpoints were located proximally to LLGL1, consistent with a heterozygous loss of this gene. In principle, any gene within the SMS common deletion region could, however, be important, because chromosomal disruptions have been shown to be associated with concomitant deletions in many instances,19 23-25 and large insert (YAC) clones, which do not detect small deletions or rearrangements, were used in the present study. Given the genetic instability of the SMS common deletion region, we also searched for possible submicroscopic interstitial deletions in six CML BC lacking chromosome 17 abnormalities. Using the YAC 481h11, which is located in proximity to LLGL1, no clearly absent or diminished signals were observed, indicating that larger deletions within the SMS common deletion region is not a generally occurring event in CML BC without chromosome 17 alterations.
The TSG TP53 at 17p13 has been shown to be mutated in several tumor types, including hematologic malignancies. As a consequence of the i(17q) formation, one copy of this gene will be lost, an event that could unmask the effect of an existing mutation in the other allele. In CML BC, the reported incidence of TP53 mutations is variable and somewhat contradictory. Several types of TP53 gene alterations have been described.26 When considering only coding TP53 point mutations, some studies have reported a relatively high incidence (17% to 28%),26-28 whereas others have found a low one (0% to 15%),29-34 with the average frequency being 11% (20 of 175). However, only a few of these studies included cytogenetic data,26,29,33,35 and when ascertaining cases of CML BC with i(17q), in which TP53mutational analysis had been performed, and excluding data on cell lines, we were able to identify 18 cases. Of these, a codingTP53 mutation was detected in four (22%).26,29,33 35 In our material, none of the seven investigated CML BC with i(17q) harbored any coding TP53mutation.
TP53 mutations have also been described at varying frequencies (0% to 25%) in MDS,36 AML,37,38ALL,39,40 and CLL.39,40 In MDS and AML,TP53 mutations have been shown to occur predominantly in cases with a complex karyotype including a 17p deletion or −17.36,41,42 As to the occurrence of codingTP53 mutations in MDS, AML, ALL, and CLL associated with i(17q), we could identify only seven cases in the literature.33,41,43 None of these showed any mutation, which is consistent with our results where 0 of 9 cases harboredTP53 mutations. One MDS (case 4) displayed a homozygous deletion of TP53, which could indicate that another closely located TSG could be of pathogenetic importance. The existence of such a TSG, located distally to TP53, has indeed been suggested in several other tumor types, eg, breast and ovarian carcinomas.44-47
Overall, we found no coding TP53 mutations in any of the cases investigated by sequence analysis. Hence, it seems clear that the formation of an i(17q) does not serve to uncover the effect of an already existing TP53 mutation on the normal-appearing chromosome 17. Whether the pathogenetically important gene(s) in i(17q)-associated leukemia is located in proximity of the SMS common deletion region—as could be suggested by the observed clustering of breakpoints in this region—or in a more distal region, remains to be established.
ACKNOWLEDGMENT
The authors thank Margareth Isaksson for expert technical assistance.
Supported by grants from the Swedish Cancer Society, the Children’s Cancer Fund of Sweden, the Swedish Society of Medicine, the IngaBritt and Arne Lundberg Foundation, and the Belgian Program and Inter-university Poles of Attraction initiated by the Belgian State, Prime Minister’s Office, Science Policy Programming.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Thoas Fioretos, MD, PhD, Department of Clinical Genetics, University Hospital, S-221 85 Lund, Sweden; e-mail:Thoas.Fioretos@klingen.lu.se.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal