Abstract
Although numerous somatic mutations that contribute to the pathogenesis of childhood acute lymphoblastic leukemia (ALL) have been identified, no specific cytogenetic or molecular abnormalities are known to be consistently associated with relapse. Thep16INK4A (p16), which encodes for both p16INK4A and p19ARF proteins, andp15INK4B (p15) genes are inactivated by homozygous deletion and/or p15 promoter hypermethylation in a significant proportion of cases of childhood ALL at the time of initial diagnosis. To determine whether alterations in these genes play a role in disease progression, we analyzed a panel of 18 matched specimen pairs collected from children with ALL at the time of initial diagnosis and first bone marrow relapse for homozygous p16 and/orp15 deletions or p15 promoter hypermethylation. Four sample pairs contained homozygous p16 and p15 deletions at both diagnosis and relapse. Among the 14 pairs that werep16/p15 germline at diagnosis, three ALLs developed homozygous deletions of both p16 and p15, and two developed homozygous p16 deletions and retained p15germline status at relapse. In two patients, p15 promoter hypermethylation developed in the interval between initial diagnosis and relapse. In total, homozygous p16 deletions were present in nine of 18 cases, homozygous p15 deletions in seven of 18 cases, and p15 promoter hypermethylation in two of eight cases at relapse. These findings indicate that loss of function of proteins encoded by p16 and/or p15 plays an important role in the biology of relapsed childhood ALL, and is associated with disease progression in a subset of cases.
RELAPSED ACUTE lymphoblastic leukemia (ALL) is the fourth most common malignancy that occurs in children.1 While approximately 75% of children with newly diagnosed ALL are expected to be cured with contemporary treatment regimens, the outcome for children who experience a bone marrow relapse is poor, with a 6-year survival rate of 20% in a recent report from the Children’s Cancer Group.1 An important challenge is to define the biologic and genetic basis for these differences in treatment outcome.
Despite identification of a wide variety of oncogenes and tumor-suppressor genes (TSGs) that encode for proteins involved in leukemogenesis, relatively little is known about the genetic features of relapsed ALL. While it is likely that mutations in genes that encode proteins involved in transport or metabolism of chemotherapy agents (eg, MDR1) play a role in the progression of ALL, several observations indicate that mutations must also develop in specific oncogenes/TSGs. Relapsed ALL is often sensitive to the same drugs with which the patient was initially treated, arguing against selection for absolute drug resistance as a sole explanation for relapse. Permanent cell lines can be established more frequently from patients at relapse than at initial diagnosis.2 Similarly, relapse ALL cells engraft in irradiated immunodeficient mice more readily than cells from diagnosis.3 Confirming the multihit model of oncogenesis, oncogene and TSG mutations have been linked to progression in specific subtypes of leukemia.4-8
Cytogenetic and molecular data suggest that TSG inactivation plays an important role in leukemogenesis; these same TSGs are also attractive candidates for the genes involved in progression of leukemia. To date,p16INK4A (p16, MTSI, CDKN2) and p15INK4B (p15, MTS2,CDKN2B) are the only TSGs identified that are inactivated in a significant percentage of leukemias at the time of initial diagnosis.9,10p16 and p15, located within 25 kb of one another on the short arm of chromosome 9 (9p21), encode proteins (p16 and p15) that function as inhibitors of the cyclin-dependent kinases CDK4 and CDK6, which, complexed with cyclin D, phosphorylate the retinoblastoma protein (Rb) to allow progression through the cell cycle.11-13p16 also encodes a second protein, p19ARF, via an alternative first exon and translation of common exons 2 and 3 in a different reading frame.14 Recent evidence suggests that p19ARF, which shares no amino acid homology with p16, enhances the functional activity of wild-type p53.15-17 Thus, homozygousp16 deletion is predicted to functionally inactivate growth-inhibitory proteins that act upstream of pathways involving both Rb and p53.18 Animal models support a critical role for proteins encoded by p16 in oncogenesis, as knockout mice that lack both p16 and p19ARF, or p19ARF alone, develop tumors, the most common of which are lymphomas, at greatly increased frequency.18 19 However, it remains uncertain whether inactivation of p16, p19ARF, or both is the critical event in human leukemogenesis.
Homozygous deletion of both p16 and p15 occurs in at least 60% of T- and 20% of B-lineage cases, and is the major means of functional inactivation of the proteins encoded by these genes in childhood ALL.20-22 In contrast to other tumors,p16 and p15 point mutations are rarely detected in childhood ALL.21p15 is also frequently inactivated by promoter hypermethylation in T-ALL, but p16 promoter methylation is uncommon in childhood ALL.22 Limited information is available regarding potential changes inp16/p15 gene status in the interval between diagnosis and relapse in childhood ALL. However, p16/p15 lesions have been associated with progression of other hematolymphoid malignancies, including chronic myelogenous leukemia (CML) in lymphoid blast crisis, high-grade and transformed non-Hodgkin’s lymphomas, and myelodysplastic syndromes.7,8,23 24
In this study, we analyzed a panel of 18 matched specimens obtained at diagnosis and first bone marrow relapse from children with ALL. We found that homozygous p16/p15 deletions and p15promoter hypermethylation are frequently acquired in the interval between initial diagnosis and relapse, suggesting that loss of function of the proteins encoded by these genes may play an important role in progression of childhood ALL.
MATERIALS AND METHODS
Specimens.
Eighteen matched diagnosis and relapse bone marrow samples from children diagnosed with ALL were available from the cell bank of The Children’s Hospital in Denver. Diagnosis specimens were collected from 1982 to 1994, and relapse specimens were collected from 1982 to 1997. Mononuclear cells were collected by density centrifugation of fresh bone marrow aspirates, resuspended in 10% dimethyl sulfoxide (DMSO) and 20% fetal bovine serum, and maintained at either −70°C or in the vapor phase of liquid nitrogen. In the current study, we analyzed all matched diagnosis/first bone marrow relapse specimen pairs collected from patients with greater than 75% blasts in their bone marrow from whom sufficient material was available that we anticipated being able to isolate at least 10 μg of DNA. Immunophenotyping was performed at the time of presentation using panels of monoclonal antibodies that changed significantly between 1982 and 1997. In general, sufficient data were available to characterize leukemias as B or T lineage. All specimens were collected as part of protocols that had been approved by the Institutional Review Board (IRB) of the University of Colorado Health Sciences Center (UCHSC) and The Children’s Hospital, and the current retrospective analyses were also approved by the IRB.
Cytogenetics.
Giemsa-banded cytogenetic studies were performed from unstimulated cultures of bone marrow aspirate obtained at diagnosis or relapse. From 1982 to 1986, overnight cultures were synchronized using methodology described by Morse et al.25 After 1986, samples were prepared using a direct technique and overnight culture methods described previously with and without giant cell tumor supernatant supplementation.26,27 All cytogenetic results were reviewed by one of the authors (L.M.) and, whenever possible, described using the International System for Cytogenetic Nomenclature (ISCN, 1995).28
Molecular analyses.
Genomic DNA was isolated by standard phenol/chloroform extraction. Southern blot analysis was performed as described previously.29 Briefly, membranes containingBamHI-digested DNAs were cohybridized with a 360-bp DNA fragment corresponding to p16 exon 2 and a previously describedMLL cDNA probe.30 This p16 probe cross-hybridizes with p15, allowing detection of deletions of either p16 or p15. Samples in which p16 and/orp15 bands were absent or were less than 10% to 20% of the intensities of control MLL bands were scored as containing homozygous deletions of the corresponding gene. For leukemias containing MLL translocations, blots were stripped and rehybridized with the p16 probe (along with a BCR cDNA probe) to insure that bands of altered migration did not representp16/p15 rearrangements.
To determine the methylation status of the p15 promoter region, membranes containing HindIII and EagI in combination with HindIII-digested DNAs were hybridized with a 270-bpp15 exon 1 probe as previously described.31Appropriate positive (DNA from the Kg1A cell line, which contains a hypermethylated p15 promoter) and negative (DNA from a healthy individual) controls were included on each blot.
RESULTS
Characteristics of study population.
We analyzed a panel of 18 paired specimens collected from children with ALL at initial diagnosis and first bone marrow relapse (Table1). Relapses occurred at a median of 36.7 months (range, 13 to 82) following initial diagnosis. Seven patients were classified as B lineage (CD10+, CD19+), three patients as T lineage (CD7+, CD2+), six patients were CD10+ but not further phenotyped, and there were two cases of CD10negative infant ALL. No major differences in immunophenotype developed in the interval between initial diagnosis and relapse.
p16INK4A andp15INK4B Gene Status in Matched Diagnosis/Relapse ALL Specimen Pairs
| Patient No. . | Phenotype . | Diagnosis Karyotype . | Relapse Karyotype* . | Diagnosis . | Relapse . | ||||
|---|---|---|---|---|---|---|---|---|---|
| p15 . | p16 . | p15M . | p15 . | p16 . | p15M . | ||||
| DR-3 | B-lineage ALL | 54,XX,+4,der(5)ins(5;?)(q11;?),del(6)(q21), +del(6)(q14), +8,+14,i(17)(q10), +18,+19,+21, +mar[13]/46,XX [12] | 54,XX,+X,+4,+6,+8, del(9)(p22p24), +14,+18,+21,+21 [16]/46,XX [2] | G/G | G/G | ND | D/D | D/D | NA |
| DR-4 | T-ALL | 55-60,XX,+X,+1,+1,+2, +2,+3,+4,+5,+6,+9, +10,+11,+12,+13, +14,+15,+17,+19, +21,+22[cp3]/46,XX[18] | 46,XX[3] incomplete analysis | D/D | D/D | NA | D/D | D/D | NA |
| DR-6 | B-lineage ALL | 52-53,XX not further characterized[3]/46,XX[17] | 52-55,XX not further characterized[5]/46,XX[15] | G/G | G/G | M | D/D | D/D | NA |
| DR-7 | B-lineage ALL | Culture failure MLL germline | 47,XY,t(2;11)(p1?1.2;q23),+mar[9]/46,XY[8] MLL germline | G/G | G/G | ND | G/G | G/G | UM |
| DR-9 | B-lineage ALL | 46,XX,der(4)t(4;?8)(p16;q2?2),del(6)(q13q21),−8, +mar[17]/45-47, idem,+8[7] | 46,XX[3] incomplete analysis | G/G | G/G | UM | G/G | G/G | UM |
| DR-10 | T-ALL | 46,XX,−18,+mar[2]/46,XX[28] | 46,XX[20] | G/G | G/G | UM | G/G | D/D | Mptl |
| DR-11 | ALL, CD10+ | 46,XX,t(6;12)(p11;q11),+mar | 46,XX[22] | D/D | D/D | NA | D/D | D/D | NA |
| DR-12 | T-ALL | 46,XY,inv(5)(p13p15)[9]/46,idem,del(6)(q21)[5]/46,XY[10] | 46X,−Y,−16,+19, +r(?)[17]/48,idem, +5,+7[2] | D/D | D/D | NA | D/D | D/D | NA |
| DR-13 | ALL, CD10+ | 45,X,−Y[5]/46,XY [13] | 46,XY[15] | D/D | D/D | NA | D/D | D/D | NA |
| DR-14 | B-lineage ALL | 47,XY,del(6)(q13), +mar[14]/46,XY[11] | 47,XY,del(12)(p12),+mar [7]/46,XY[4] | G/G | G/G | ND | G/G | G/G | UM |
| DR-15 | ALL, CD10+ | 46,XY,del(11)(p13)[6]/46,XY[14] | 46,XY[20] | G/G | G/G | ND | G/G | G/G | UM |
| DR-16 | Infant ALL, CD10− | 46,XY,t(4;11)(q21;q23)[9] MLL rearranged | 47,XY,t(4;11)(q21;q23), +15[19] MLL rearranged | G/G | G/G | ND | G/G | G/G | UM |
| DR-17 | ALL, CD10+ | 54,XX,+add(1)(q1?2),+6, +10,+14,+14,+17, +21,+21[13]/46,XX[6] | 53,XX,+add(1)(q1?2), +6,+10,+14, +14,+21,+21[21] | G/G | G/G | UM | G/G | G/G | ND |
| DR-18† | ALL, CD10+ | 53,XY,+X,+6,+10,+18,+19, +21,+21[12]/46,XY[2] | 53,XY,+X,+Y, +add(1)(p1?3)+6,+10, +21,+21[10]/46,XY[2] | G/G | G/G | ND | G/G | D/D | ND |
| DR-19 | ALL, CD10+ | 54,XY,+4,+5,+8,+12,+D, +E,+21,+22 | 54,XY,+X,+Y,+4,+5,+6, +14,+21, +21[17]/46,XY[9] | G/G | G/G | UM | G/G | G/G | Mptl |
| DR-20 | Infant ALL, CD10− | No record MLL rearranged | 47,XX,+X,t(11;19)(q23;p13)[12]/46,XX[8] MLL rearranged | G/G | G/G | M | G/G | G/G | UM |
| DR-21 | B-lineage ALL | 46,XX[16] | 46,XX[10] | G/G | G/G | UM | D/D | D/D | NA |
| DR-22 | B-lineage ALL | 46,XX,add(5)(q31)[10]/46,XX[10] | 46,XX,add(5)(q31), add(9)(q22), del(20)(q11.2q13.1)[8]/47,idem,+?22[2]/46, XX[19] | G/G | G/G | M | G/G | G/G | ND |
| Patient No. . | Phenotype . | Diagnosis Karyotype . | Relapse Karyotype* . | Diagnosis . | Relapse . | ||||
|---|---|---|---|---|---|---|---|---|---|
| p15 . | p16 . | p15M . | p15 . | p16 . | p15M . | ||||
| DR-3 | B-lineage ALL | 54,XX,+4,der(5)ins(5;?)(q11;?),del(6)(q21), +del(6)(q14), +8,+14,i(17)(q10), +18,+19,+21, +mar[13]/46,XX [12] | 54,XX,+X,+4,+6,+8, del(9)(p22p24), +14,+18,+21,+21 [16]/46,XX [2] | G/G | G/G | ND | D/D | D/D | NA |
| DR-4 | T-ALL | 55-60,XX,+X,+1,+1,+2, +2,+3,+4,+5,+6,+9, +10,+11,+12,+13, +14,+15,+17,+19, +21,+22[cp3]/46,XX[18] | 46,XX[3] incomplete analysis | D/D | D/D | NA | D/D | D/D | NA |
| DR-6 | B-lineage ALL | 52-53,XX not further characterized[3]/46,XX[17] | 52-55,XX not further characterized[5]/46,XX[15] | G/G | G/G | M | D/D | D/D | NA |
| DR-7 | B-lineage ALL | Culture failure MLL germline | 47,XY,t(2;11)(p1?1.2;q23),+mar[9]/46,XY[8] MLL germline | G/G | G/G | ND | G/G | G/G | UM |
| DR-9 | B-lineage ALL | 46,XX,der(4)t(4;?8)(p16;q2?2),del(6)(q13q21),−8, +mar[17]/45-47, idem,+8[7] | 46,XX[3] incomplete analysis | G/G | G/G | UM | G/G | G/G | UM |
| DR-10 | T-ALL | 46,XX,−18,+mar[2]/46,XX[28] | 46,XX[20] | G/G | G/G | UM | G/G | D/D | Mptl |
| DR-11 | ALL, CD10+ | 46,XX,t(6;12)(p11;q11),+mar | 46,XX[22] | D/D | D/D | NA | D/D | D/D | NA |
| DR-12 | T-ALL | 46,XY,inv(5)(p13p15)[9]/46,idem,del(6)(q21)[5]/46,XY[10] | 46X,−Y,−16,+19, +r(?)[17]/48,idem, +5,+7[2] | D/D | D/D | NA | D/D | D/D | NA |
| DR-13 | ALL, CD10+ | 45,X,−Y[5]/46,XY [13] | 46,XY[15] | D/D | D/D | NA | D/D | D/D | NA |
| DR-14 | B-lineage ALL | 47,XY,del(6)(q13), +mar[14]/46,XY[11] | 47,XY,del(12)(p12),+mar [7]/46,XY[4] | G/G | G/G | ND | G/G | G/G | UM |
| DR-15 | ALL, CD10+ | 46,XY,del(11)(p13)[6]/46,XY[14] | 46,XY[20] | G/G | G/G | ND | G/G | G/G | UM |
| DR-16 | Infant ALL, CD10− | 46,XY,t(4;11)(q21;q23)[9] MLL rearranged | 47,XY,t(4;11)(q21;q23), +15[19] MLL rearranged | G/G | G/G | ND | G/G | G/G | UM |
| DR-17 | ALL, CD10+ | 54,XX,+add(1)(q1?2),+6, +10,+14,+14,+17, +21,+21[13]/46,XX[6] | 53,XX,+add(1)(q1?2), +6,+10,+14, +14,+21,+21[21] | G/G | G/G | UM | G/G | G/G | ND |
| DR-18† | ALL, CD10+ | 53,XY,+X,+6,+10,+18,+19, +21,+21[12]/46,XY[2] | 53,XY,+X,+Y, +add(1)(p1?3)+6,+10, +21,+21[10]/46,XY[2] | G/G | G/G | ND | G/G | D/D | ND |
| DR-19 | ALL, CD10+ | 54,XY,+4,+5,+8,+12,+D, +E,+21,+22 | 54,XY,+X,+Y,+4,+5,+6, +14,+21, +21[17]/46,XY[9] | G/G | G/G | UM | G/G | G/G | Mptl |
| DR-20 | Infant ALL, CD10− | No record MLL rearranged | 47,XX,+X,t(11;19)(q23;p13)[12]/46,XX[8] MLL rearranged | G/G | G/G | M | G/G | G/G | UM |
| DR-21 | B-lineage ALL | 46,XX[16] | 46,XX[10] | G/G | G/G | UM | D/D | D/D | NA |
| DR-22 | B-lineage ALL | 46,XX,add(5)(q31)[10]/46,XX[10] | 46,XX,add(5)(q31), add(9)(q22), del(20)(q11.2q13.1)[8]/47,idem,+?22[2]/46, XX[19] | G/G | G/G | M | G/G | G/G | ND |
Abbreviations: G, germline; D, deleted; ND, not done; NA, not applicable because of deletion; UM, unmethylated; M, methylated; Mptl, partially methylated.
All relapse specimens were collected at the time of first marrow relapse.
Patient with isolated CNS relapse before bone marrow relapse.
Cytogenetic analyses.
Clonal karyotypic abnormalities were identified in 15 cases at initial diagnosis. Six ALLs were hyperdiploid with modal chromosome numbers of 52 to 60. Several cases contained recognized, nonrandom cytogenetic abnormalities, including del(6q) (DR-3, DR-9, DR-14), and t(4;11) (DR-16). At relapse, the clone observed at initial diagnosis was observed in eight of 15 cases. Additional numerical and structural changes were present within this clone in several cases. Of note, a del(9)(p22p24) developed in the interval between diagnosis and relapse in case DR-3. Two of 18 cases had incomplete analyses at relapse, and four of 18 had a 46,XX or 46,XY karyotype at relapse. Clonal abnormalities were observed at relapse in two of three cases that did not have clonal abnormalities detected at initial diagnosis. Cytogenetic analyses were unsuccessful due to culture failure for case DR-7 at initial diagnosis, but a t(2;11)(p1?12;q23) was identified at relapse. This translocation did not affect the 11q23 gene MLL, which was germline at both diagnosis and relapse (data not shown). Cytogenetic results were not available from DR-20 at initial diagnosis, when she was 2 months old. At relapse, a t(11;19)(q23;p13) was observed. Identical MLL rearrangements were present in both the diagnosis and relapse samples (data not shown), indicating that the t(11;19) was also present at initial diagnosis.
p16 and p15 gene deletions.
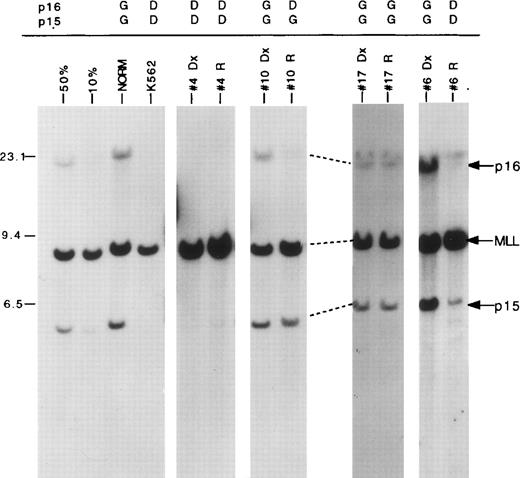
Southern blot analysis was used to determine p16 andp15 gene status (Fig 1 and Table1). Four sample pairs contained homozygous p16 and p15deletions at both diagnosis and relapse. Two of these patients had T-ALL, and the other two were classified as CD10+ ALLs. Among the 14 patients who were p16/p15 germline at diagnosis, three B-lineage ALLs exhibited homozygous deletions of bothp16 and p15 at relapse. In two additional patients (DR-10, a T-ALL, and DR-18, a CD10+ ALL), homozygous deletion of p16 was present in the relapse specimens, whilep15 retained germline status. In total, homozygous p16deletions were present in nine of 18 cases and homozygous p15deletions in seven of 18 cases at relapse.
p16/p15 deletions in matched diagnostic and relapse ALL specimen pairs. Autoradiogram of a blot containingBamH1-digested DNAs cohybridized with p16 and MLLcDNA probes. The locations of the germline p16,p15, and MLL bands are indicated by arrows at right. The migration of molecular size markers are shown in kilobases on the left. Samples include DNA from dilutions of the K562 cell line, which has homozygous p16 and p15 deletions, into normal DNA to simulate deletions in 50% and 10% of the cell population; a healthy control (NORM); K562; and four matched diagnostic (Dx) and relapse (R) patient samples (no. 4, 10, 17, 6). The gene status ofp16 and p15 is listed above each patient sample (G, germline; D, deleted). Faint residual p16 and p15 bands (<10%) in no. 6 relapse patient sample are due to contamination with small amounts of normal cells.
p16/p15 deletions in matched diagnostic and relapse ALL specimen pairs. Autoradiogram of a blot containingBamH1-digested DNAs cohybridized with p16 and MLLcDNA probes. The locations of the germline p16,p15, and MLL bands are indicated by arrows at right. The migration of molecular size markers are shown in kilobases on the left. Samples include DNA from dilutions of the K562 cell line, which has homozygous p16 and p15 deletions, into normal DNA to simulate deletions in 50% and 10% of the cell population; a healthy control (NORM); K562; and four matched diagnostic (Dx) and relapse (R) patient samples (no. 4, 10, 17, 6). The gene status ofp16 and p15 is listed above each patient sample (G, germline; D, deleted). Faint residual p16 and p15 bands (<10%) in no. 6 relapse patient sample are due to contamination with small amounts of normal cells.
p15 promoter hypermethylation.
Using the strategy outlined in Fig 2, we determined whether or not the p15 promoter region was hypermethylated, which correlates with silencing of gene expression, in samples that did not contain p15 deletions. Sufficient DNA was available to perform these analyses for eight diagnostic samples.p15 exon 1 was homozygously hypermethylated in three of these eight patients, and unmethylated in the remaining five (Table 1). Sufficient DNA was available to perform these analyses for eight of 11p15-germline relapse specimens. Two cases reproducibly showed partial p15 promoter hypermethylation (Fig 2 and Table 1), suggesting either that one allele was hypermethylated in all cells, or that a subpopulation, accounting for about half the cells, was homozygously hypermethylated. We observed several different patterns ofp15 methylation status in the matched specimens. Particularly interesting is case DR-10, which had no evident p16/p15abnormalities at initial diagnosis, and contained homozygousp16 deletion and heterozygous p15 promoter hypermethylation at relapse (Figs 1 and 2).
p15 promoter hypermethylation. (A) Schematic representation of p15 exon 1 and surrounding HindIII (H3) and EagI (E) sites. The p15 probe used (indicated above) hybridizes to exon 1. A 2.8-kb band is seen when the samples are digested with HindIII alone. When the samples are digested withEagI (a methylation-sensitive enzyme) in combination withHindIII, a 0.6-kb band is recognized by the probe if the gene is unmethylated (cuts at EagI) and a 2.8-kb band is present if the sample is methylated (does not cut at EagI) (note that a 2.2-kb band is not seen, because the exon 1 probe does not hybridize to this region of DNA). (B) Autoradiogram of a blot containing HindIII and EagI-in combination with HindIII-digested DNAs hybridized with the p15 exon 1 probe. The locations of the expected 2.8-kb and 0.6-kb bands are indicated by arrows at the left. Samples include DNA from Kg1A cell line, a healthy control (NORM), DR#10 at diagnosis (Dx) and at relapse (R). Homozygous methylation is seen in Kg1A, and hemizygous methylation in DR#10 at relapse, but not at initial diagnosis.
p15 promoter hypermethylation. (A) Schematic representation of p15 exon 1 and surrounding HindIII (H3) and EagI (E) sites. The p15 probe used (indicated above) hybridizes to exon 1. A 2.8-kb band is seen when the samples are digested with HindIII alone. When the samples are digested withEagI (a methylation-sensitive enzyme) in combination withHindIII, a 0.6-kb band is recognized by the probe if the gene is unmethylated (cuts at EagI) and a 2.8-kb band is present if the sample is methylated (does not cut at EagI) (note that a 2.2-kb band is not seen, because the exon 1 probe does not hybridize to this region of DNA). (B) Autoradiogram of a blot containing HindIII and EagI-in combination with HindIII-digested DNAs hybridized with the p15 exon 1 probe. The locations of the expected 2.8-kb and 0.6-kb bands are indicated by arrows at the left. Samples include DNA from Kg1A cell line, a healthy control (NORM), DR#10 at diagnosis (Dx) and at relapse (R). Homozygous methylation is seen in Kg1A, and hemizygous methylation in DR#10 at relapse, but not at initial diagnosis.
DISCUSSION
In the current study, we found that homozygous p16 deletions were common at the time of first bone marrow relapse of childhood ALL (9 of 18 cases). While four of these cases contained homozygous deletions at both diagnosis and relapse, 5 of the 14 cases (36%) that were p16/p15 germline at diagnosis acquired homozygousp16 deletions in the interval between diagnosis and relapse. Three of these five cases also acquired homozygous p15deletions, and one acquired a partial p15 exon 1 hypermethylation. One additional case acquired partial p15promoter hypermethylation at relapse in the absence of p16abnormalities. These results suggest that acquisition of p16and p15 abnormalities may be a critical event associated with disease progression in children with ALL. As all deletions involvedp16 exon 2, which is included in both p16 andp19ARF transcripts,14,19 it is not certain whether the critical protein target was p16, p19ARF, or both. Experimental data indicate that inactivation of these two proteins profoundly alters cell-cycle progression and growth arrest via loss of a negative regulator of the Rb pathway (p16) and a positive regulator of wild-type p53 activity (p19ARF),11-13 15-17 suggesting that these alterations may contribute to the clinical chemotherapy refractoriness observed in relapsed ALL.
It is important to emphasize that our results do not distinguish between two different potential explanations for the presence ofp16/p15 abnormalities in relapse, but not diagnosis, samples. First, the p16/p15 abnormalities may not have been present in any of the leukemic cells at diagnosis, but rather developed during (and perhaps instigated) the process of disease recurrence. Alternatively, a small subclone, that was below the limits of detection of Southern blot analysis, containing these genetic abnormalities could have been present at the time of initial diagnosis, and loss of p16/p15/p19ARF function could have conferred a proliferative and/or survival advantage that allowed this to become the dominant clone at relapse.
To our knowledge, this is the largest series of childhood ALLs in whichp16 and p15 gene status has been determined in matched diagnostic and relapse specimen pairs. Ohnishi et al reported thatp16 status was unchanged (one deleted, six germline) between diagnosis and relapse in seven paired samples.21 Takeuchi et al used microsatellite analysis to identify deletions within 9p in six matched pairs of childhood ALLs and found that no differences developed in the pattern of allelic loss between diagnosis and relapse.32 In contrast, Ogawa et al found that at least hemizygous loss of p16 developed between initial diagnosis and relapse in two of three paired samples.33 To determine whether there is any cytogenetic data to support our findings that acquisition of p16/p15 abnormalities are likely to play an important role in progression of childhood ALL, we reviewed the published literature on karyotypes from children with relapsed ALL, focusing on 9p abnormalities. In five series, complete karyotypes were available from 34 cases of childhood ALL that had an abnormal karyotype at diagnosis, and had additional changes within this clone at relapse.34-38 Deletions of 9p evolved between diagnosis and relapse in four of these 34 (12%) cases. As the majority of ALLs containing p16/p15 deletions do not have visible 9p abnormalities (and only one of the five cases that acquiredp16/p15 deletions in our study also acquired visible 9p abnormalities), these cytogenetic data likely underestimate the true incidence of new p16/p15 abnormalities.
Taken together with data from other studies,7,8,23,24 39these results strongly suggest that p16/p15 genetic abnormalities play an important role in the biology of relapsed hematologic and lymphoid malignancies, and may be directly related to disease progression. Our sample is too small, and the clinical characteristics and treatment of the patients too heterogeneous, to draw any conclusions regarding the potential prognostic import ofp16/p15 abnormalities at relapse. In the future, it will be important to analyze a larger, more homogeneous group of patients to accurately define the percentage of childhood ALLs that acquire p16/p15 abnormalities in the interval between initial diagnosis and relapse, and to determine whether this has prognostic significance. Increased understanding of the role ofp16/p15 inactivation in the initiation and progression of lymphoid malignancies should provide important insights into leukemia biology and lay the groundwork for rationally designed therapeutic interventions.
K.W.M. is the recipient of the Greg and Laura Norman Fellowship Award from the National Childhood Cancer Foundation and is also supported in part by a University of Colorado Cancer Center seed grant. S.P.H. was supported by a BLOOD/ASH Scholar Award and a Professional Development Award from The Children’s Hospital Research Institute, Denver, CO, and is a Leukemia Society Translational Research Awardee. Research supported by a grant from the Cancer League of Colorado to S.P.H. and a Cancer Center Core Grant (CA 46934).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Stephen P. Hunger, MD, UCHSC Campus Box C229, 4200 E Ninth Ave, Denver, CO 80262; e-mail:Stephen.Hunger@UCHSC.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal