THE ASSOCIATION of several characteristic chromosomal translocations with various subtypes of acute and chronic myelogenous and lymphocytic leukemias has facilitated the cloning of a number of unique leukemia-associated fusion genes. Characterization of these fusion genes has in turn led to the discovery of previously unrecognized genes. Some of these have now been extensively characterized in vitro, and many encode transcription factors that play important roles in normal hematopoietic development.1Recently, the function of several leukemia-associated fusion genes has been investigated using various transgenic murine systems. Despite the generation of acute leukemias in some cases, inconsistent results have made comparisons among different transgenic models problematic. Consequently, fundamental questions remain unanswered about the precise nature of the events leading to the generation of acute leukemias, and the hematopoietic cell type(s) in which such events can occur. The purpose of this perspective is to discuss briefly the potential advantages and disadvantages inherent in various transgenic approaches that are used to model leukemias at the present time, and to discuss future applications of these approaches for achieving a better understanding of the molecular events leading to acute leukemia.
WHAT IS THE TRANSFORMED HEMATOPOIETIC COMPARTMENT (THE CELLULAR “SOIL”) OF HUMAN MYELOID LEUKEMIAS?
Bonnet and Dick2 have recently demonstrated that transmission of human acute myeloid leukemias (AML) to nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice requires transplantation of a phenotypically immature CD34+/38− cell population that comprise a small minority of circulating cells (“SCID-leukemia initiating cells [SL-ICs]). These studies used primary leukemia cells from a variety of AML French-American British (FAB) phenotypic subclassifications (M1, M2, M3, M4, M5). However, cells from patients with the M3 subtype (acute promyelocytic leukemia [APML]) were different, in that M3 SL-ICs were not present in any progenitor fraction (or in unfractionated peripheral blood). These observations suggest that for most AML subtypes, a relatively primitive cell is the target of leukemic transformation, regardless of the extent and lineage of differentiation observed in the majority of leukemic cells present in the peripheral circulation. Given the inability to transplant the M3 subtype using the NOD/SCID system, however, this model cannot be extended at present to include APML.2 Another study attempted to define the transformed compartment in APML.3 Fluorescence-activated cell sorting (FACS) of primary APML cells was used to show that the APML-associated fusion gene PML/RARA was expressed in CD34−/38+, but not CD34+/38− cells, suggesting that in APML, the transformation process may involve a more differentiated cell type than the pluripotent progenitor and/or stem cell compartments implicated in other myeloid leukemia subtypes.2 3
CURRENT APPROACHES FOR TARGETING ONCOGENE EXPRESSION (THE “SEED”) TO SPECIFIC HEMATOPOIETIC COMPARTMENTS IN MICE
To investigate the leukemogenic capacity of various leukemia-associated fusion genes in vivo, murine transgenic models have been used. A gain-of-function transgenic model for leukemogenesis should ideally result in the expression of a specific potential oncogene, without perturbing the expression of other endogenous mouse genes. It should cause the transgene to be expressed in a tissue- and development-specific fashion, mirroring the pattern observed in human disease. The regulation of expression of the oncogene should be such that normal hematopoiesis can occur before (or in parallel with) the development of leukemia.
Among the currently available methods for generating gain-of-function models of leukemia in vivo, “standard” transgenesis has been in use for the longest period of time. This method involves microinjection of transgene DNA into murine oocytes, which are subsequently implanted into pseudopregnant female mice to generate transgenic offspring. The transgene consists of two essential components: the cDNA of interest, and genomic sequences linked to the transgene that regulate its pattern of expression (for examples, see Fig 1). Regulatory sequences may be those normally located near the gene of interest itself, or may be derived from an unrelated gene whose temporal and tissue-specific expression conforms to the pattern of expression desired for the study of the transgene. With this approach, transgene integration into the host genome is thought to be random. As a result, the level and pattern of transgene expression may vary significantly between different founders. Furthermore, random transgene integration into the host genome may have unanticipated consequences on the expression of normal mouse genes. For these reasons, phenotypic analyses require the characterization of multiple founder lines before conclusions can be reliably drawn. Finally, extensive prior characterization of the linked regulatory sequences is required, to ensure that all the elements essential for the expression of the target gene are included. An advantage of the standard transgenesis approach is that the differing levels of transgene expression among founders affords an oppurtunity to study the effects of gene dosage, which may be a critical parameter for the development of many leukemias.
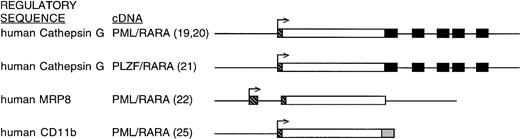
Regulatory sequences used to control cDNA expression in various published murine APML models. cDNA sequences are represented as open boxes. Cathepsin G coding exons, included downstream of the PML/RARA or PLZF/RARA cDNAs, are represented by filled boxes. The 5′ untranslated regions of the respective targeting loci are represented by hatched boxes. The remainder of the targeting loci (upstream regulatory regions, intronic sequences, and 3′ untranslated regions) are represented by plain lines. The MRP8 targeting construct includes 3′ untranslated sequences placed downstream of the inserted cDNA. The CD11b targeting construct consists of upstream CD11b regulatory sequence and an SV40 polyadenylation signal (shaded box) located downstream of the cDNA. These representations are not drawn precisely to scale.
Regulatory sequences used to control cDNA expression in various published murine APML models. cDNA sequences are represented as open boxes. Cathepsin G coding exons, included downstream of the PML/RARA or PLZF/RARA cDNAs, are represented by filled boxes. The 5′ untranslated regions of the respective targeting loci are represented by hatched boxes. The remainder of the targeting loci (upstream regulatory regions, intronic sequences, and 3′ untranslated regions) are represented by plain lines. The MRP8 targeting construct includes 3′ untranslated sequences placed downstream of the inserted cDNA. The CD11b targeting construct consists of upstream CD11b regulatory sequence and an SV40 polyadenylation signal (shaded box) located downstream of the cDNA. These representations are not drawn precisely to scale.
More recently, approaches using homologous recombination have been employed to circumvent many of the problems inherent in standard transgenesis (Fig 2). With this approach, a fusion gene cDNA is targeted directly to a predefined locus in the mouse genome, which is selected based on a desired pattern of expression. The homologous recombination event is mediated by DNA arms derived from the targeted murine locus. These arms flank the desired site of transgene insertion, typically in the 5′ untranslated region between the transcriptional and translational start sites. Targeting arms are placed upstream and downstream from the cDNA in the targeting construct, together with a selectable marker cassette (eg, PGK-neo) which facilitates the positive selection of stably transfected embryonic stem (ES) cell clones. ES cell clones that have the desired homologous recombination event are expanded and microinjected into murine blastocysts, which are implanted in pseudopregnant females to generate chimeric offspring. These chimeras are then bred to wild-type mice to transmit the targeted allele to the germline.
Targeting an endogenous murine locus using homologous recombination. Coding exons of the targeted locus are represented as solid boxes. The cDNA of interest and PGK-neo selectable marker cassette are represented as open boxes. Transcriptional start sites for the targeted gene locus and PGK-neo selectable marker cassette are indicated by arrows. The 5′ untranslated region is represented by a hatched box. LoxP sites used for CRE recombinase-mediated selection cassette excision are represented as open ovals.
Targeting an endogenous murine locus using homologous recombination. Coding exons of the targeted locus are represented as solid boxes. The cDNA of interest and PGK-neo selectable marker cassette are represented as open boxes. Transcriptional start sites for the targeted gene locus and PGK-neo selectable marker cassette are indicated by arrows. The 5′ untranslated region is represented by a hatched box. LoxP sites used for CRE recombinase-mediated selection cassette excision are represented as open ovals.
There are several potential advantages of the “knock-in” approach. Because a single transgene is inserted into the mouse genome at a discrete, predefined locus, variegation in transgene expression patterns among founder lines should be eliminated. In theory, the expression pattern of the transgene should mirror that of the targeted locus, because all regulatory elements will usually be left intact. However, there are also potential problems with the knock-in transgenesis approach. These include: (1) the requisite disruption of one allele of the targeted locus, which can result in heterozygous loss-of-function and may affect the ultimate phenotype, independent of transgene expression. (2) Insertion of the transgene into the targeted locus can potentially alter the desired regulation of the locus by disrupting normally juxtaposed regulatory elements, or by introducing new regulatory elements located within the transgene itself. (3) There is compelling evidence that selectable marker cassettes can negatively affect the expression of other nearby genes,4-9 as well as that of the targeted gene itself (unpublished observations, June 1998) (“neighborhood effects”). This problem has been successfully addressed by removal of the selectable marker in targeted ES cell clones, using a recombinase-mediated excision event (Fig 2)5,6,8 9 (and unpublished observations, June 1998).
A variation of the “knock-in” approach has been used to evaluate the role of various leukemia-associated fusion genes in murine leukemia models. With this approach, the endogenous murine locus from the upstream partner in the human leukemia-associated fusion gene is used as the locus targeted for homologous recombination; the test cDNA is inserted at a point in the endogenous locus corresponding to the breakpoint. For example, Corral et al10 inserted the 3′ portion of a human AF9 cDNA into the murine MII locus in ES cells, recreating the MII/AF9 fusion gene formed as a result of the human acute lymphoblastic leukemia–associated translocation t(4;11), and observed the generation of acute leukemias in chimeric mice. The advantage of this approach is that it recreates almost exactly the fusion gene as it occurs in human leukemia, since the regulatory sequences used are those of the murine homologue of the upstream fusion partner. The major potential disadvantage of this approach is that many of the genes involved in leukemia-associated translocations may play essential roles in early hematopoietic development or may be expressed in nonhematopoietic tissues. By directing the expression of a leukemia-associated fusion gene to all primitive hematopoietic cells, normal hematopoiesis may be sufficiently disrupted as to result in embryonic lethality, precluding analysis of its capacity to cause leukemia. For example, when a portion of a human ETO cDNA was targeted to the murine AML-1 locus, or a portion of a human MYH11 cDNA was targeted to the CBF-β locus, the resultant phenotypes in both cases resembled those of the AML-1 and CBF-β knockout animals, respectively, suggesting a dominant negative effect of the fusion gene on normal hematopoiesis.11 12
Another potential disadvantage of all the transgenic approaches described above is that in each, the transgene is carried in the germline of the transgenic animal, unlike in human leukemia, in which the translocation resulting in the formation of the fusion gene is an acquired mutation that occurs in somatic cells. To circumvent the problem of universal expression of the cDNA of interest in hematopoietic tissues, one alternative approach could use CRE-mediated recombination in vivo. With this approach, LoxP sites would be sequentially targeted in ES cells to the breakpoints in the respective upstream and downstream fusion gene partners of interest. Expression of CRE recombinase in a desired hematopoietic compartment would be achieved by breeding these mice with a line in which a CRE cDNA was expressed in that compartment (as discussed above) to recreate the relevant chromosomal translocation by means of a somatic recombination event. Although attractive in theory, such an approach would ultimately depend on an intermolecular recombination event between different chromosomes, which may or may not occur with meaningful efficiency. Furthermore, the continuous expression of CRE recombinase within the hematopoietic compartment could potentially lead to unexpected recombination and/or other unwanted events.
TRANSGENIC MURINE MODELS OF APML: A DEMONSTRATION OF THE IMPORTANCE OF THE TARGETED COMPARTMENT
The PML/RARA fusion gene, resulting from the t(15:17) balanced reciprocal translocation characteristic of APML, is among the most thoroughly studied of the leukemia-associated fusion genes.13-17 Several transgenic models of APML have been generated using an identical breakpoint 1–derived PML/RARA cDNA14 expressed in different hematopoietic compartments (Fig 1). Grisolano et al,18 in our laboratory, developed an expression construct that directed human cathepsin G (hCG) transgene expression exclusively to the promyelocyte compartment in hematopoietic cells, and subsequently used this construct to direct PML/RARA expression to the murine promyelocyte compartment in a C57BL/6 × C3H/He background.19 Despite a low level of PML/RARA expression, 100% of transgene-expressing animals displayed altered myeloid development, manifest by myeloid expansion in their bone marrow and spleens and splenic extramedullary hematopoiesis, but normal peripheral blood counts and myeloid differentiation. Over the course of 6 to 13 months, 30% of the transgenic animals from three different founder lines developed AML. These leukemias were characterized by profound leukocytosis, anemia and/or thrombocytopenia, and extensive organ infiltration by leukemic cells with disruption of normal histologic architecture. Morphologic differential analysis of bone marrow and peripheral blood demonstrated markedly increased numbers of promyelocytes, but full myeloid maturation was observed. Transplantation of leukemia cells into SCID recipients resulted in fatal leukemias in 100% of the recipients within 5 weeks. Treatment of leukemic splenocytes in vitro with ATRA (10−6 mol/L) resulted in nuclear changes that were morphologically consistent with apoptosis, but these cells exhibited no morphologic or flow cytometric evidence of differentiation. He et al20 reported similar results using a nearly identical hCG-PML/RARA targeting construct in a C57BL/6 × CBACa background. Evidence of myeloid expansion in the bone marrow, spleen, lymph nodes, and thymus, but not peripheral blood, was observed in 100% of transgenic animals by 12 months of age. Between 12 and 14 months, 10% of animals developed overt acute leukemia characterized by peripheral leukocytosis, organ infiltration, and an accumulation of blasts and promyelocytes in bone marrow and peripheral blood. Furthermore, in agreement with the findings of Grisolano et al, terminal myeloid maturation was shown.
Brown et al22 used a human MRP8 expression cassette to direct expression of a PML/RARA cDNA to the myeloid lineage in an inbred FVB/N background. The MRP8 gene, which encodes an intracellular calcium-binding protein, is expressed in early myeloid and monocytic cells and throughout myeloid differentiation, based on coexpression with GR-1 and Mac-1 differentiation markers. The precise stage of myeloid development at which MRP8 expression is activated has not been clearly defined, although expression during embryonic development has been detected in whole embryo suspensions as early as day E11, and abundant MRP8-expressing myeloid cells were observed in fetal liver at day E14. Low levels of MRP8 expression have also been observed in nonhematopoietic tissues including lung, spinal cord, muscle, and thymus; expression in the skin was originally not reported (see below).23 24 PML/RARA expression in transgenic animals was detected by Western blotting in bone marrow and peripheral blood in 8 of 9 founder lines, and a diffuse pattern of PML/RARA distribution was demonstrated by immunofluorescence staining in the granulocytic series. Subtle decreases in cell surface GR-1 expression, a marker of myeloid differentiation, and decreased neutrophil granularity were evident by fluorescence-activated cell sorter (FACS) analysis, but normal hematopoietic cell numbers and morphologic differentiation were observed in bone marrow and peripheral blood. Assessment of the latency and incidence of leukemias was complicated by a retinoid-responsive epidermal papillomatosis syndrome which developed in the highest expressing lines. Acute myeloid leukemias developed in 12 animals from five different founders (5% of surviving mice at the time of publication). Leukemias were characterized by an accumulation of large numbers of myeloperoxidase-positive promyelocytes with prominent azurophilic granules. However, unlike the observations of Grisolano et al and He et al, no evidence of granulocytic maturation was seen in the leukemic animals. Treatment of leukemia cells with ATRA in vitro led to morphologic and flow cytometric evidence of differentiation; treatment of a limited number of leukemic animals with ATRA in vivo led to variable results, although at least one animal appeared to enter a complete remission. Transplantation of leukemic versus preleukemic bone marrow cells from transgenic animals into irradiated recipients led to 100% versus 13% incidences of leukemia, respectively.
Early et al25 used CD11b regulatory sequences to direct expression of a PML/RARA cDNA to a more differentiated granulocytic compartment in transgenic mice in an ICR Swiss or non-Swiss background. In this regulatory context, no leukemias were observed, despite demonstration of PML/RARA expression in bone marrow by reverse transcriptase-polymerase chain reaction (RT-PCR) assays. The only apparent abnormality in these animals was a decrease in myeloid progenitors and a prolonged period of bone marrow suppression after sublethal irradiation, suggesting a subtle defect in myelopoiesis.
Finally, Greer et al attempted to direct PML/RARA expression to the murine hematopoietic stem cell compartment by inserting a PML/RARA cDNA (linked to an internal ribosomal entry site) into the second coding exon of a c-fes gene targeting construct (cited in 26). No leukemias were observed in these studies. Transgene mRNA expression was detected in bone marrow using RT-PCR, but protein expression within the hematopoietic compartment was not clearly detected. As such, the significance of these results is unclear (Peter Greer, personal communication, July 1998).
He et al21 also targeted the expression of PLZF/RARA, a variant APML-associated fusion cDNA, to the promyelocyte compartment using the same hCG targeting construct in a C57BL/6 × B6CBACa background. Similar to the PML/RARA-CG mice, 100% of these animals developed a myeloproliferative preleukemic state. Acute leukemias subsequently developed following a similar 6- to 18-month latency period; unlike the PML/RARA mice, however, the penetrance of the leukemia phenotype was also 100%. These mice appeared to have a less pronounced differentiation block than was observed in the PML/RARA-CG mice, with a decreased percentage of blasts and promyelocytes, and a higher percentage of terminally differentiated myeloid cells in the bone marrow and peripheral blood. Finally, treatment of PLZF/RARA leukemic animals with ATRA resulted in a minimal response, compared with transient complete responses observed in their PML/RARA animals. The potential effects of random integration on transgene expression patterns and/or transgene dose cannot be formally excluded as contributing the differences between PML/RARA- and PLZF/RARA-expressing animals. The analysis of multiple founder lines and semi-quantitative analysis of transgene expression levels, however, suggests that differences among individual leukemia-associated fusion genes can, in fact, contribute to the different leukemia phenotypes in the same cellular compartment.
SEED VERSUS SOIL: FUTURE DIRECTIONS IN MURINE LEUKEMIA MODELS
From the composite analysis of these studies, it is clear that both the PML/RARA and PLZF/RARA fusion genes are leukemogenic when expressed in the appropriate developmental context. The prolonged latency and variable penetrance of the leukemia phenotype suggest that both PML/RARA and PLZF/RARA expression can cause APML, but neither is sufficient to do so. However, the precise role of the hematopoietic developmental context of PML/RARA expression in the generation of leukemia remains incompletely defined. The importance of the cellular “soil” is emphasized by the failure of PML/RARA to cause leukemias when expressed late in myeloid differentiation (under control of the CD11b promotor), and by differences in the phenotypes resulting from CG- and MRP8-directed PML/RARA expression. For example, despite an accumulation of immature myeloid cells, terminal differentiation was observed in leukemias derived from the PML/RARA-CG mice, but not PML/RARA-MRP8 mice. These differences in differentiation phenotype may reflect the fact that CG expression is limited to the promyelocytic compartment, whereas MRP8 expression begins at a similar point in myeloid differentiation, but remains active throughout terminal differentiation (Table 1). The fact that a similar, but not identical, leukemia phenotype was observed when a different APML-associated fusion gene (PLZF/RARA) was expressed in the promyelocyte compartment highlights the contribution of the individual leukemia-associated cDNA (the “seed”) for the determination of the leukemia phenotype. As discussed above, however, the potential effects of random integration on transgene expression pattern and/or “dose” cannot be dismissed as a potential explanation for the difference in leukemia phenotype or penetrance between the PML/RARA and PLZF/RARA lines. Moreover, the potential role of mouse strain differences contributing to the different observed phenotypes cannot be formally excluded at this time.
Comparison of PML/RARA and PLZF/RARA Transgenic Phenotypes
| cDNA . | Regulatory Element . | Compartment of Expression . | Preleukemic State . | Leukemia Penetrance (%) . | Leukemia Latency (mo) . | Differentiation Block . | ATRA Response . |
|---|---|---|---|---|---|---|---|
| PML/RARA | hCG19 20 | Promyelocytes | Yes | 10-30 | 6-13 | Partial | Yes |
| PML/RARA | MRP822 | Pros → PMNs | Yes | 5 | 3-8 | Complete | Yes |
| PML/RARA | CD11b25 | Myelocyte → PMN | No | 0 | — | No | Not done |
| PLZF/RARA | hCG21 | Promyelocyte | Yes | 100 | 6-18 | Partial* | Minimal |
| cDNA . | Regulatory Element . | Compartment of Expression . | Preleukemic State . | Leukemia Penetrance (%) . | Leukemia Latency (mo) . | Differentiation Block . | ATRA Response . |
|---|---|---|---|---|---|---|---|
| PML/RARA | hCG19 20 | Promyelocytes | Yes | 10-30 | 6-13 | Partial | Yes |
| PML/RARA | MRP822 | Pros → PMNs | Yes | 5 | 3-8 | Complete | Yes |
| PML/RARA | CD11b25 | Myelocyte → PMN | No | 0 | — | No | Not done |
| PLZF/RARA | hCG21 | Promyelocyte | Yes | 100 | 6-18 | Partial* | Minimal |
Less pronounced accumulation of immature cells noted, compared with hCG-PML/RARA phenotype.
It is not known what effect PML/RARA expression would exert at an earlier stage of myeloid development, or in a different hematopoietic lineage. One possibility is that the intracellular regulatory milieu of the compartment in which PML/RARA is expressed determines the phenotypic characteristics of the resultant leukemia. In this scenario, targeting PML/RARA expression to a more primitive myeloid compartment might cause a less differentiated leukemia phenotype, whereas expression in the erythroid or megakaryocytic compartments might result in the generation of acute leukemias with phenotypic features of those lineages. Conversely, targeting the expression of a different fusion gene characteristically associated with a less differentiated leukemia phenotype (such as an AML/ETO cDNA) to the promyelocyte compartment under the control of CG regulatory sequences might, in turn, cause a promyelocytic leukemia phenotype. Another possibility is that PML/RARA itself may direct an APML phenotype exclusively, regardless of the hematopoietic cell type in which it is expressed. Alternative outcomes of targeting PML/RARA expression to other hematopoietic compartments include (1) a failure to cause leukemias, if the myeloid precursor stage at which CG and MRP8 expression is activated represents the earliest hematopoietic compartment sensitive to transformation by PML/RARA; or (2) embryonic lethality, if expression in other compartments disrupts normal hematopoiesis, as seen in the case of the AML/ETO and CBFβ/MYH11 knock-in experiments.
To definitively address the role of the target cell “soil” in a murine model of PML/RARA-mediated leukemogenesis, PML/RARA expression must be targeted to discrete hematopoietic developmental compartments. A critical component in the design and ultimate interpretation of these experiments will be the choice of loci whose expression patterns are well characterized and limited to hematopoietic cells, such as those of developmentally regulated hematopoietic growth factor receptors. As discussed above, the targeted knock-in transgenesis approach (Fig 2) should prove to be ideally suited for this purpose, since it eliminates the potential variables of gene dosage and locus-specific effects on transgene expression due to random integration, and allows valid comparison of the phenotypes arising from the expression of different fusion genes from the same locus. We anticipate that over the next few years, the application of these recent technological advances in the generation of gain-of-function transgenic models will enable investigators to elucidate the potentially important role of the target cell in leukemic transformation.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Timothy J. Ley, MD, Washington University School of Medicine, Division of Bone Marrow Transplantation and Stem Cell Biology, 660 S Euclid Ave, Campus Box 8007, St Louis, MO 63110-1093; e-mail: timley@im.wustl.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal