Hypoxia regulates expression of erythropoietin (EPO), a glycoprotein that stimulates erythrocytosis, at the level of transcription and also possibly at the level of messenger RNA (mRNA) stability. A pyrimidine-rich region within the EPO mRNA 3′ untranslated region was implicated in regulation of EPO mRNA stability element and shown to bind protein factors. In the present study we wished to identify the protein factor binding to the pyrimidine-rich sequence in the EPO mRNA stability element. Using mobility shift assays, ultraviolet light cross-linking, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and electroelution of protein factors from the gel slices corresponding to the ribonucleoprotein complexes, we found that two isoforms of a 40 kD poly(C) binding protein (PCBP, also known as CP or hnRNPE), PCBP1, and PCBP2 are present in that complex. In Hep3B or HepG2 cells hypoxia induces neither expression of PCBP nor formation of the ribonucleoprotein complex associated with EPO mRNA that involves PCBP.
ERYTHROPOIETIN (EPO) IS A GLYCOPROTEIN synthesized and released from the kidney, which regulates differentiation and maturation of progenitor cells toward erythrocytes, leading to polycythemia during long-term adaptation to hypoxia.1 EPO expression under normoxic conditions is minimal, but increases during exposure to hypoxia in the fibroblast-like type-I interstitial cells of the kidney cortex and outer medulla,2 and also in the two cell lines derived from liver tumors (Hep3B, HepG2) that express EPO in an O2-regulated manner.3 EPO synthesis during hypoxia is induced at the level of gene expression; in addition to transcriptional induction,4 it may involve regulation of EPO mRNA stability.5 Transcriptional regulation of EPO induction by hypoxia has been studied extensively.6 In contrast, much less is known about regulation of EPO messenger RNA (mRNA) stability, primarily because EPO mRNA stability is affected both by ongoing transcription and by protein synthesis.5
Analysis of the 3′ untranslated region (3′ UTR) of EPO mRNA shows putative stability and instability elements. Deletion of the 186 bases of the conserved sequence from the distal 3′ UTR of EPO mRNA increased the t1/2 from 2 hours to 15 hours.7 This indicates that the deleted fragment of 3′ UTR may contain an RNA instability element, whereas the remaining region, located 5′ from the deleted region, may contain an RNA stability element.7 Deletion of the 104 bases immediately downstream from the EPO translation stop codon results in destabilization of EPO mRNA from 7 hours to 2.6 hours, an indication that this fragment may contain an RNA stability element.8 These experiments, however, were performed in the presence of actinomycin D, which nonspecifically stabilizes EPO mRNA.5,8 Thus it is difficult to estimate the role of this fragment of EPO 3′ UTR in regulating EPO mRNA stability. This putative EPO mRNA stability region previously was found to bind cytoplasmic proteins.9,10 Ultraviolet (UV) light cross-linking and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of the EPO RNA-protein complexes showed two bands of 70 and 135 to 140 kD.9 Except in the brain and the spleen, however, the binding activity in most cell lines and tissues studied was not induced by hypoxia.9 The binding of these proteins to EPO mRNA was redox-sensitive and required reduced thiol groups.10
Regulation at the level of mRNA stability during hypoxia occurs in the case of tyrosine hydroxylase (TH), the rate-limiting enzyme in catecholamine synthesis in pheochromocytoma-derived PC12 cells. Hypoxia augments TH mRNA half-life twofold.11,15 This increase in TH mRNA stability during hypoxia involves enhanced formation of a ribonucleoprotein complex associated with an RNA stability element in the 3′ UTR of TH mRNA (hypoxia-inducible protein-binding sequence, HIPBS).12-15 The binding proteins are two isoforms of a poly(C) binding protein (PCBP, also known as αCP or hnRNPE),15-20 and expression of an isoform PCBP1 is induced by hypoxia in PC12 cells.15HIPBS sequence is analogous to a pyrimidine-rich sequence located within the EPO RNA–protein-binding region.9 Both sequences are pyrimidine-rich and contain short stretches of cytidines interrupted by one or two uridines.13 In view of these sequence similarities between the TH and the EPO mRNA, we hypothesized that the same PCBP protein should bind to the pyrimidine-rich HIPBS-like element of the EPO mRNA. Indeed, we determined that two isoforms of PCBP are present in the ribonucleoprotein complex associated with the EPO mRNA 3′ UTR. Expression of PCBP isoforms, however, is not induced by hypoxia; nor is formation of the ribonucleoprotein complex associated with pyrimidine-rich sequence in the EPO 3′ UTR.
MATERIALS AND METHODS
Cell culture and preparation of cytoplasmic extracts.
PC12, Hep3B, and HepG2 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM)/F12 medium containing 10% fetal bovine serum, 100 units/mL penicillin, and 100 μg/mL streptomycin, as described previously.12-14 Cells reaching 75% to 85% confluence were exposed to either normoxia or hypoxia (1% O2) in an oxygen-regulated incubator (Forma Scientific Marietta, OH) for 24 hours.12-14 Cytosolic protein extracts were obtained as described previously. 12-14
Plasmid constructs and RNA transcript.
A 210 bp StuI-BglII fragment of human EPO complementary DNA (cDNA) (a gift from Dr H.F. Bunn, Fig1A) corresponding to the last 22 bases of the coding region and the first 188 bases of the 3′ UTR, which contains the HIPBS-like site, was subcloned into SmaI-BglII sites of transcription vector pSP73. Templates for transcription were obtained by linearizing pSP73-EPO vector with either DdeI, NcoI, or BglII (Fig 1A). The 93–base-long transcript, obtained from the template linearized with DdeI, included only the pyrimidine-rich stretch. Comparison of this sequence in different species, starting at the translation stop codon, is shown in Fig 1B. In vitro transcriptions were performed using T7 RNA polymerase in the presence of 50 μCi of [α-32P] uridine triphosphate (UTP) (3000 Ci/mmol), 2.5 mmol/L of all four unlabeled nucleotides, 20 mmol/L dithiothreitol (DTT), and RNasin at 42°C for 60 minutes, as described previously.12 TH transcripts were prepared as described previously.12-14 The sequences of the synthetic wild-type and mutated EPO oligoribonucleotides are shown in Fig 1C.
(A) Schematic representation of human EPO cDNA, and location of restriction sites DdeI, NcoI, andBglII within the 3′ UTR used to generate EPO transcripts. The hatched area represents the HIPBS-like region. TGA - translation stop codon. Bracket marks the cDNA used to generate the riboprobe. (B) Alignment of pyrimidine-rich tracts in the 3′ UTR of EPO mRNA from different species. Sequences start with the translation stop codon. The first pyrimidine-rich tract conserved in primates is indicated by italics and underlined. The second motif conserved in various species is indicated by bold type and underlined. Gene bank accession numbers for each EPO mRNA are X02157 (human), M18189(monkey), L10608 (rat), M12482 (mouse), L10607 (swine), U44762(bovine). (C) Sequences of wild-type TH and EPO HIPBS elements, and localization of mutations (MUT). The G residue within the protein binding site is underlined.
(A) Schematic representation of human EPO cDNA, and location of restriction sites DdeI, NcoI, andBglII within the 3′ UTR used to generate EPO transcripts. The hatched area represents the HIPBS-like region. TGA - translation stop codon. Bracket marks the cDNA used to generate the riboprobe. (B) Alignment of pyrimidine-rich tracts in the 3′ UTR of EPO mRNA from different species. Sequences start with the translation stop codon. The first pyrimidine-rich tract conserved in primates is indicated by italics and underlined. The second motif conserved in various species is indicated by bold type and underlined. Gene bank accession numbers for each EPO mRNA are X02157 (human), M18189(monkey), L10608 (rat), M12482 (mouse), L10607 (swine), U44762(bovine). (C) Sequences of wild-type TH and EPO HIPBS elements, and localization of mutations (MUT). The G residue within the protein binding site is underlined.
RNA-protein binding reactions and electrophoresis of complexes.
Gel shift reactions and UV light cross-linking electrophoresis of RNA-protein complexes were performed essentially as described previously.12-14 Briefly, cytoplasmic protein extracts (20 to 40 μg) were incubated in the binding buffer (10 mmol/L HEPES, pH 7.9, 5 mmol/L MgCl2, 50 mmol/L KCl, and 10% glycerol, 200 ng/ml Escherichia coli transfer RNA [tRNA], and 1 mmol/L DTT) at room temperature for 15 minutes. Labeled RNA transcripts or oligoribonucleotides (100,000 cpm/reaction) were added, and the reaction mixture was incubated for 10 minutes. After completion of the binding reaction, RNase T1 (10 units) and heparin sulfate (final concentration 2.5 mg/mL) were added sequentially to the reaction mixture for 5 minutes each. In competition experiments the cold competitor probe was added before the radioactive probe. For UV light cross-linking, binding reactions were exposed to 1 × 106 J/cm2. The cross-linked reaction was separated on 9% acrylamide gel under reducing conditions (100 mmol/L DTT in the sample buffer). When TH and EPO RNA-protein interactions were compared in the same experiment four times less of the PC12 protein extract was used with the TH probe to obtain autoradiographically comparable signal.
For electroelution of protein factors, the binding reactions were performed using 10 μg of unlabeled EPO DdeI transcript and 1.5 to 2 mg of protein extract from HepG2 cells in a 300 μL reaction volume. The control reactions were identical except for the RNA. Complexes were visualized by corunning one reaction that contained32P-labeled RNA. Identified complexes were excised, macerated, and loaded into the elution cell in the inside cup buffer, which contained 5 mmol/L Tris-acetate, pH 8, 0.1% SDS, 0.1 mmol/L EDTA. Proteins were eluted overnight at 4°C with a constant voltage of 100 mV in the outside cup buffer (50 mmol/L Tris-acetate, pH 8, 0.1% SDS, 0.1 mmol/L EDTA). The inside and outside compartments were separated by dialysis membrane with molecular weight cut-off (MWCO) 12,000 to 14,000 (Spectra/por*2, Spectrum Medical Inc). The eluted proteins were concentrated using Microcon-10 microconcentrators, resuspended in the sample buffer, and subjected to 9% SDS-PAGE analysis. Proteins then were transferred onto the nitrocellulose with a semidry blotter (Bio-Rad, Hercules, CA). The blots were blocked in Tris-buffered saline, Tveen 20 (TBST) with 5% nonfat milk, and were first probed with a specific anti-PCBP1 antibody,15 and then stripped, and reprobed with a specific anti-PCBP2 antibody (gift from A.V. Gamarnik and R. Andino) in TBST with 5% dry milk. The signal was visualized by exposing blots to chemiluminescence reagents (Amersham, Arlington Heights, IL).
RESULTS
Similarities in the formation of ribonucleoprotein complex associated with EPO and TH HIPBS-like elements.
Analysis of the 3′ UTR of the EPO mRNA showed sequences highly homologous to the HIPBS element in TH mRNA (Fig 1B). The 3′ UTR of primate EPO mRNA contains two repeats of cytidine/uridine-rich motifs, separated by a sequence richer in purines. The first motif is present only in the 3′ UTR of primates (Fig 1B, underlined and italicized sequences), whereas the second motif is conserved in various species (Fig 1B, underlined bold sequences).
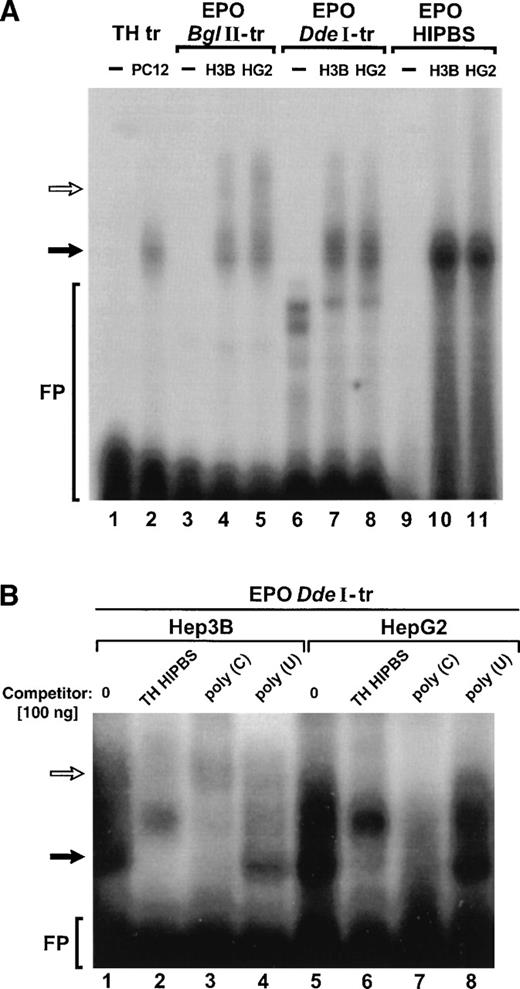
Because of these similarities in the sequence we hypothesized that the pyrimidine-rich motifs in EPO and TH mRNA bind the same protein factors. Binding reactions were performed using EPO or TH transcripts containing HIPBS-like sequences and cytoplasmic protein extracts from Hep3B, HepG2, or PC12 cells (Fig 2). The two EPO transcripts (BglII and DdeI) formed essentially the same two complexes with protein extracts from either Hep3B or HepG2 cells (Fig 2A, lanes 3 to 8). This indicates that the protein-binding sites are restricted within the 93 bases (bases 738 to 860) of the EPODdeI transcript. The faster-migrating complex (solid arrow) migrated with the same mobility as the complex formed by TH transcript with protein factors from PC12 cells (Fig 2A, lane 2) or the complex formed by EPO oligoribonucleotide with protein factors from Hep3B or HepG2 cells (Fig 2A, lanes 10 and 11). This EPO oligoribonucleotide corresponds to the pyrimidine/cytidine-rich sequence (HIPBS-like motif) in EPO mRNA conserved in various species. A second, slower-migrating complex (open arrow) was usually visible in the binding reactions with both EPO transcripts, but not with the EPO HIPBS-like oligoribonucleotide. These data indicate that the faster-migrating ribonucleoprotein complex is associated with the conserved 818 to 830 base fragment of EPO 3′ UTR, which contains the HIPBS-like motif, whereas the slower-migrating complex is most likely associated with the less conserved more proximal region corresponding to bases 738 to 818 of the EPO 3′ UTR.
EPO transcripts containing HIPBS-like sequence form ribonucleoprotein complexes with protein extracts from Hep3B and HepG2 that migrate with the same mobility as the TH mRNA-associated complex. (A) RNA gel-shift assay with TH transcript (TH-tr, lanes 1 to 2) and PC12 cells extract (10 μg), EPO BglII transcript (EPOBglII-tr, lanes 3 to 5), EPO DdeI transcript (EPO DdeI-tr, lanes 6 to 8), and EPO oligoribonucleotide (EPO-HIPBS, lanes 9 to 11) and with proteins (40 μg) from Hep3B (H3B) and HepG2 (HG2) cells. Solid arrow indicates the main, faster-migrating complex. Open arrow indicates the second, slower-migrating complex, visible only when EPO transcripts are used. FP - free probe. Free probes migrate only in lanes 1, 3, 6, and 9. Note that less PC12 protein extract was used in the binding reaction with the TH transcript to obtain autoradiographically comparable signal between TH and EPO complexes. (B) Competition of the complexes formed by EPO DdeI transcript with proteins (40 μg) from Hep3B (lanes 1 to 4) or Hep2G (lanes 5 to 8) by TH HIPBS (lanes 2 and 6) or poly(C) RNA (lanes 3 and 7) but not by poly(U) RNA (lanes 4 and 8). Solid arrow indicates specific complex. Open arrow indicates slower-migrating complex.
EPO transcripts containing HIPBS-like sequence form ribonucleoprotein complexes with protein extracts from Hep3B and HepG2 that migrate with the same mobility as the TH mRNA-associated complex. (A) RNA gel-shift assay with TH transcript (TH-tr, lanes 1 to 2) and PC12 cells extract (10 μg), EPO BglII transcript (EPOBglII-tr, lanes 3 to 5), EPO DdeI transcript (EPO DdeI-tr, lanes 6 to 8), and EPO oligoribonucleotide (EPO-HIPBS, lanes 9 to 11) and with proteins (40 μg) from Hep3B (H3B) and HepG2 (HG2) cells. Solid arrow indicates the main, faster-migrating complex. Open arrow indicates the second, slower-migrating complex, visible only when EPO transcripts are used. FP - free probe. Free probes migrate only in lanes 1, 3, 6, and 9. Note that less PC12 protein extract was used in the binding reaction with the TH transcript to obtain autoradiographically comparable signal between TH and EPO complexes. (B) Competition of the complexes formed by EPO DdeI transcript with proteins (40 μg) from Hep3B (lanes 1 to 4) or Hep2G (lanes 5 to 8) by TH HIPBS (lanes 2 and 6) or poly(C) RNA (lanes 3 and 7) but not by poly(U) RNA (lanes 4 and 8). Solid arrow indicates specific complex. Open arrow indicates slower-migrating complex.
To determine the specificity of the binding, competition reactions were performed (Fig 2B). Formation of the complex between EPO DdeI transcript and cytoplasmic protein extracts from either Hep3B or HepG2 cells, which migrated with the same mobility as the TH mRNA-protein complex (solid arrow), was abolished when TH HIPBS oligoribonucleotide or poly(C) RNA was added to the binding reaction, but not when poly(U) RNA was added (Fig 2B). On the other hand, formation of the slower migrating complex (open arrow) was resistant to the competition with TH HIPBS, although poly(C) RNA also competed with it. These results indicate that the faster-migrating complex associated with EPODdeI transcript has an affinity for the same poly(C)-binding protein factors as TH HIPBS, whereas the slower-migrating complex involves other protein(s) specific to the EPO transcript.
To further identify EPO RNA-protein complexes, binding reactions containing TH probe or EPO NcoI or DdeI transcripts and cytoplasmic protein extracts from either PC12, Hep3B, or HepG2 cells were UV light cross-linked and analyzed by SDS-PAGE (Fig 3). The binding reactions were performed in the presence or absence of poly(U) RNA as a nonspecific competitor. In the absence of poly(U), TH transcript formed a 50 kD complex and additional complexes migrating between 50 and 80 kD (lane 1) with proteins from PC12 cells. In the presence of poly(U), TH transcript formed only one complex migrating at 50 kD (lane 2), as we described previously.15 In the absence of poly(U), both EPO transcripts formed two complexes with protein factors from PC12 cell line (lanes 3 and 5) migrating at approximately 50 and 60 kD. Addition of poly(U) RNA, which does not affect formation of the complexes visible in the gel retardation assays (Fig 2B), completely abolished formation of the 60 kD complex with EPO transcripts, leaving only the 50 kD complex (lanes 4 and 6). Importantly, formation of the 50 kD complex with EPO DdeI transcript was specifically abolished by addition of poly(C) RNA or TH HIPBS oligoribonucleotide (lanes 7 to 10). This indicates that the same complex is formed with protein extracts by both TH and EPO DdeI transcripts. The same 50 kD complex was formed with the EPODdeI transcript in the presence of poly(U) RNA when protein extracts were used from either Hep3B or HepG2 cells (Fig 3B). In the case of protein extracts from both Hep3B and HepG2 cells, an additional, fainter band was visible at 70 kD. The presence of this band was less consistent as compared with the 50 kD complex.
SDS-PAGE analysis of the UV light cross-linked RNA-protein complexes. (A) Characterization of the complexes formed by TH transcript (TH-tr), and by two EPO transcripts (NcoI-tr andDdeI-tr) and proteins (40 μg) from PC12 cells. Poly(U) RNA, TH HIPBS, or poly(C) RNA were added to the binding reaction as competitors (+ 50 ng, ++ 100 ng). Solid arrow points to the 50 kD complex. FP - free probe. (B) Comparison of complexes formed with the EPO DdeI transcript and protein factors from PC12 (lane 2), Hep3B (H3B, lane 3), and HepG2 (HG2, lane 4) before (lanes 2, 4, and 6) and after (lanes 3, 5, and 7) addition of 100 ng of poly(U) RNA. Lane 1 - free DdeI EPO probe. Solid arrow indicates the 50 kD complex. Open arrow indicates the 70 kD complex formed with extracts from Hep3B and HepG2 cells.
SDS-PAGE analysis of the UV light cross-linked RNA-protein complexes. (A) Characterization of the complexes formed by TH transcript (TH-tr), and by two EPO transcripts (NcoI-tr andDdeI-tr) and proteins (40 μg) from PC12 cells. Poly(U) RNA, TH HIPBS, or poly(C) RNA were added to the binding reaction as competitors (+ 50 ng, ++ 100 ng). Solid arrow points to the 50 kD complex. FP - free probe. (B) Comparison of complexes formed with the EPO DdeI transcript and protein factors from PC12 (lane 2), Hep3B (H3B, lane 3), and HepG2 (HG2, lane 4) before (lanes 2, 4, and 6) and after (lanes 3, 5, and 7) addition of 100 ng of poly(U) RNA. Lane 1 - free DdeI EPO probe. Solid arrow indicates the 50 kD complex. Open arrow indicates the 70 kD complex formed with extracts from Hep3B and HepG2 cells.
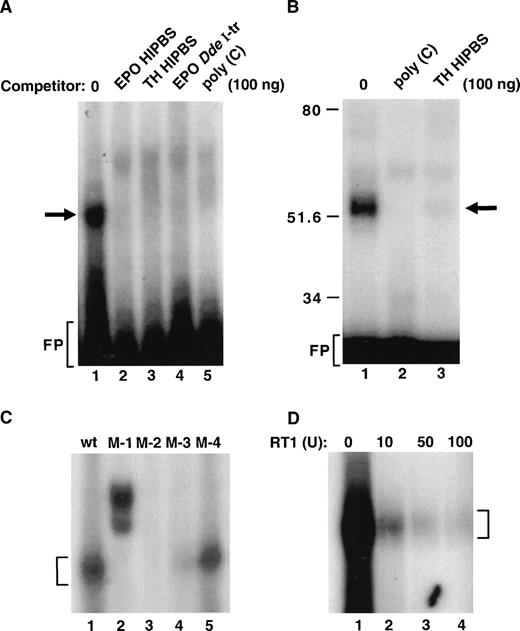
Because the EPO-HIPBS is the binding site for the protein factors, we attempted to characterize binding of protein factors to this fragment of EPO RNA (Fig 4). EPO HIPBS formed a single complex identified in the RNA gel shift assay (Fig 4A, lane 1); formation of this complex was abolished by adding either TH or EPO HIPBS oligoribonucleotides, EPO DdeI transcript, or poly(C) RNA (Fig 4A, lanes 2 to 5). This complex, identified in a gel-shift assay, corresponded to a single 50 kD complex identified after UV light cross-linking of the binding reaction and SDS-PAGE analysis of the complexes (Fig 4B, lane 1). Again, formation of this complex was abolished by either poly(C) or TH HIPBS RNA (Fig 4B, lanes 2 and 3).
Formation of the ribonucleoprotein complex associated with EPO HIPBS-like oligoribonucleotide. (A) Gel-shift analysis of the binding reaction of EPO oligoribonucleotide and protein factors from HepG2 cells. EPO oligoribonucleotide is sufficient to form a complex with proteins from HepG2 cells (lane 1). Formation of this complex is prevented by competition with 100 ng of cold EPO (lane 2) or TH HIPBS (lane 3) oligoribonucleotides, EPO DdeI transcript (lane 4), or poly(C) RNA (lane 5). (B) Analysis of the EPO oligoribonucleotide-protein complex after UV light cross-linking and SDS-PAGE. EPO oligoribonucleotide forms a 50 to 55 kD complex with cytoplasmic proteins from HepG2 cells (lane 1). Formation of this complex is abolished by competition with 100 ng of poly(C) RNA (lane 2) or TH HIPBS (lane 3). (C) Gel-shift analysis of complexes formed by proteins from HepG2 cells and wild-type (wt) or mutated (M-1, 2, 3, 4) EPO HIPBS. Bracket indicates the specific complex. (D) Gel-shift analysis of the HepG2 protein complex associated with EPO HIPBS in the presence of increasing concentrations of RT1 (lanes 1 to 4). Bracket indicates the specific complex. FP-freeprobe
Formation of the ribonucleoprotein complex associated with EPO HIPBS-like oligoribonucleotide. (A) Gel-shift analysis of the binding reaction of EPO oligoribonucleotide and protein factors from HepG2 cells. EPO oligoribonucleotide is sufficient to form a complex with proteins from HepG2 cells (lane 1). Formation of this complex is prevented by competition with 100 ng of cold EPO (lane 2) or TH HIPBS (lane 3) oligoribonucleotides, EPO DdeI transcript (lane 4), or poly(C) RNA (lane 5). (B) Analysis of the EPO oligoribonucleotide-protein complex after UV light cross-linking and SDS-PAGE. EPO oligoribonucleotide forms a 50 to 55 kD complex with cytoplasmic proteins from HepG2 cells (lane 1). Formation of this complex is abolished by competition with 100 ng of poly(C) RNA (lane 2) or TH HIPBS (lane 3). (C) Gel-shift analysis of complexes formed by proteins from HepG2 cells and wild-type (wt) or mutated (M-1, 2, 3, 4) EPO HIPBS. Bracket indicates the specific complex. (D) Gel-shift analysis of the HepG2 protein complex associated with EPO HIPBS in the presence of increasing concentrations of RT1 (lanes 1 to 4). Bracket indicates the specific complex. FP-freeprobe
The predicted protein-binding site within the EPO HIPBS is the cytidine-rich motif CCCUCCCCCGCC (Fig 1B). Synthetic oligoribonucleotides with mutations within this predicted protein-binding motif were used in the binding reactions (Fig 4C). Mutations within the EPO that affected this cytidine-rich motif abolished protein binding (Fig 4C, lanes 2 to 4). Mutations outside this motif allowed for some decreased residual binding (Fig 4C, lane 5). This indicates that the predicted cytidine-rich motif is the protein-binding site. Because of the guanidine residue within the cytidine-rich region, formation of the complex was tested for sensitivity to the RNAse T1 (Fig 4D). Clearly, formation of this complex was abolished by increasing concentrations of RNase T1 (RT1, Fig 4D). This proves, as predicted, that the G residue in this EPO HIPBS-like element is located in the protein-binding site and to some extent is protected from RT1 activity by the protein binding. The higher doses of RT1, however, overcame the protective effects of bound proteins.
The ribonucleoprotein complex associated with EPO HIPBS-like RNA contains 40 kD PCBP.
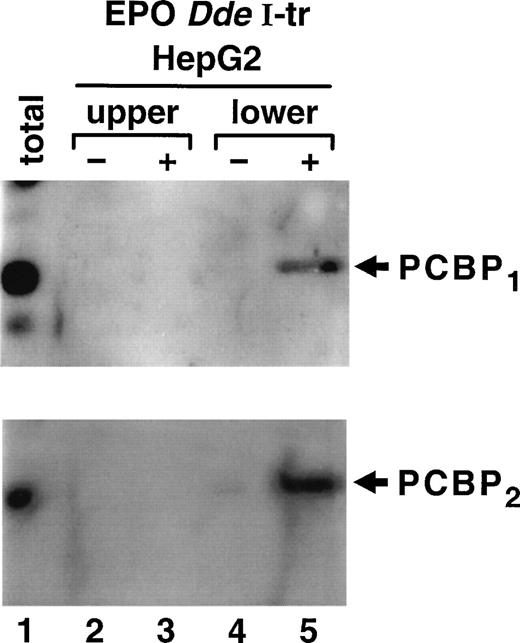
The protein factor binding to the TH HIPBS in a hypoxia-inducible manner is the 40 kD PCBP.15 To determine whether PCBP participates in formation of the complex associated with the EPO HIPBS-like element, binding reactions were performed using EPODdeI transcript and proteins from HepG2 cells. Gel-shift assays were performed, and protein factors were electroeluted from a complex identified as migrating with the same mobility as the TH HIPBS-associated complex (lower complex). Proteins were also eluted from the EPO mRNA-protein complexes that migrated with lower mobility (upper complex) than the TH-protein complexes, and from gel slices containing binding reactions without RNA. The eluted proteins were analyzed by Western blot with specific anti-PCBP1 or PCBP2 antibodies (Fig 5). The lower EPO mRNA protein complex specifically contained both isoforms of PCBP (Fig 5, lane 5). This protein was absent in the control binding reactions performed without RNA (Fig 4, lane 4), and in the upper complex and its control (lanes 2 and 3). Note that PCBP2migrates slightly slower, most likely because of the posttranslational modifications.
Identification of PCBP as part of the ribonucleoprotein complex associated with EPO HIPBS-like element in HepG2 cells Proteins electroeluted from the EPO DdeI transcript-associated complexes were analyzed by Western blot technique. The same membrane was first probed with specific anti-PCBP1 antibody,and then stripped and reprobed with anti-PCBP2 antibody. The lower complex (corresponding to the TH-HIPBS–associated complex) contains both isoforms of PCBP (lane 5), which is absent in the eluates from gel slices containing binding reaction without RNA (lane 4) or in the upper complex (lanes 2 and 3). Lane 1, 100 μg of HepG2 protein extract.
Identification of PCBP as part of the ribonucleoprotein complex associated with EPO HIPBS-like element in HepG2 cells Proteins electroeluted from the EPO DdeI transcript-associated complexes were analyzed by Western blot technique. The same membrane was first probed with specific anti-PCBP1 antibody,and then stripped and reprobed with anti-PCBP2 antibody. The lower complex (corresponding to the TH-HIPBS–associated complex) contains both isoforms of PCBP (lane 5), which is absent in the eluates from gel slices containing binding reaction without RNA (lane 4) or in the upper complex (lanes 2 and 3). Lane 1, 100 μg of HepG2 protein extract.
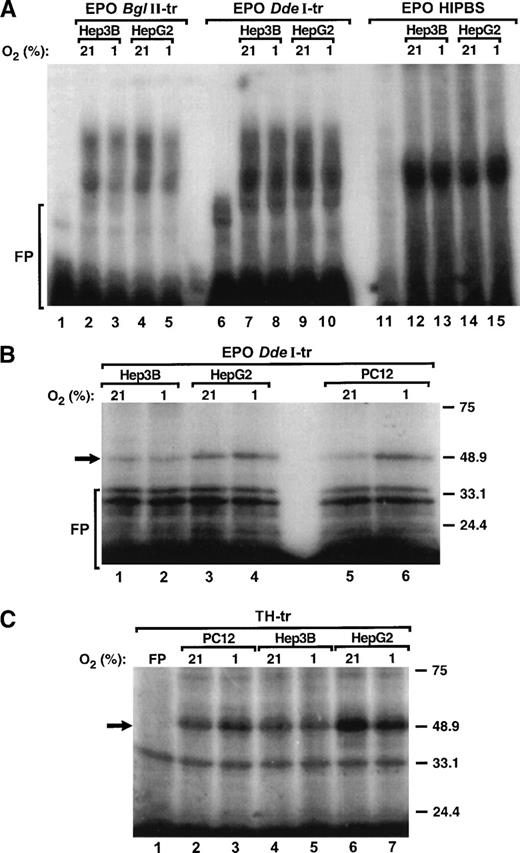
To determine whether binding of protein factors to the HIPBS-like element within the 3′ UTR of EPO mRNA is regulated by hypoxia, binding reactions were performed with protein extracts from PC12, Hep3B, and HepG2 cells exposed to 21% or 1% O2 and EPOBglII, DdeI transcripts, EPO HIPBS oligoribonucleotide, or TH transcript and analyzed by gel-shift assay of UV light cross-linking and SDS-PAGE. Clearly, hypoxia did not induce formation of the complexes associated with any of the EPO or TH probes and protein extracts from either Hep3B or HeG2 cells (Fig 6A, 6B, lanes 1 to 4, 6C, lanes 4 to 7). However, hypoxia induced formation of the complexes with either EPODdeI (Fig 6B, lanes 6 to 7) or TH (Fig 6C, lanes 2 to 3) transcripts when protein extracts were used from PC12 cells. In contrast to PC12 cells,15 in HepG2 or Hep3B cells hypoxia did not induce expression of the PCBP (not shown).
Formation of the ribonucleoprotein complex associated with EPO mRNA is not hypoxia-inducible. (A) Gel-shift analysis of the protein complexes associated with EPOBglII (lanes 1 to 5), EPO DdeI (lanes 6 to 10) transcripts or EPO HIPBS (lanes 11 to 15) and protein extracts (40 μg) from either Hep3B or HepG2 cells during normoxia (21% O2) and hypoxia (1% O2). (B) SDS-PAGE analysis of UV light cross-linked complexes formed by EPO DdeI-transcript and protein factors (40 μg) from Hep3B (lanes 1, 2), HepG2 (lanes 3, 4), and PC12 cells (lanes 5, 6) that were exposed to normoxia (21% O2) or hypoxia (1% O2). (C) SDS-PAGE analysis of UV light cross-linked complexes formed by TH transcript and protein factors from PC12 (10 μg, lanes 2, 3), Hep3B (40 μg, lanes 4, 5), and HepG2 cells (40 μg, lanes 6, 7) that were exposed to normoxia (21% O2) or hypoxia (1% O2). FP - free probe.
Formation of the ribonucleoprotein complex associated with EPO mRNA is not hypoxia-inducible. (A) Gel-shift analysis of the protein complexes associated with EPOBglII (lanes 1 to 5), EPO DdeI (lanes 6 to 10) transcripts or EPO HIPBS (lanes 11 to 15) and protein extracts (40 μg) from either Hep3B or HepG2 cells during normoxia (21% O2) and hypoxia (1% O2). (B) SDS-PAGE analysis of UV light cross-linked complexes formed by EPO DdeI-transcript and protein factors (40 μg) from Hep3B (lanes 1, 2), HepG2 (lanes 3, 4), and PC12 cells (lanes 5, 6) that were exposed to normoxia (21% O2) or hypoxia (1% O2). (C) SDS-PAGE analysis of UV light cross-linked complexes formed by TH transcript and protein factors from PC12 (10 μg, lanes 2, 3), Hep3B (40 μg, lanes 4, 5), and HepG2 cells (40 μg, lanes 6, 7) that were exposed to normoxia (21% O2) or hypoxia (1% O2). FP - free probe.
DISCUSSION
In this study we identified two isoforms of a 40 kD PCBP17as a part of a ribonucleoprotein complex associated with a cytidine-rich sequence located within the 3′ UTR of EPO mRNA. This is the first identification of protein factors that are associated with EPO mRNA. The conserved cytidine-rich sequence is located within the putative EPO mRNA stability region. Direct evidence regarding the role of this region in regulation of EPO mRNA stability is still missing. However, deletion of the EPO 3′ UTR downstream from the pyrimidine-rich region results in stabilization of the EPO mRNA.7 In contrast, deletion of the 100 bases including the pyrimidine-rich region results in destabilization of the mRNA, although under conditions of prolonged EPO mRNA half-life due to the use of actinomycin D.8 In addition, some other mRNA, such as α2 globin,16-19 collagen α1(I),20 and TH15 mRNA, were shown to use cytidine-rich motifs as determinants of their stability and to bind the PCBP. Thus, it is likely that the cytidine-rich motif represents the EPO mRNA stabilizing element and that PCBP plays a stabilizing role in maintaining constitutive stability of EPO mRNA.
The role of this cytidine-rich motif in potential hypoxic regulation of EPO mRNA stability is less clear. On the basis of functional studies, this regulatory stability element in the 3′ UTR of TH mRNA is necessary but not sufficient for the hypoxic regulation of TH mRNA stability.15 The same may be true in the case of EPO mRNA. In contrast to TH mRNA, however, formation of the ribonucleoprotein complexes associated with the HIPBS-like element is not inducible by hypoxia in the EPO-synthesizing cell lines such as Hep3B or HepG2. This finding is consistent with inability to induce expression of PCBP by hypoxia in HepG2 or Hep3B cells, and with the previously reported lack of hypoxic inducibility of erythropoietin RNA binding proteins (ERBP) binding to EPO mRNA.9 This may indicate that if EPO mRNA stability is regulated by hypoxia, protein factors other than the PCBP may be involved in this regulation. In that respect, several studies have implicated that hypoxia-inducible protein binding to the AU-rich instability elements within the 3′ UTR of the vascular endothelial growth factor mRNA may regulate stability of this mRNA during hypoxia.21,22 In this respect the 3′ UTR of EPO mRNA may contain instability elements because deletion of the conserved distal part of EPO 3′ UTR results in stabilization of the remaining EPO mRNA.8 In addition, a single instability element, an AUUUA motif (bases 1100 to 1105), is present in the distal part of EPO 3′ UTR. Thus, perhaps the role of the EPO mRNA instability elements located downstream from the EPO mRNA stability element should be examined in hypoxic regulation of EPO mRNA.
We have clearly showed that there are tissue-specific differences in regulation of PCBP expression by hypoxia. In PC12 cells expression of PCBP1 but not PCBP2 is induced by hypoxia.15 In contrast, neither of the PCBP is induced by hypoxia in Hep3B or HepG2 cells. This finding is not surprising because, in addition to universal mechanisms exist specific mechanisms for oxygen-sensing and regulating gene expression. For example, regulation of TH gene expression by reduced oxygen tension is tissue specific for carotid body and PC12 cells but not for other catecholaminergic cells.23 In addition, oxygen-sensing mechanisms in PC12 cells or carotid body cells are more sensitive than in other tissues. In this respect, TH gene expression is induced by even mild (5%) hypoxia, whereas induction of EPO requires stronger reduction in oxygen tension, usually to 1%.24 Finally, different molecular mechanisms are involved in transcriptional regulation of TH and of EPO gene expression by hypoxia. Induction of EPO gene transcription involves predominantly Hif-1,25whereas induction of TH gene transcription involves binding of c-fos and jun-b to the Ap1 site on the TH promoter.26
Analysis of the UV light cross-linked complexes showed that the main specific complex associated with EPO DdeI transcript in different cell lines is 50 kD, which corresponds to the binding protein PCBP of 40 kD. The presence of this 50 kD complex was not reported previously. The previous studies identified the EPO mRNA-associated complexes as 70 and 135 kD (ERBP).9 In our experiments we have inconsistently observed formation of a 70 kD complex with proteins from Hep3B and HepG2 cells. It is possible that this 70 kD complex corresponds with the one reported previously by others.9 We have not identified the 135 kD complex, however. One possible reason for this discrepancy is that the 135 kD complex may contain more than one protein factor because analysis of the EPO RNA-protein complexes in that study was performed in the absence of reducing agents in the sample buffer.9 In our experiments the cross-linked reactions are analyzed in the presence of 100 mmol/L DTT, that is, under reducing conditions. In this study we did not attempt to identify other protein factors binding to the EPO mRNA, that may be involved in formation of the slower-migrating complex identified in the gel shift assays. It is possible that this complex corresponds to the 70 kD complex identified by SDS-PAGE .
In this study we did not determine whether the PCBP are the only proteins present in the ribonucleoprotein complex associated with the EPO mRNA pyrimidine-rich sequence or whether other proteins also interact with PCBP. Recently it was proposed that ERBP interacts with the heat-shock protein, hsp70.27 During normoxia, ERBP is bound to the hsp 70, and its binding to the EPO mRNA is low. During hypoxia, hsp 70 is sequestered; this allows the ERBP to bind fully to the EPO mRNA.27 Further studies are required to identify such potential interactions.
Identification of the PCBP protein in the complex associated with EPO mRNA stability represents a significant step in understanding posttranscriptional regulation of EPO gene expression.
ACKNOWLEDGMENT
We thank A. Gamarnik and R. Andino for anti-PCBP2 antibody, H.F. Bunn for EPO cDNA, and G. Doerman for preparation of the figures.
Supported by NIH grants no. NIH HL51078, HL58687, and American Heart Association Grant-in-Aid 9750110N.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal