Interferon- (IFN-) is a pleiotropic cytokine that has antiviral, antiproliferative, and immunoregulatory functions. There is increasing evidence that IFN- has an important role in T-cell biology. We have analyzed the expression ofIL-2R, c-myc, and pim-1 genes in anti-CD3–activated human T lymphocytes. The induction of these genes is associated with interleukin-2 (IL-2)–induced T-cell proliferation. Treatment of T lymphocytes with IFN-, IL-2, IL-12, and IL-15 upregulated IL-2R, c-myc, andpim-1 gene expression. IFN- also sensitized T cells to IL-2–induced proliferation, further suggesting that IFN- may be involved in the regulation of T-cell mitogenesis. When we analyzed the nature of STAT proteins capable of binding to IL-2R,pim-1, and IRF-1 GAS elements after cytokine stimulation, we observed IFN-–induced binding of STAT1, STAT3, and STAT4, but not STAT5 to all of these elements. Yet, IFN- was able to activate binding of STAT5 to the high-affinity IFP53 GAS site. IFN- enhanced tyrosine phosphorylation of STAT1, STAT3, STAT4, STAT5a, and STAT5b. IL-12 induced STAT4 and IL-2 and IL-15 induced STAT5 binding to the GAS elements. Taken together, our results suggest that IFN-, IL-2, IL-12, and IL-15 have overlapping activities on human T cells. These findings thus emphasize the importance of IFN- as a T-cell regulatory cytokine.
INTERFERON-α (IFN-α) has antiviral, antiproliferative, and immunomodulatory functions.1-3IFN-α is produced in the course of immune response, especially during viral infections. Macrophages and other antigen-presenting cells are the main cellular sources of IFN-α. Immunomodulatory functions of IFN-α include the enhancement of natural killer (NK) cell and T-cell cytotoxicity,2,3 and knockout mice lacking a functional type I IFN receptor were unable to generate a cytotoxic T-lymphocyte response to lymphocytic choriomeningitis virus.4 IFN-α has been shown to promote Th1-type immune responses, eg, by inducing interleukin-12 (IL-12) receptor β2 chain and IFN-γ gene expression in human T cells.5-7
IL-2 is the major growth factor for T lymphocytes. IL-2 stimulation of T cells is associated with the activation of several signaling pathways, including the Janus tyrosine kinase-signal transducer and activator of transcription (Jak-STAT) pathway.8-11 Binding of IL-2 to its receptor activates Jak1 and Jak3 tyrosine kinases, leading to tyrosine phosphorylation, dimerization, and nuclear translocation of STAT5 proteins. The role of STAT5 in IL-2–induced T-cell proliferation is controversial,12,13 but recent studies with STAT5a-deficient mice suggest that STAT5a is involved in T-cell proliferation. Splenocytes from STAT5a-deficient mice show decreased proliferative response to IL-2, a result of impaired expression of IL-2 receptor α (IL-2Rα) chain.14
IL-2 signals via a specific receptor, the high-affinity form of which is composed of three subunits, α (CD25), β (CD122), and γ (γcommon) chains. Whereas the β and γ chains are constitutively expressed in resting T lymphocytes, the α chain is expressed only in activated T lymphocytes.15 Both T-cell receptor stimulation and IL-2 treatment are able to induce α chain gene expression. IL-2Rα chain is required for the high-affinity binding of IL-2 and it is essential for optimal proliferative responses of the mature T cells.14,15 A human immune disorder, characterized by decreased numbers of peripheral T cells, arises from a mutation in the gene coding for the IL-2Rα chain.16
IL-2–induced T-cell proliferation is associated with the induction of several IL-2–responsive genes, such as c-myc andpim-1.15,17 The c-myc gene is expressed in a wide variety of proliferating cells, and it is activated in many types of human and animal neoplasia, including leukemias and lymphomas.18 The pim-1 gene encodes a serine/threonine kinase that is expressed predominantly in hematopoietic cells.19,20 Studies with transgenic mice have demonstrated that pim-1 can also act as an oncogene in synergy with c-myc, N-myc, or bcl-2 to induce B- and T-cell lymphomas.21-23 In addition, there is evidence that overexpression of pim-1 can accelerate lymphoid and myeloid cell proliferation by enhancing cell survival.24-26 Both c-Myc and Pim-1 proteins may thus have important roles in mediating cytokine-induced mitogenic signals.
Although IFN-α is an important T-cell regulatory cytokine, most studies so far regarding IFN-α signaling and gene activation have been performed using nonlymphoid cells. In this report, we have studied the cytokine-dependent gene expression in anti-CD3–activated human T lymphocytes focusing on genes that are involved in T-cell mitogenesis. We show that, in addition to IFN-α, also IL-2, IL-12, and IL-15 induce IL-2Rα, c-myc, and pim-1 gene expression. We also show that IFN-α induces tyrosine phosphorylation of STAT1, STAT3, STAT4, STAT5a, and STAT5b and enhances their binding to the IFN-γ activated sequence (GAS) elements of the IFN-α–responsive genes.
MATERIALS AND METHODS
T-cell culture.
Leukocyte-rich buffy coats were obtained from healthy blood donors (Finnish Red Cross Blood Transfusion Service, Helsinki, Finland). Mononuclear cells were isolated by density gradient centrifugation using Ficoll-Paque (Pharmacia Biotech AB, Uppsala, Sweden). Monocytes were removed by adherence, and T cells were further purified by nylon wool columns. Purified T cells were activated with a 1:1,000 dilution of anti-CD3 monoclonal antibodies containing ascites fluid (kindly provided by Dr Matti Kaartinen, University of Helsinki, Helsinki, Finland) and cultured in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS; Integro, Zaandam, The Netherlands) and 100 IU/mL IL-2 for 5 to 6 days. T cells were further expanded for 3 to 5 days with RPMI supplemented with IL-2. As determined by flow cytometry, more than 99% of cells were CD3+, consisting of CD4+(30%) and CD8+ (70%) cells (data not shown). In all experiments, T cells were removed from IL-2–containing medium 12 to 16 hours before cytokine stimulation. In each experiment, T cells from two to four donors were used.
Cytokines.
Highly purified human leukocyte IFN-α (13 × 106IU/mL) was provided by Dr Hannele Tölö (Finnish Red Cross Blood Transfusion Service, Helsinki, Finland). IFN-β (0.2 × 106 IU/mL) was from Bioferon GmbH & Co (Laupheim, Germany). Human recombinant IL-12 (rIL-12) and rIL-15 were purchased from R&D Systems (Abingdon, UK) and rIL-2 from Chiron Corp (Emeryville, CA). The cytokine concentrations used were as follows, unless otherwise indicated: IFN-α (100 IU/mL), IL-2 (100 IU/mL), IL-12 (5 ng/mL), and IL-15 (5 ng/mL).
RNA isolation and Northern blot analysis.
T cells were treated with different cytokines and cells were harvested 3 hours after stimulation. Total cellular RNA was isolated by guanidium isothiocyanate lysis followed by centrifugation through CsCl cushion.27 28 Total cellular RNA was quantitated photometrically and samples containing equal amounts of RNA (20 μg) were size fractionated on a 1.0% formaldehyde-agarose gel, transferred to a nylon membrane (Hybond; Amersham, Buckinghamshire, UK), and hybridized with the cDNA probes encoding human IL-2Rα,c-myc, and pim-1. EtBr staining of rRNA bands was used to ensure equal RNA loading. The probes were labeled with [α-32P] dCTP (3,000 Ci/mmol; Amersham) using a random primed DNA labeling kit (Boehringer Mannheim, Mannheim, Germany). The membranes were hybridized under conditions of high stringency (50% formamide, 5× Denhardt’s solution, 5× SSPE, and 0.5% sodium dodecyl sulfate [SDS]). The membranes were washed twice at room temperature and once at 60°C in 1× SSC/0.1% SDS for 30 minutes each time. The membranes were exposed to Kodak AR X-Omat films (Eastman Kodak, Rochester, NY) at −70°C using intensifying screen.
Electrophoretic mobility shift assay (EMSA).
Nuclear extracts29 and nuclear protein/DNA binding reactions30 were performed as described previously. IFP 53 GAS (5′-GATCAATCACCCAGATTCTCAGAAACACTT-3′),IRF-1 GAS (5′-AGCTTCAGCCTGATTTCCCCGAAATGACGGA-3′),pim-1 GAS (5′-ACACACATCCCTTCCCAGAAATCAGGATTC-3′), and IL-2Rα GAS-c/GAS-n (5′-TTTCTTCTAGGAAGTACCAAACATTTCTGATAATAGAA-3′) oligonucleotides were synthesized with an IBI oligonucleotide synthesizer (Foster City, CA) and purified on polyacrylamide gel electrophoresis (PAGE) in the presence of 8 mol/L urea. The probes were labeled by T4 polynucleotide kinase. The binding reaction was performed at room temperature for 30 minutes. Samples were analyzed by elecrophoresis on 6% nondenaturing low-ionic strength polyacrylamide gels in 0.25× TBE. The gels were dried and visualized by autoradiography. Antibodies used in supershift experiments were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The following antibodies were used: anti-STAT1 (sc-345X), anti-STAT2 (sc-839X), anti-STAT3 (sc-482X), anti-STAT4 (sc-486X), anti-STAT5b (sc-835X; recognizes both STAT5a and STAT5b), and anti-STAT6 (sc-621X). In supershift experiments, antibodies (1/20 dilution) were incubated with nuclear extracts for 1 hour on ice.
Immunoprecipitation, gel electrophoresis, and Western blotting.
T cells were stimulated with different cytokines for 15 minutes, washed once with phosphate-buffered saline (PBS), and lysed in immunoprecipitation buffer (50 mmol/L Tris, pH 7.4, containing 150 mmol/L NaCl, 5 mmol/L EDTA, and 1% Triton X-100) on ice for 15 minutes. Cell lysates were cleared by microfuge centrifugation and immunoprecipitated with anti-STAT1 (Santa Cruz), anti-STAT3 (Santa Cruz), anti-STAT4 (Santa Cruz), anti-STAT5a (71-2400; ZYMED, San Francisco, CA), or anti-STAT5b (71-2500; ZYMED) antibodies.
The proteins were separated on 10% SDS-PAGE using the Laemmli buffer system31 and transferred electrophoretically onto Immobilon membranes (Millipore, Bedford, MA). Biotinylated antihuman IL-2Rα polyclonal antibody (0.2 μg/mL; BAF223; R&D Systems) was allowed to bind in PBS containing 3% bovine serum albumin (BSA) for 1 hour at room temperature, followed by peroxidase-conjugated streptavidin (1/2,000 dilution; 016-030-084; Jackson ImmunoResearch Laboratories, Inc, West Grove, PA) for 1 hour at room temperature. Primary antiphosphotyrosine antibody (sc-7020; 1/1,000 dilution; Santa Cruz) was allowed to bind in PBS containing 3% BSA for 1 hour at room temperature, followed by secondary antibody binding with Biotin-SP–conjugated goat antimouse IgG (115-066-072; 1/10,000 dilution; Jackson) for 1 hour at room temperature. After this, peroxidase-conjugated streptavidin (Jackson) was allowed to bind for 1 hour at room temperature. The bands were visualized by the ECL chemiluminescence system (Amersham). The membranes were reprobed with anti-STAT1 (Santa Cruz), anti-STAT3 (Santa Cruz), anti-STAT4 (Santa Cruz), anti-STAT5a (ZYMED), or anti-STAT5b (ZYMED) antibodies in PBS containing 5% nonfat milk for 2 hours at room temperature, followed by secondary antibody binding with peroxidase-conjugated goat antirabbit IgG (Bio-Rad, Richmond, CA) for 1 hour at room temperature and visualized by the ECL chemiluminescence system (Amersham).
T-cell proliferation assay.
Anti-CD3–activated T cells were removed from IL-2–containing media before the proliferation assay. Cell cultures with greater than 90% viability were used. A total of 10 × 106 cells (106 cells/mL) were left untreated or treated with IFN-α for 24 hours. The cells were then washed twice with PBS, counted, and resuspended into IFN-free RPMI. Fifty microliters of cell suspension (50,000 cells) was seeded in triplicates into a round-bottom 96-well plate (Greiner, Nürtingen, Germany). IL-2 was added at a final concentration of 0, 10, 30, or 100 IU/mL. After thorough mixing, cells were incubated in 5% CO2 at 37°C for 18 hours. The cells were incubated for another 6 hours in the presence of 1 μCi/well of 3H-labeled thymidine (Amersham). The cells were collected using a harvester (Tomtec Harvester 96 Mach IIIM, Tomtec Inc, Hamden, CT) and counted in a microplate counter (Wallac 1450 Microbeta Liquid Scintillation Counter, Wallac, Turku, Finland). The proliferation index was calculated as follows: index = (IL-2 induced incorporation − background incorporation)/background incorporation.
RESULTS
IFN-α, IL-2, IL-12, and IL-15 enhance IL-2Rα, c-myc, and pim-1 mRNA expression in human T lymphocytes.
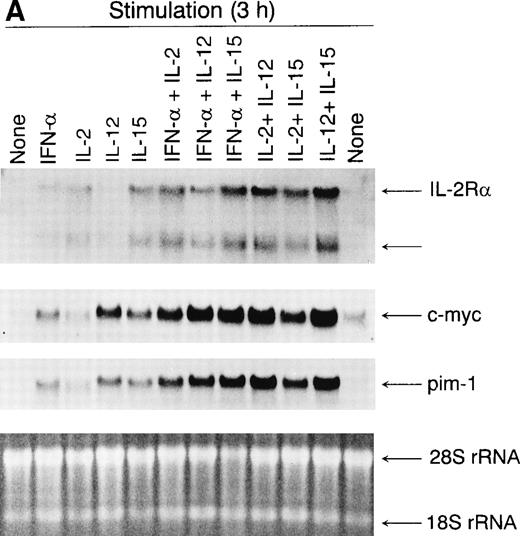
IFN-α is an important T-cell regulatory cytokine that, eg, enhances T-cell cytotoxicity2,3 and stimulates T-cell growth.32 We therefore studied whether IL-2Rα,c-myc, and pim-1, genes involved in T-cell proliferation previously shown to be induced by IL-2 are also upregulated by IFN-α, IL-12, and IL-15. We used activated T cells to ensure that the cells were maximally responsive to cytokine treatment. T cells were incubated with IFN-α, IL-2, IL-12, or IL-15 or with different combinations of these cytokines. After 3 hours of stimulation, the cells were collected and total cellular RNA was prepared for Northern blot analysis. IFN-α, IL-2, IL-12, and IL-15 all enhanced IL-2Rα, c-myc, and pim-1 mRNA expression (Fig 1A). However, there were certain differences between the cytokines in inducing IL-2Rα,c-myc, and pim-1 genes. IL-2Rα was induced by IFN-α, IL-2, and IL-15, whereas only a weak induction was seen by IL-12 (Fig 1A). IFN-α, IL-2, IL-12, and IL-15 all inducedc-myc and pim-1 gene expression, with IL-12 having the strongest and IL-2 the weakest effect. IL-12 combined with IL-2 or IL-15 most strongly enhanced IL-2Rα, c-myc, andpim-1 gene expression. In addition, IFN-α combined with IL-2 or IL-15 had a synergistic effect on the expression of all genes studied (Fig 1A).
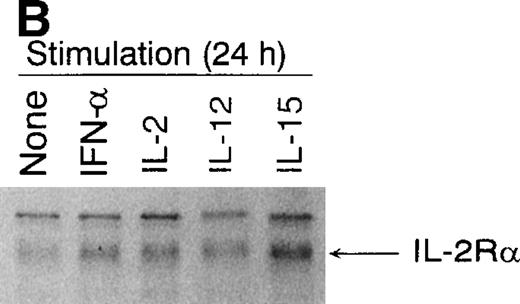
(A) Induction of IL-2R, c-myc, andpim-1 gene expression by IFN-, IL-2, IL-12, and IL-15 in human T lymphocytes. T cells were stimulated with different cytokines for 3 hours, the cells were collected, and the total cellular RNA was isolated. RNA samples (20 μg) were size-fractionated on agarose gels, transferred to nylon membranes, and hybridized with specificIL-2R, c-myc, and pim-1 cDNA probes. EtBr-stained gel is shown to verify equal RNA loading. The result shown is from one experiment but is representative of three individual experiments. (B) Induction of IL-2R protein expression by IFN-, IL-2, IL-12, and IL-15. T cells were stimulated with different cytokines for 24 hours, and cell lysates were prepared. Proteins were separated on 10% SDS-PAGE, transferred to membranes, and immunoblotted with anti–IL-2R antibody.
(A) Induction of IL-2R, c-myc, andpim-1 gene expression by IFN-, IL-2, IL-12, and IL-15 in human T lymphocytes. T cells were stimulated with different cytokines for 3 hours, the cells were collected, and the total cellular RNA was isolated. RNA samples (20 μg) were size-fractionated on agarose gels, transferred to nylon membranes, and hybridized with specificIL-2R, c-myc, and pim-1 cDNA probes. EtBr-stained gel is shown to verify equal RNA loading. The result shown is from one experiment but is representative of three individual experiments. (B) Induction of IL-2R protein expression by IFN-, IL-2, IL-12, and IL-15. T cells were stimulated with different cytokines for 24 hours, and cell lysates were prepared. Proteins were separated on 10% SDS-PAGE, transferred to membranes, and immunoblotted with anti–IL-2R antibody.
Because IFN-α, IL-2, IL-12, and IL-15 upregulated IL-2Rα mRNA expression in activated T cells, we also studied IL-2Rα protein expression by Western blotting. T cells were stimulated for 24 hours with IFN-α, IL-2, IL-12, and IL-15, after which cells were collected and lysates were prepared for Western blot analysis. As shown in Fig1B, IL-2Rα was expressed at basal level in activated T cells. In accordance with mRNA data, IL-15 was the strongest and IL-12 the weakest inducer of IL-2Rα protein expression. Also, IFN-α and IL-2 clearly upregulated IL-2Rα protein expression (Fig 1B).
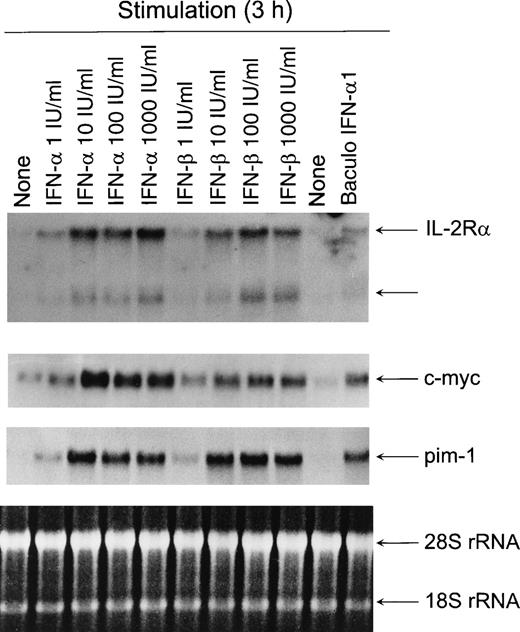
Because IFN-α was able to turn on genes associated with T-cell proliferation, we wanted to study the dose-dependent effect of IFN-α and IFN-β on IL-2Rα, c-myc, and pim-1 gene expression. Therefore, T cells were treated with different concentrations of IFN-α or IFN-β. Very low doses (1 IU/mL) of IFN-α and IFN-β were able to induce IL-2Rα, c-myc, andpim-1 mRNA expression (Fig 2), suggesting that the induction takes place with physiologically relevant IFN concentrations. High expression levels of these genes were detected with 10 IU/mL of both IFN-α and IFN-β.
Dose-dependent activation of IL-2R, c-myc, andpim-1 gene expression by type I IFNs. T cells were stimulated with different doses of IFN- or IFN-β for 3 hours, the cells were collected, and total cellular RNA was isolated. RNA samples (20 μg) were size-fractionated on agarose gels, transferred to nylon membranes, and hybridized with IL-2R, c-myc, and pim-1 cDNA probes. EtBr-stained gel is shown to verify equal RNA loading.
Dose-dependent activation of IL-2R, c-myc, andpim-1 gene expression by type I IFNs. T cells were stimulated with different doses of IFN- or IFN-β for 3 hours, the cells were collected, and total cellular RNA was isolated. RNA samples (20 μg) were size-fractionated on agarose gels, transferred to nylon membranes, and hybridized with IL-2R, c-myc, and pim-1 cDNA probes. EtBr-stained gel is shown to verify equal RNA loading.
IFN-α priming enhances IL-2–induced T-cell proliferation.
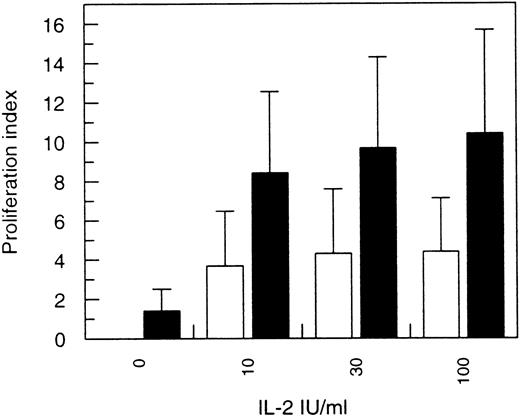
Because IFN-α was able to enhance IL-2Rα gene expression, we studied whether IFN-α pretreatment would sensitize T cells to IL-2–induced cell proliferation. The cells were left untreated or pretreated with IFN-α for 24 hours and restimulated with IL-2, and the T-cell proliferation was analyzed by 3H-labeled thymidine incorporation assay. Proliferation indexes were clearly higher in IFN-α–primed cells compared with unprimed ones. This phenomenon was observed with all IL-2 concentrations tested (Fig 3).
Effect of IFN- pretreatment on IL-2–induced T-cell proliferation. T lymphocytes were activated with anti-CD3-antibodies and expanded in the presence of IL-2, after which IL-2–containing medium was removed. The cells were then left untreated or treated with 100 IU/mL of IFN- for 24 hours. The cells were collected, and an equal number of (□) untreated or (▪) IFN-–primed T cells was applied in microtiter plates. Different doses of IL-2 were added for 18 hours, followed by further incubation of 6 hours in the presence of 1 μCi/well of 3H-labeled thymidine. After harvesting the cells, the proliferation index was determined as described in Materials and Methods. The mean proliferation index (±SD) of six individual donors is shown.
Effect of IFN- pretreatment on IL-2–induced T-cell proliferation. T lymphocytes were activated with anti-CD3-antibodies and expanded in the presence of IL-2, after which IL-2–containing medium was removed. The cells were then left untreated or treated with 100 IU/mL of IFN- for 24 hours. The cells were collected, and an equal number of (□) untreated or (▪) IFN-–primed T cells was applied in microtiter plates. Different doses of IL-2 were added for 18 hours, followed by further incubation of 6 hours in the presence of 1 μCi/well of 3H-labeled thymidine. After harvesting the cells, the proliferation index was determined as described in Materials and Methods. The mean proliferation index (±SD) of six individual donors is shown.
Cytokine-induced STAT DNA-binding to the IL-2Rα GAS-c/GAS-n element.
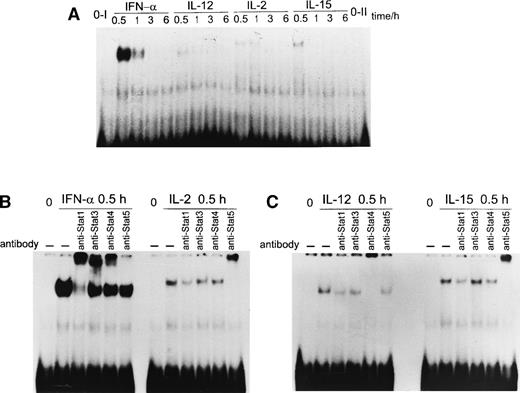
Many cytokines use the Jak/STAT pathway in their signaling and activate genes via GAS elements.10,33 Recently, an essential upstream IL-2 response element of the IL-2Rα gene was characterized. This region contains a consensus and a nonconsensus GAS motif, both of which are required for IL-2–induced IL-2Rαgene expression.34 35 To characterize the proteins capable of binding to the GAS-c/GAS-n element (−3778 to −3740) of the IL-2Rα gene, we stimulated T cells with different cytokines, prepared nuclear extracts, and performed EMSA. EMSAs with the GAS-c/GAS-n oligonucleotide showed the induction of one major complex in response to IFN-α, IL-2, IL-12, and IL-15 stimulation (Fig 4A). However, the mobility of IL-2– and IL-15–induced complexes differed from the ones induced by IFN-α or IL-12, suggesting that these cytokines activate a different range of STAT proteins. The intensity of the IFN-α–induced complex was clearly the strongest. This complex was formed already at 0.5 hours after IFN-α stimulation, and it was detectable at 1 hour after stimulation but disappeared thereafter. Similarly, IL-2–, IL-12–, and IL-15–induced complexes were detectable only at 0.5 hours and 1 hour after stimulation. Next, we identified the proteins within these complexes using specific anti-STAT antibodies in supershift experiments. Antibodies against STAT1 almost completely abolished IFN-α–induced DNA binding to GAS-c/GAS-n. Surprisingly, the addition of anti-STAT3 and anti-STAT4 antibodies resulted in a clearly detectable supershift (Fig 4B). The data suggest that IFN-α–induced complexes consist of at least STAT1, STAT3, and STAT4 proteins. IL-12–induced DNA binding complex reacted only with anti-STAT4 antibodies (Fig 4C). As expected, IL-2 and IL-15 both induced STAT5 DNA binding to GAS-c/GAS-n as detected by supershift experiments (Fig 4B and C).
(A) STAT DNA binding to the IL-2R GAS-c/GAS-n in response to cytokine stimulation. T cells were stimulated with IFN-, IL-2, IL-12, or IL-15 as indicated; nuclear extracts were prepared from the cells; and the STAT DNA binding was analyzed by EMSA. (B) STAT DNA binding to the IL-2R GAS-c/GAS-n in response to IFN- and IL-2. T cells were stimulated with IFN- or IL-2 for 30 minutes, after which nuclear extracts were prepared. Nuclear extracts were incubated for 1 hour on ice with STAT antibodies indicated, followed by binding to 32P-labeled IL-2RGAS-c/GAS-n probe. (C) STAT DNA binding to the IL-2RGAS-c/GAS-n in response to IL-12 and IL-15. T cells were stimulated with IL-2 or IL-15 for 30 minutes, and nuclear extracts were prepared and incubated for 1 hour on ice with different anti-STAT antibodies followed by binding to 32P-labeled IL-2RGAS-c/GAS-n probe. The results are representative of three separate experiments.
(A) STAT DNA binding to the IL-2R GAS-c/GAS-n in response to cytokine stimulation. T cells were stimulated with IFN-, IL-2, IL-12, or IL-15 as indicated; nuclear extracts were prepared from the cells; and the STAT DNA binding was analyzed by EMSA. (B) STAT DNA binding to the IL-2R GAS-c/GAS-n in response to IFN- and IL-2. T cells were stimulated with IFN- or IL-2 for 30 minutes, after which nuclear extracts were prepared. Nuclear extracts were incubated for 1 hour on ice with STAT antibodies indicated, followed by binding to 32P-labeled IL-2RGAS-c/GAS-n probe. (C) STAT DNA binding to the IL-2RGAS-c/GAS-n in response to IL-12 and IL-15. T cells were stimulated with IL-2 or IL-15 for 30 minutes, and nuclear extracts were prepared and incubated for 1 hour on ice with different anti-STAT antibodies followed by binding to 32P-labeled IL-2RGAS-c/GAS-n probe. The results are representative of three separate experiments.
IFN-α induces STAT1, STAT3, and STAT4 DNA binding toIRF-1 and pim-1 GAS elements.
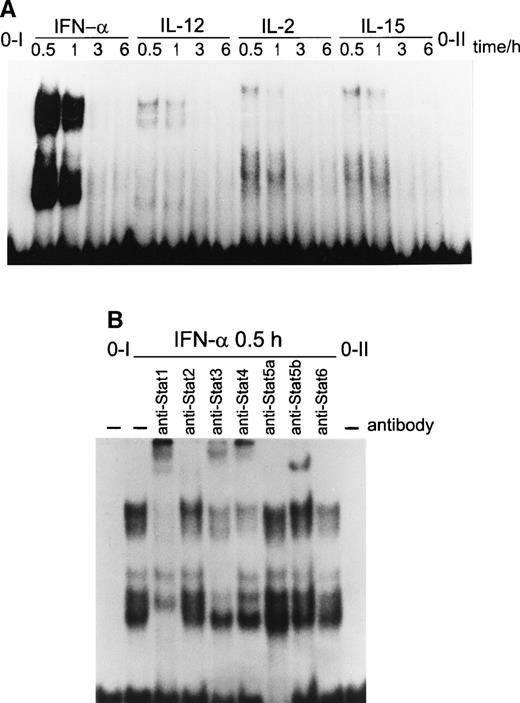
Analysis of pim-1 gene promoter has shown a functional GAS element that binds STAT1 homodimer in response to IFN-γ stimulation.36 We studied STAT binding to pim-1 GAS in T lymphocytes in response to cytokine stimulation. EMSA indicated that several STAT complexes were able to bind to pim-1 GAS element (−934 to −905) in response to IFN-α stimulation (Fig 5A). These complexes were detectable at 0.5 hours after IFN-α treatment and disappeared within 3 hours. IL-12 induced two different DNA binding complexes when pim-1GAS was used as a probe (Fig 5A). Both IL-2 and IL-15 induced three different DNA binding complexes. Supershift experiments with specific anti-STAT antibodies indicated that IFN-α–induced complexes consisted of STAT1, STAT3, and STAT4 (Fig 5B). The lowermost complex induced by IFN-α supershifted with anti-STAT1 antibody only. In addition, complexes that supershifted with anti-STAT1 and/or anti-STAT3 antibodies suggested that STAT1-STAT3 heterodimers and STAT3 homodimers were formed after IFN-α stimulation (Fig 5B). The mobility of IL-12–induced complex was comparable to the uppermost complex seen in IFN-α–treated T cells. These complexes supershifted with anti-STAT4 antibodies, suggesting that IFN-α is also able to induce STAT4 DNA binding. In addition, the uppermost complex was also supershifted with anti-STAT1 antibody (Fig 5B), suggesting that heterodimers consisting of STAT1 and STAT4 are formed in response to IFN-α stimulation.
(A) Multiple STAT complexes bind to the pim-1 GAS element in response to cytokine stimulation. T cells were stimulated with IFN-, IL-2, IL-12, or IL-15 for the indicated times, and nuclear extracts were prepared from the cells. The extracts were incubated with 32P-labeled pim-1 GAS probe, and the STAT DNA binding was analyzed by EMSA. (B) IFN- induced STAT1, STAT3, and STAT4 DNA binding to pim-1 GAS. T cells were stimulated with IFN- for 30 minutes, and nuclear extracts were prepared. The extracts were incubated for 1 hour on ice with anti-STAT antibodies, followed by binding to 32P-labeledpim-1 GAS probe. Comparable data were obtained in three independent experiments, each consisting of pooled T cells from two different donors.
(A) Multiple STAT complexes bind to the pim-1 GAS element in response to cytokine stimulation. T cells were stimulated with IFN-, IL-2, IL-12, or IL-15 for the indicated times, and nuclear extracts were prepared from the cells. The extracts were incubated with 32P-labeled pim-1 GAS probe, and the STAT DNA binding was analyzed by EMSA. (B) IFN- induced STAT1, STAT3, and STAT4 DNA binding to pim-1 GAS. T cells were stimulated with IFN- for 30 minutes, and nuclear extracts were prepared. The extracts were incubated for 1 hour on ice with anti-STAT antibodies, followed by binding to 32P-labeledpim-1 GAS probe. Comparable data were obtained in three independent experiments, each consisting of pooled T cells from two different donors.
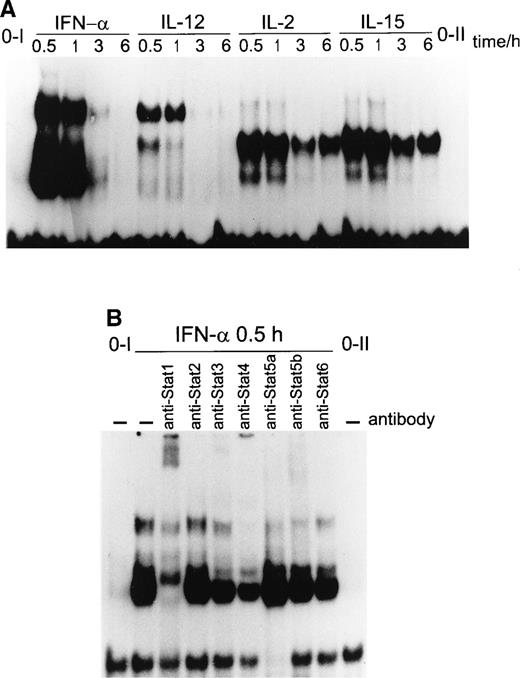
Because IRF-1 GAS element binds STATs 1, 3, 4, 5, and 6, albeit with different strengths,33 we wanted to analyze the T-cell–specific STAT binding to this element. Again, IFN-α induced the DNA binding of multiple complexes (Fig6A). IFN-α induced the DNA binding of STAT1, STAT3, and STAT4 toIRF-1 GAS (Fig 6B). IFN-α–induced STAT3 DNA binding toIRF-1 GAS was very weak compared with STAT1 or STAT4 DNA binding, probably due to the low affinity of STAT3 to IRF-1 GAS. IL-12 induced two complexes, of which the upper complex supershifted with anti-STAT4 antibody only (data not shown). Both IL-2 and IL-15 stimulation resulted in the formation of only one major DNA binding complex that was detectable at least up to 6 hours (Fig 6A). This complex was supershifted with anti-STAT5 antibody (data not shown).
(A) STAT binding to the IRF-1 GAS by IFN-, IL-2, IL-12, or IL-15. T cells were stimulated with IFN-, IL-2, IL-12, or IL-15 for the times indicated, and nuclear extracts were prepared. The extracts were incubated with 32P-labeledIRF-1 GAS probe, and the DNA binding activity was analyzed by EMSA. (B) IFN-–induced STAT1, STAT3, and STAT4 DNA binding toIRF-1 GAS. T cells were stimulated with IFN- for 30 minutes, and nuclear extracts were prepared and incubated for 1 hour on ice with anti-STAT antibodies, followed by binding to 32P-labeledIRF-1 GAS. The experiment was repeated three times with similar results.
(A) STAT binding to the IRF-1 GAS by IFN-, IL-2, IL-12, or IL-15. T cells were stimulated with IFN-, IL-2, IL-12, or IL-15 for the times indicated, and nuclear extracts were prepared. The extracts were incubated with 32P-labeledIRF-1 GAS probe, and the DNA binding activity was analyzed by EMSA. (B) IFN-–induced STAT1, STAT3, and STAT4 DNA binding toIRF-1 GAS. T cells were stimulated with IFN- for 30 minutes, and nuclear extracts were prepared and incubated for 1 hour on ice with anti-STAT antibodies, followed by binding to 32P-labeledIRF-1 GAS. The experiment was repeated three times with similar results.
IFN-α enhances STAT1, STAT3, STAT4, STAT5a, and STAT5b tyrosine phosphorylation in anti-CD3–activated human T lymphocytes.
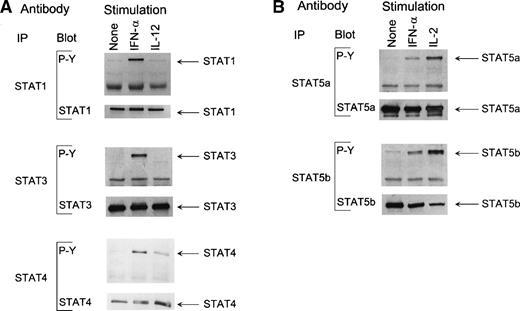
Because IFN-α induced STAT1, STAT3, and STAT4 DNA binding to GAS elements, we also analyzed the tyrosine phosphorylation of these proteins. T cells were stimulated with IFN-α or IL-12, STATs were immunoprecipitated with specific antibodies, and their tyrosine phosphorylation was analyzed by antiphosphotyrosine immunoblotting. In unstimulated cells, STAT1 and STAT3 were weakly phosphorylated on tyrosine residues, but IFN-α strongly enhanced STAT1 and STAT3 tyrosine phosphorylation (Fig 7A). Both IFN-α and IL-12 strongly enhanced STAT4 tyrosine phosphorylation (Fig7A). The blots were stripped of detecting antibody and reprobed for STAT1, STAT3, and STAT4 to confirm equal loading of each lane (lower panel). Because it has previously been shown that IFN-α activates STAT5a in promonocytic U937 and STAT5b in HeLa cells,37 we wanted to study whether IFN-α is able to induce tyrosine phosphorylation of STAT5 also in human T lymphocytes. T cells were stimulated with IFN-α or IL-2, and STAT5a and STAT5b immunoprecipitates were analyzed by antiphosphotyrosine immunoblotting. IFN-α enhanced tyrosine phosphorylation of both STAT5a and STAT5b (Fig 7B). IL-2 was a much more potent in inducing STAT5a and STAT5b tyrosine phosphorylation than IFN-α. Again, the blots were reblotted with STAT5a and STAT5b antibodies to determine equal loading (lower panel).
(A) IFN- induces tyrosine phosphorylation of STAT1 and STAT4 in human T lymphocytes. T cells were treated with IFN- or IL-12 for 15 minutes, and T-cell lysates were prepared and immunoprecipitated with anti-STAT1, anti-STAT3, or anti-STAT4 antibodies. Proteins were separated on 10% SDS-PAGE, transferred to membranes, and immunoblotted with antiphosphotyrosine antibody. The membranes were stripped and reblotted with anti-STAT antibodies. (B) IFN- induces tyrosine phosphorylation of STAT5a and STAT5b. T cells were treated with IFN- or IL-2 for 15 minutes and T-cell lysates were prepared. The lysates were immunoprecipitated with anti-STAT5a or anti-STAT5b antibodies, followed by immunoblotting with antiphosphotyrosine antibody. Membranes were then stripped and reblotted with anti-STAT antibodies, as indicated.
(A) IFN- induces tyrosine phosphorylation of STAT1 and STAT4 in human T lymphocytes. T cells were treated with IFN- or IL-12 for 15 minutes, and T-cell lysates were prepared and immunoprecipitated with anti-STAT1, anti-STAT3, or anti-STAT4 antibodies. Proteins were separated on 10% SDS-PAGE, transferred to membranes, and immunoblotted with antiphosphotyrosine antibody. The membranes were stripped and reblotted with anti-STAT antibodies. (B) IFN- induces tyrosine phosphorylation of STAT5a and STAT5b. T cells were treated with IFN- or IL-2 for 15 minutes and T-cell lysates were prepared. The lysates were immunoprecipitated with anti-STAT5a or anti-STAT5b antibodies, followed by immunoblotting with antiphosphotyrosine antibody. Membranes were then stripped and reblotted with anti-STAT antibodies, as indicated.
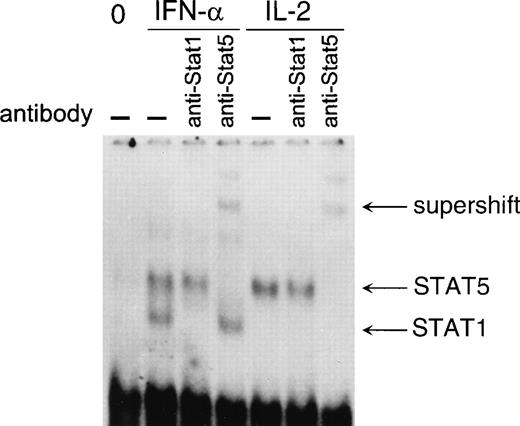
Although we observed IFN-α–induced tyrosine phosphorylation of STAT5a and STAT5b, we were not able to detect any clear STAT5 DNA binding to IL-2Rα, pim-1, or IRF-1 GAS elements. To further analyze whether IFN-α is able to activate STAT5 DNA binding in T cells, we performed EMSA with IFP53 GAS element that is known to bind STAT5 with high affinity.38 T cells were stimulated with IFN-α and IL-2 for 0.5 hours, and nuclear extracts were prepared and analyzed in EMSA. IFN-α induced both STAT1 and STAT5 DNA binding to IFP53 GAS, whereas IL-2 was able to induce the DNA binding of STAT5 (Fig 8).
IFN-–induced STAT5 DNA binding to the IFP53GAS element. T cells were stimulated with IFN- or IL-2 for 30 minutes, and nuclear extracts were prepared and incubated with anti-STAT1 or anti-STAT5 antibodies for 1 hour on ice, followed by EMSA with 32P-labeled IFP53 GAS probe.
IFN-–induced STAT5 DNA binding to the IFP53GAS element. T cells were stimulated with IFN- or IL-2 for 30 minutes, and nuclear extracts were prepared and incubated with anti-STAT1 or anti-STAT5 antibodies for 1 hour on ice, followed by EMSA with 32P-labeled IFP53 GAS probe.
IFN-α priming enhances IL-2–induced STAT1 and STAT5 DNA binding.
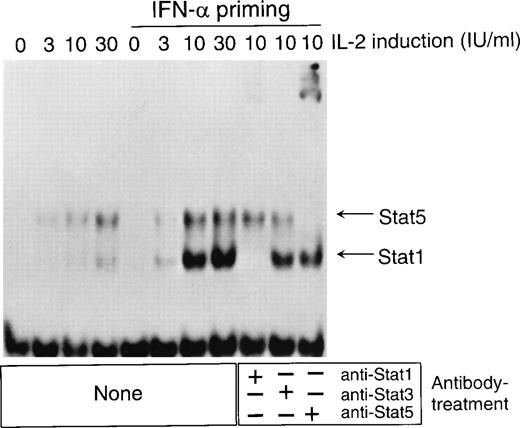
Because IFN-α priming enhanced IL-2–induced T-cell proliferation (Fig 3), we studied whether IFN-α has any effect on IL-2–induced STAT DNA binding. T cells were left untreated or treated with IFN-α for 24 hours, followed by stimulation with different concentrations of IL-2 (0, 3, 10, or 30 IU/mL) for 30 minutes. In unprimed T cells, IL-2 induced one major and one minor IRF-1 GAS binding complex. In IFN-α–primed, IL-2–stimulated cells, IRF-1 GAS DNA binding was markedly enhanced. The upper complex supershifted with anti-STAT5 antibody and the lower complex with anti-STAT1 antibody (Fig 9). The data suggest that in T cells IL-2 can activate STAT1 and that the IFN-α pretreatment strongly enhances IL-2–induced STAT1 (and STAT5) DNA binding (Fig 9). Enhanced STAT1 and STAT5 complex formation in IFN-α–primed T cells in response to IL-2 stimulation was also detected when pim-1 GAS was used as a probe in EMSA (data not shown). In contrast, IFN-α did not enhance IL-2–induced STAT5 DNA binding to IL-2Rα GAS-c/GAS-n (data not shown).
IFN- enhances IL-2–induced STAT1 and STAT5 binding to the IRF-1 GAS. T cells were left untreated or treated with IFN- (100 IU/mL) for 24 hours. The cells were washed and resuspended in fresh medium, and different concentrations of IL-2 (0, 3, 10, or 30 IU/mL) were added. After 30 minutes of incubation, nuclear extracts were prepared and analyzed in EMSA with IRF-1 GAS probe. The experiment was repeated three times with similar results.
IFN- enhances IL-2–induced STAT1 and STAT5 binding to the IRF-1 GAS. T cells were left untreated or treated with IFN- (100 IU/mL) for 24 hours. The cells were washed and resuspended in fresh medium, and different concentrations of IL-2 (0, 3, 10, or 30 IU/mL) were added. After 30 minutes of incubation, nuclear extracts were prepared and analyzed in EMSA with IRF-1 GAS probe. The experiment was repeated three times with similar results.
DISCUSSION
The gene knock-out studies have shown that a functional IFN-α/β system is essential for the clearance of viral infection.4,39,40 It is also well accepted that IFN-α can directly inhibit proliferation of normal and tumor cells in vitro and in vivo.41 However, there is some evidence that IFN-α would enhance T-cell growth. Tough et al32 have shown that IFN-α–stimulated CD8+ cells proliferate and survive in vivo for longer periods of time than their unstimulated counterparts. In the present study, we demonstrate that IFN-α priming can augment IL-2–induced human T-cell proliferation in vitro, and this phenomenon was associated with the induction of IL-2Rα, c-myc, andpim-1 gene expression. Especially, IFN-α–induced upregulation of IL-2Rα expression could directly contribute to enhanced responsiveness of T cells to IL-2. Enhanced pim-1gene expression may also contribute to increased T-cell survival. There is evidence that overexpression of pim-1 can accelerate lymphoproliferation by inhibiting apoptosis.24,25 Recent results from studies on cytokine-mediated signaling suggest that pim-1 expression correlates with the viability, but not with the proliferation potential of the cells.42 43
Not only IFN-α and IL-2, but also IL-12 and IL-15 as well induced the expression of the IL-2Rα gene in anti-CD3–activated T lymphocytes. Induction of IL-2Rα gene expression by IL-15 was expected, because IL-15 also activates STAT5 and uses β and γ chains of the IL-2 receptor during downstream signaling.11However, the finding that IFN-α and IL-12 induced the upregulation of IL-2Rα gene expression was more surprising. An IL-2–responsive element in the IL-2Rα gene promoter has recently been characterized.34,35 This element binds at least STAT5, Elf-1, HMG-I(Y), and GATA family transcription factors. The STAT5 binding site consists of two adjacent GAS elements, a consensus GAS (GAS-c) and a nonconsensus one (GAS-n), which are well-conserved in human and mouse promoters. Single GAS elements of IL-2Rαpromoter appear to function only as low-affinity STAT5 binding sites and therefore both of these elements are required for IL-2–induced STAT5 binding.44 Previously, cooperative binding of STAT complexes to weak STAT binding sites has been observed, eg, in the first intron of IFN-γ45 gene and in the promoter of mig chemokine gene.46 In our experiments, by using nuclear extracts from cytokine-stimulated T cells, we were able to detect not only IL-2– and IL-15–induced STAT5 binding, but also IL-12–induced STAT4 binding to the IL-2RαGAS-c/GAS-n element. More interestingly, the IFN-α–induced complexes consisted of STAT1, STAT3, and STAT4 proteins, suggesting similar cooperative STAT binding mechanisms as described for the other genes mentioned above.45 46
The expression of c-myc and pim-1 genes was induced in human T cells by IL-2, which is the primary growth factor of mature T cells. We observed that both IFN-α and IL-12 were able to upregulate the expression of c-myc and pim-1 genes in anti-CD3–activated T lymphocytes. pim-1 gene promoter contains a functional GAS element.36 It is also presumable that IFN-α– and IL-12–activated STAT proteins are involved in the upregulation of pim-1 gene expression. Indeed, we were able to demonstrate that IFN-α induced STAT1, STAT3, and STAT4 DNA binding topim-1 GAS. Similarly, IL-12 induced STAT4 binding topim-1 GAS. In addition, it has been shown that IL-12 upregulates c-myc gene expression in NK and T cells.47 It is also known that Jak3 tyrosine kinase is essential for IL-2–induced expression of c-mycgene,48 but there is little information about downstream signal transducing molecules and no evidence of STAT protein involvement. Further analyses of the molecular mechanisms of IFN-α–, IL-2–, and IL-12–induced c-myc gene expression are clearly needed.
Previously, IFN-α was observed to be able to specifically activate IFN-stimulated gene factor complex (STAT1, STAT2, and p48 heterotrimer).49 Our results suggest that IFN-α can also induce the DNA binding of STAT3, STAT4, and STAT5 to the specific GAS elements in human T lymphocytes. IFN-α–induced STAT3 activation takes place in certain cell types,50,51 and this has been suggested to be due to a direct phosphotyrosine-dependent interaction between IFNAR-1 and the SH2 domain of STAT3.52 Whether similar interactions between STAT4 or STAT5 with the type I IFN receptor exist remains to be studied. Previously, it was demonstrated that both IL-12 and IFN-α were able to activate DNA binding of STAT4 to FcγRI GAS in T lymphocytes.53 In the present study, we clearly demonstrate that in our T-cell system both IFN-α and IL-12 can induce STAT4 tyrosine phosphorylation and subsequent binding of STAT4 to IRF-1 and pim-1 GAS elements. This is consistent with the Northern blot data that demonstrated thatpim-1 (Fig 1) as well as IRF-1 (data not shown) genes were upregulated by IFN-α and IL-12.
We also observed that IFN-α enhanced tyrosine phosphorylation of STAT5a and STAT5b in T cells. It was previously shown that IFN-α selectively enhances tyrosine phosphorylation and DNA binding of STAT5a and STAT5b in U937 cells and in HeLa cells, respectively, although both cell types are able to express STAT5a and STAT5b.37 This suggests that cell-type–specific mechanisms regulate IFN-α–induced activation of different STAT5 isoforms. Our results show that, in human T cells, IFN-α induces the tyrosine phosphorylation of both STAT5a and STAT5b. IFN-α–induced STAT5a and STAT5b tyrosine phosphorylation was much weaker compared with IL-2 induction, and no clear IFN-α–induced STAT5 DNA binding to IRF-1, pim-1, orIL-2Rα GAS elements was detected. However, IFN-α induced STAT5 DNA binding to IFP53 GAS element, which is known to bind STAT5 with high affinity.38 This may suggest that IFN-α–induced STAT5 activation has physiological significance in the activation of genes that contain high-affinity STAT5 binding sites.
Because IFN-α was able to enhance IL-2Rα gene expression and T-cell proliferation, we studied the role of IFN-α in IL-2–induced STAT activation. Pretreatment of T cells with IFN-α resulted in a twofold to threefold increase in IL-2–induced STAT5 DNA binding, which may contribute to the enhanced T-cell proliferation. In IFN-α–pretreated T cells, IL-2–induced STAT1 DNA binding was strongly enhanced (Fig 9). This could be due to the upregulation of intracellular STAT1 levels or, alternatively, to enhanced IL-2–induced signaling in IFN-α pretreated cells. The former possibility may be the more likely one, because IFN-α strongly enhances STAT1, STAT2, and p48 mRNA and protein expression in peripheral blood mononuclear cells and in human macrophages54 as well as in T lymphocytes (data not shown). However, STAT1 was expressed in relatively high levels in T cells and it is possible that other mechanisms, such as STAT1 tyrosine or serine/threonine phosphorylation by IFN-α, may also explain enhanced IL-2–induced STAT1 DNA binding in IFN-α–primed T cells.
IFN-α was first described as a substance inhibiting virus growth in infected cells.55 Thereafter, much of the research work focused on the antiviral and antiproliferative properties of IFN-α, and its role as an important T-cell regulatory molecule was neglected.2 In the present study, we provide evidence that IFN-α/β is able to activate the expression of IL-2Rα, c-myc,and pim-1 genes that are involved in T-cell proliferation or survival. Pretreatment of activated T cells with IFN-α also enhanced IL-2–induced T-cell proliferation, suggesting that, in T cells, IFN-α is rather growth stimulatory than inhibitory. In addition, in T cells, IFN-α was able to activate STAT4 and STAT5, the signaling molecules used by IL-12 and IL-2, respectively, which further emphasizes the role of IFN-α/β as important molecules in T-cell biology.
ACKNOWLEDGMENT
The authors are grateful to Dr Hannele Tölö for the IFN-α and Dr Matti Kaartinen for the anti-CD3 antibodies and to Marika Yliselä, Mari Tapaninen, and Valma Mäkinen for their expert technical assistance.
Supported by the Medical Research Council of the Academy of Finland, the Sigrid Juselius Foundation, the Technology Development Center of Finland, the Jenny and Antti Wihuri Foundation, and the Finnish Cancer Organizations.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Sampsa Matikainen, PhD, Department of Virology, National Public Health Institute, Mannerheimintie 166, FIN-00300 Helsinki, Finland; e-mail: sampsa.matikainen@ktl.fi.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal