Abstract
The 2β1 integrin, a collagen receptor on platelets and megakaryocytes, is required for normal platelet function. Transcriptional regulation of the 2 integrin gene in cells undergoing megakaryocytic differentiation requires a core promoter between bp −30 and −92, a silencer between bp −92 and −351, and megakaryocytic enhancers in the distal 5′ flank. We have now identified a 229-bp region of the distal 5′ flank of the 2 integrin gene required for high-level enhancer activity in cells with megakaryocytic features. Two tandem AP1 binding sites with dyad symmetry are required for enhancer activity and for DNA-protein complex formation with members of the c-fos/c-jun family. The requirement for AP1 activation suggested a role for the mitogen-activated protein kinase (MAPK) signaling pathway in regulating 2 integrin gene expression. Inhibition of the MAP kinase cascade with PD98059, a specific inhibitor of MAPK kinase 1, prevented the expression of the 2 integrin subunit in cells induced to become megakaryocytic. We provide a model of megakaryocytic differentiation in which expression of the 2 integrin gene requires signaling via the MAP kinase pathway to activate two tandem AP1 binding sites in the 2 integrin enhancer.
DISTINCT MEMBERS OF the integrin family of cell adhesion receptors are expressed at different times during hematopoietic differentiation. The expression of some integrin receptors, such as the α5β1 integrin on erythroid cells, is required for differentiation; the expression of other receptors, such as α2β1 and αIIbβ3 on megakaryocytes and platelets, is required for normal function.1-3 The α2β1 integrin mediates platelet adhesion to collagen in vessel walls and is essential for the formation of a normal platelet plug.3 Lack of α2β1integrin expression by platelets, either due to congenital abnormalities or the development of myelodysplastic/myeloproliferative disorders, is associated with bleeding.4-9 The regulation of integrin gene expression during hematopoietic differentiation, specifically megakaryocytic differentiation, has been a major focus of our laboratory.
Expression of the α2β1 integrin is exquisitely regulated by distinctly different transcriptional regulatory elements in epithelial cells, fibroblasts, megakaryocytes, and platelets.10-15 To evaluate the molecular mechanisms by which differentiation-dependent expression of the α2β1 integrin is regulated during megakaryocytic differentiation, we have used several models of megakaryocytic differentiation including K562 cells, a pluripotent hematopoietic cell line, and DAMI cells, a megakaryocytic cell line.10,14 K562 cells cultured in the presence of 40 nmol/L phorbol dibutyrate (PDB) acquired megakaryocytic features, including the expression of the platelet/megakaryocyte integrins αIIbβ3 and α2β1. The increased α2β1 integrin expression was a result of increased steady-state levels of α2 mRNA due to transcriptional activation of the α2 integrin gene.11 Characterization of the 5′ flanking region of the α2 integrin gene showed three distinct regions of the 5′ flank that are responsible for the expression of the α2 integrin gene.13,14,16 Regulation of the α2 integrin gene in both the K562 and DAMI cell models undergoing megakaryocyte differentiation requires a core promoter between bp −30 and −92, a silencer between bp −92 and −351, and additional megakaryocytic enhancers within the distal 5′ flank.14 16
Our earlier studies localized the strong megakaryocytic enhancer to the region between bp −1426 and −2592.14 We now verify that the 1,166-bp region serves as an enhancer in the irrelevant promoter construct SV40pCAT in both the K562 and DAMI models of megakaryocytic differentiation. Enhancer activity requires two tandem AP1 binding sites with dyad symmetry. The short enhancer fragment containing the tandem AP1 consensus sites binds members of the c-fos and c-jun family. Inhibition of the mitogen activation protein kinase (MAPK) cascade prevents the upregulation of α2β1 integrin expression induced by PDB. We present a model in which activation of the MAPK cascade signals via c-fos/c-jun family members to activate α2 integrin gene expression.
MATERIALS AND METHODS
Cell cultures and transfection assays.
The K562 cell line obtained from the American Type Culture Collection (Rockville, MD) and the DAMI cell line obtained from Dr Robert I. Handin (Harvard Medical School, Boston, MA) were propagated in RPMI 1640 medium. Megakaryocytic differentiation was induced by the addition of 40 nmol/L PDB in dimethyl sulfoxide, as previously described.10 In experiments using the MAPK kinase 1 inhibitor, PD98059, cells were treated with PD98059 (50 μmol/L) in dimethyl sulfoxide for 15 minutes before the addition of PDB.
The K562 cell line was transfected by electroporation using a BTX Electro Cell Manipulator 600 (BTX Inc, San Diego, CA).17Approximately 1.0 × 107 cells were transfected in RPMI medium containing 100 μg/mL salmon sperm DNA, 30 μg of plasmid DNA, and 3 μg of RSV-luciferase DNA by electroporation at 275 V and 600 μF. Cell extracts were harvested after 48 hours. Luciferase activity was analyzed using a Monolight 2010 luminometer (Analytical Luminescence Laboratory, San Diego, CA), as described previously,18 and was used to normalize for transfection efficiency. Cell extracts containing identical amounts of luciferase activity were then assayed for chloramphenicol acetyl transferase (CAT) activity using the standard method of Gorman et al.19Acetylation of chloramphenicol was quantified by thin-layer chromatography.
Gel mobility shift analysis.
Nuclear extracts were obtained from uninduced and phorbol-induced K562 cells by isolating nuclei, as previously described.20 The cells were washed with phosphate-buffered saline (PBS) and lysed in 0.5% Triton X-100 in 10 mL of STKM buffer (30% sucrose, 40 mmol/L Tris, pH 7.5, 37 mmol/L KCI, and 12 mmol/L MgC12). After pelleting, the nuclei were washed in TNM buffer (100 mmol/L Tris, pH 7.5, 10 mmol/L NaCl, and 30 mmol/L MgCl2). The proteins were extracted from the nuclei using a buffer containing 20 mmol/L HEPES, 20% glycerol, 0.35 mol/L KCl, 0.1 mmol/L EGTA, 0.5 mmol/L EDTA, 10 μg/mL aprotinin, and 10 μg/mL leupeptin. After incubation with gentle rotation for 1 hour at 4°C, the sample was centrifuged at 100,000g for 2 hours. The supernatant was collected and the concentration of nuclear proteins in the extracts was determined by Bradford assay (Bio-Rad Laboratory, Hercules, CA). The gel-purified double-stranded 75-bp DNA fragment extending from bp −1503 to −1578 or the double-stranded oligonucleotides were end-labeled with 32P-dCTP using Klenow DNA polymerase. Oligonucleotides containing a single AP1 consensus element (5′-CGCTTGATGACTCAGCCGGAA-3′), a consensus element with a 2-bp mutation of the AP1 site (AP1m; 5′-CGCTTGATGACTTGGCCGGAA-3′), a single Sp1 consensus element (5′-ATT CGATCGGGGCGGGGCGAGC-3′), or an early growth response (Egr) consensus element (5′-GGATCCAGCGGGGGCGAGCGGGGGCGA-3′) were obtained from Santa Cruz Biotech, Inc (Santa Cruz, CA). Approximately 1 to 2 μg of nuclear protein extract was added to 20,000 counts per minute of labeled DNA in 15 μL of binding buffer (10% glycerol, 25 mmol/L Tris HCl, pH 7.5, 100 mmol/L KCI, 5 mmol/L spermidine, 5 mmol/L EDTA, 1 mmol/L dithiothreitol, and 2 mmol/L phenylmethylsulfonyl fluoride) and incubated on ice for 15 minutes. Some incubations contained an excess of unlabeled competitor DNA fragment, or oligonucleotide, as indicated. In antibody inhibition experiments, the indicated amount of anti–c-fos or anti–c-jun antibody (Santa Cruz Biotech, Inc) was preincubated with the nuclear extract for 18 hours at 4°C before the addition of32P-labeled probe. The reactions were then analyzed by polyacrylamide gel electrophoresis using 6% acrylamide/Bis (19:1) in Tris, pH 8.8.
α2-CAT fusion constructs.
The pα2 2592-CAT and pα2 1426-CAT constructs were generated by restriction enzyme digestion of the original pα2 5000-CAT construct at the restriction enzyme sites Stu I (−2592) and Sac I (−1426), respectively, as described earlier.14 The StuI/Sac I fragment was then inserted into the blunted SalI site within the multiple cloning site of the SV40pCAT plasmid (Promega, Madison, WI). Two SV40pCAT constructs containing the bp −1426 to −2592 region in opposite orientations were selected by restriction enzyme and sequence analyses. A series of deletion mutants of the 1166 bp (bp −1426 to −2592) were generated by restriction enzyme digestion of the −1426/−2592 SV40pCAT plasmid at the restriction sitesXmn (−1842), Nsi (−1655), MnlI(−1578) and Mva I (−1503). The −1426/−1655 SV40pCAT plasmid was used as a template for polymerase chain reaction (PCR) to generate the site-specific mutations that disrupt the AP2, GATA, or both binding sites in the construct. The −1503/−1578 SV40pCAT construct was used as a template for PCR-directed mutagenesis to generate the constructs containing site-specific mutations that disrupt the two AP1 binding sites, including 5′ AP1m −1503/−1578 SV40pCAT, 3′ AP1m −1503/−1578 SV40pCAT, and Dbl AP1m −1503/−1578 SV40pCAT. The following 5′ primers were used for the GATA and AP2 mutant constructs: GATAm (5′-GTT ATT TCC CCC CAC CCC CAG ATT TAA AAC AC-3′), AP2m (5′-GTT ATT TCC CCC TTT CCC CAG AGA TAA AAC AC-3′), or the AP2-GATAm (5′-GTT ATT TCC CCC TTT CCC CAG ATT TAA AAC AC-3′). The following primers were used for the AP1 mutant constructs: 5′AP1m −1503/−1578 SV40pCAT (5′-GAA ATG TAT GGG AGT GTT TGG-3′), 3′ AP1m −1503/−1578 SV40pCAT (5′-CCT TC TAT CGG ACT GAA ATG-3′), and Dbl AP1m −1503/−1578 SV40pCAT (5′-CCA AAC ACT CCC ATA CAT TTC AGT CCG ATA GAA-3′). A common 3′ primer from the M13 vector sequence was used in all reactions. All constructs were completely sequenced by the dideoxynucleotide chain termination method of Sanger et al21 to insure that the site-specific mutations were incorporated with no error. Mutations in the full-length CAT construct pα2 2592-CAT were produced by site-directed mutagenesis.22
Flow cytometric analysis.
Flow cytometric analysis was performed on uninduced K562 cells, K562 cells induced for 6 days with PDB, or K562 cells incubated with PD98059 for 15 minutes before induction with PDB. Single cells (1 × 106) in PBS with 1.5% horse serum were incubated with the monoclonal anti-α2 antibody (P1E6) at a concentration of 2 μg/mL for 45 minutes at 4°C. Cells were washed three times and incubated with 2.5 μg/mL of a secondary goat antimouse coupled to fluorescein (Tago, Inc, Burlingame, CA) for 45 minutes at 4°C, washed twice, and resuspended in PBS. Fluorescein-labeled cells were analyzed using a FACScan instrument (Becton Dickinson, Mountain View, CA).
RESULTS
A 229-bp region of the distal 5′ flank functions as an α2 integrin subunit enhancer.
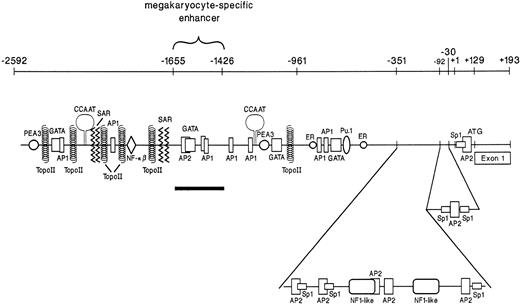
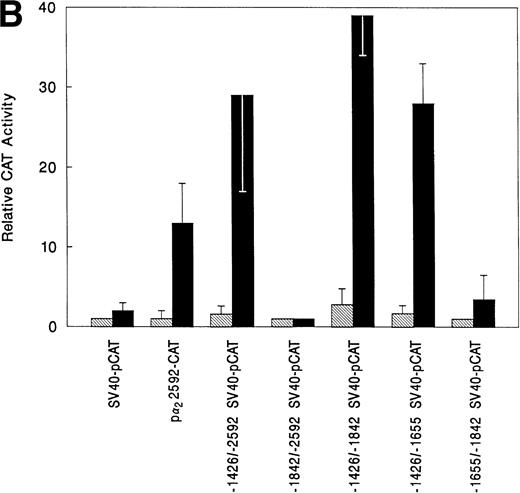
Three distinct regions of the 5′ flank of the α2integrin gene were required for high-level gene expression in cells with megakaryocytic features (Fig1).14 To verify that the region between bp −1426 and −2592 served enhancer function in cells with megakaryocytic features, the 1,166-bp fragment was placed in the irrelevant promoter construct SV40pCAT in both 5′ to 3′ and 3′ to 5′ orientations (Fig 2A). Both the 5′ to 3′ and 3′ to 5′ −1426/−2592 SV40pCAT constructs, independent of orientation, directed high-level, reporter gene activity in K562 cells induced with phorbol dibutyrate to undergo megakaryocytic differentiation (Fig 2B and data not shown). The −1426/−2592 SV40pCAT constructs directed only low-level CAT activity in uninduced K562 cells. The enhancer activity of the 1,166-bp region in the irrelevant SV40pCAT construct responded to induction in a manner similar to the activity of the original intact promoter/enhancer construct containing the entire 5′ flank of the α2integrin gene, as shown in Fig 2A and B. The orientation-independent activity of the region between bp −1426 and −2592 in an irrelevant promoter construct established its role as an enhancer.
A schematic diagram of the distal 5′ flank of the 2 integrin gene. Identification of numerous potential binding sites for ubiquitous as well as megakaryocyte-specific transcription factors are shown. Three distinct regions are responsible for 2 integrin gene expression in cells with megakaryocytic origin. These include a core promoter between bp −30 and −92, a silencer/repressor between bp −92 and −351, and megakaryocyte-specific enhancers in the distal 5′ flank between bp −1426 and −1655. The megakaryocyte-specific enhancer is underlined and bracketed. Data from Zutter et al.14
A schematic diagram of the distal 5′ flank of the 2 integrin gene. Identification of numerous potential binding sites for ubiquitous as well as megakaryocyte-specific transcription factors are shown. Three distinct regions are responsible for 2 integrin gene expression in cells with megakaryocytic origin. These include a core promoter between bp −30 and −92, a silencer/repressor between bp −92 and −351, and megakaryocyte-specific enhancers in the distal 5′ flank between bp −1426 and −1655. The megakaryocyte-specific enhancer is underlined and bracketed. Data from Zutter et al.14
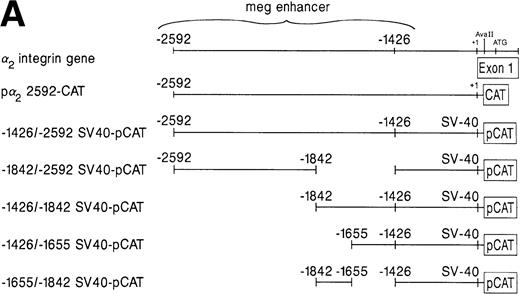
A schematic diagram of the 2 promoter-CAT construct and the megakaryocytic enhancer in the irrelevant promoter construct SV40pCAT. (A) The construct p22592-CAT contains the entire 5′ flanking region from bp −2592 to +109 of the 5′ untranslated region of the 2 integrin gene upstream to the CAT structural gene. The construct −1426/−2592 SV40pCAT consists of the megakaryocyte enhancer region extending from bp −1426 to −2592 in the irrelevant promoter construct SV40pCAT. Constructs −1842/−2592 SV40pCAT, −1426/−1842 SV40pCAT, −1426/−1655 SV40pCAT, and −1426/−1542 SV40pCAT were derived from the construct −1426/−2592 SV40pCAT. (B) The 1,166-bp region of the distal 5′ flank serves as a megakaryocytic enhancer. The constructs p22592-CAT, −1426/−2592 SV40pCAT, −1842/−2592 SV40pCAT, −1426/−1842 SV40pCAT, −1426/−1655 SV40pCAT, and −1655/−1842 SV40pCAT were transfected in parallel with the SV40pCAT plasmid that contains the strong viral promoter SV40 without an enhancer into either uninduced (⊠) or induced (▪) K562 cells. Cotransfection with RSV-luciferase was used to control for transfection efficiency. After 48 hours of incubation, cell extracts were assayed. After normalization for transfection efficiency, CAT activity of the constructs was determined using thin-layer chromatography and differential extraction. The mean and standard deviation of CAT activity of the mutated constructs in uninduced or induced K562 cells from three separate experiments was determined relative to the activity of SV40pCAT in uninduced K562 cells, which was assigned a value of 1.0.
A schematic diagram of the 2 promoter-CAT construct and the megakaryocytic enhancer in the irrelevant promoter construct SV40pCAT. (A) The construct p22592-CAT contains the entire 5′ flanking region from bp −2592 to +109 of the 5′ untranslated region of the 2 integrin gene upstream to the CAT structural gene. The construct −1426/−2592 SV40pCAT consists of the megakaryocyte enhancer region extending from bp −1426 to −2592 in the irrelevant promoter construct SV40pCAT. Constructs −1842/−2592 SV40pCAT, −1426/−1842 SV40pCAT, −1426/−1655 SV40pCAT, and −1426/−1542 SV40pCAT were derived from the construct −1426/−2592 SV40pCAT. (B) The 1,166-bp region of the distal 5′ flank serves as a megakaryocytic enhancer. The constructs p22592-CAT, −1426/−2592 SV40pCAT, −1842/−2592 SV40pCAT, −1426/−1842 SV40pCAT, −1426/−1655 SV40pCAT, and −1655/−1842 SV40pCAT were transfected in parallel with the SV40pCAT plasmid that contains the strong viral promoter SV40 without an enhancer into either uninduced (⊠) or induced (▪) K562 cells. Cotransfection with RSV-luciferase was used to control for transfection efficiency. After 48 hours of incubation, cell extracts were assayed. After normalization for transfection efficiency, CAT activity of the constructs was determined using thin-layer chromatography and differential extraction. The mean and standard deviation of CAT activity of the mutated constructs in uninduced or induced K562 cells from three separate experiments was determined relative to the activity of SV40pCAT in uninduced K562 cells, which was assigned a value of 1.0.
To determine which elements within the 1,166-bp region were required for enhancer activity in cells undergoing megakaryocytic differentiation, a series of deletion mutants of the −1426/−2592 SV40pCAT construct were constructed as shown in Fig 2A. This series of deletion mutants localized strong enhancer activity to the region between bp −1426 and −1655 relative to the activity of SV40pCAT in uninduced K562 cells (Fig 2B). The construct −1426/−1655 SV40pCAT exhibited enhancer activity comparable to that of the longer enhancer constructs −1426/−1842 SV40pCAT and −1426/−2592 SV40pCAT (Fig 2A and B). These findings suggested that the enhancer was located in the 229-bp region between bp −1426 and −1655.
AP2 and GATA elements do not mediate enhancer function.
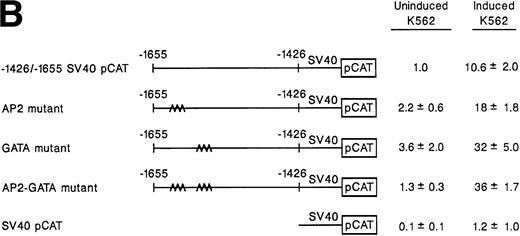
Sequence analysis of this 229-bp region showed a single GATA box between bp −1628 to −1684 adjacent to an AP2 site between bp −1637 to −1644 and two tandem, but inversely oriented AP1 binding sites at bp −1543 and −1557 (Fig 1). Based on data from other laboratories suggesting that members of the GATA family are involved in erythro-megakaryocytic differentiation, we initially focused on the role of the GATA and AP2 sites in megakaryocytic enhancer activity.23-29 To evaluate the role of these two potential binding sites in enhancer activity, mutations in the AP2 site, the GATA site, or both sites that are known to eliminate protein binding were constructed by PCR-directed mutagenesis of the −1426/−1655 SV40pCAT construct (Fig 3A).30-35 Activity of the mutant constructs in uninduced and induced K562 cells was compared with the activity of the intact −1426/−1655 SV40pCAT construct in uninduced K562 cells (Fig 3B). The activity of the intact −1426/−1655 SV40pCAT construct in uninduced K562 cells was assigned a value of 1.0. Results are represented as the mean and standard deviation of at least three separate experiments. The −1426/−1655 SV40pCAT construct directed at least 10-fold greater enhancer activity in K562 cells induced to become megakaryocytic than in uninduced K562 cells. In contrast to the loss of enhancer activity we expected upon mutation of the AP2 or GATA sites, mutation of either the AP2 site, the GATA site, or both sites increased enhancer activity twofold to threefold in both uninduced and induced K562 cells. Clearly, the AP2 and GATA sites do not mediate enhancer activity for the α2 integrin gene and may serve to diminish enhancer activity in hematopoietic cells.
Site-directed mutagenesis of the AP2 and GATA consensus sites. (A) A diagram demonstrates the sequence of a 32-bp region within the megakaryocytic enhancer extending between bp −1623 and −1655 containing the intact AP2 and GATA binding sites and the sequence of 3 mutant constructs prepared by PCR to produce mutations of either the AP2 site, the GATA site, or both AP2 and GATA recognition sites. (B) Mutations in either the AP2 site, the GATA site, or both AP2 and GATA sites were incorporated into the construct −1426/−1655 SV40pCAT. The enhancer activity of the mutant constructs containing either a single mutation of the AP2 site, GATA site, or the AP2 and GATA sites was compared with the activity of the intact −1426/−1655 SV40pCAT in uninduced and induced K562 cells. Cotransfection with RSV-luciferase was used to control for transfection efficiency. After 48 hours of incubation, cell extracts were assayed. After normalization for transfection efficiency, CAT activity of the constructs was determined by thin-layer chromatography and differential extraction. The mean and standard deviation of CAT activity of the mutant constructs from at least three separate electroporations was determined relative to −1426/−1655 SV40pCAT in uninduced K562 cells, which was assigned the value of 1.0.
Site-directed mutagenesis of the AP2 and GATA consensus sites. (A) A diagram demonstrates the sequence of a 32-bp region within the megakaryocytic enhancer extending between bp −1623 and −1655 containing the intact AP2 and GATA binding sites and the sequence of 3 mutant constructs prepared by PCR to produce mutations of either the AP2 site, the GATA site, or both AP2 and GATA recognition sites. (B) Mutations in either the AP2 site, the GATA site, or both AP2 and GATA sites were incorporated into the construct −1426/−1655 SV40pCAT. The enhancer activity of the mutant constructs containing either a single mutation of the AP2 site, GATA site, or the AP2 and GATA sites was compared with the activity of the intact −1426/−1655 SV40pCAT in uninduced and induced K562 cells. Cotransfection with RSV-luciferase was used to control for transfection efficiency. After 48 hours of incubation, cell extracts were assayed. After normalization for transfection efficiency, CAT activity of the constructs was determined by thin-layer chromatography and differential extraction. The mean and standard deviation of CAT activity of the mutant constructs from at least three separate electroporations was determined relative to −1426/−1655 SV40pCAT in uninduced K562 cells, which was assigned the value of 1.0.
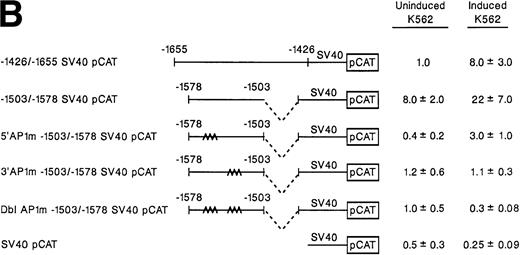
Two tandem, inversely oriented AP1 sites demonstrate full enhancer activity.
We then turned our attention to the potential role of the two tandem AP1 sites with dyad symmetry located between bp −1543 and −1557. To focus only on the two tandem AP1 sites, a shorter double-stranded DNA fragment extending from bp −1503 to −1578 that included both AP1 binding sites but eliminated 77 bp of the 5′ region including the GATA and AP2 sites and 77 bp of 3′ sequence was prepared by PCR and placed in the enhancer location of the SV40pCAT vector (Fig 4A and B). As shown in Fig 4B, the short construct −1530/−1578 SV40pCAT that lacked 5′ and 3′ elements including the GATA and AP2 sites contained all of the enhancer activity of the longer construct −1426/−1655 SV40pCAT in induced K562 cells. To determine the role of one or both AP1 sites, mutations of either the 5′ AP1 site, the 3′ AP1 site, or both sites were prepared in the shorter construct −1503/−1578 SV40pCAT, as shown in Fig 4A. The ability of the mutant constructs to direct reporter gene activity in the irrelevant promoter construct SV40pCAT was compared with the activity of the nonmutated construct −1503/−1578 SV40pCAT and the longer −1426/−1655 SV40pCAT construct in both induced and uninduced K562 cells. In contrast to the high level of reporter gene activity generated by the intact −1503/−1578 SV40pCAT construct, the activity of the constructs with mutations of either the 5′ AP1 site, the 3′ AP1 site, or both AP1 sites was markedly reduced to levels only slightly above background in both uninduced and induced K562 cells (Fig 4B). These results indicate that the two adjacent but inversely oriented AP1 binding sites play an important role in enhancer activity of the α2 integrin gene in K562 cells induced to become megakaryocytic. The tandem AP1 binding sites appear responsible for both enhancer gene activity in K562 cells induced to differentiate along the megakaryocyte lineage and for the low level of reporter gene activity in uninduced K562 cells.
Site-directed mutagenesis of tandem AP1 consensus binding sites. (A) The diagram demonstrates the sequence of the 40-bp region of the 2 enhancer extending from bp −1530 bp to −1570 containing the two intact but inversely oriented AP1 binding sites. The sequence of the three mutant constructs prepared by PCR containing mutations of the 5′ AP1 site, the 3′ AP1 site, or both AP1 binding sites is demonstrated. (B) To determine the role of one or both AP1 sites in enhancer activity, mutations of either the 5′ AP1 site, the 3′ AP1 site, or both sites were introduced into the shorter construct −1503/−1578 SV40pCAT. This shorter construct includes both AP1 binding sites but eliminates 77 bp of the 5′ region including the GATA and AP2 site and 77 bp of 3′ sequence. The enhancer activity of the original −1426/−1655 SV40pCAT construct was compared with the shorter intact construct −1503/−1578 SV40pCAT and the three mutant constructs, 5′ AP1m −1503/−1578 SV40pCAT, 3′ AP1m −1503/−1578 SV40pCAT, and Dbl AP1m −1503/−1578 SV40pCAT in uninduced and induced K562 cells after transient transfection. Cotransfection with RSV-luciferase was used to control for transfection efficiency. After 48 hours of incubation, cell extracts were assayed after normalization for transfection efficiency. CAT activity of the constructs was determined by thin-layer chromatography and differential extraction. The mean and standard deviation of CAT activity of the mutant constructs from at least three separate electroporations was determined relative to the −1426/−1655 SV40pCAT construct in uninduced K562 cells, which was assigned a value of 1.0.
Site-directed mutagenesis of tandem AP1 consensus binding sites. (A) The diagram demonstrates the sequence of the 40-bp region of the 2 enhancer extending from bp −1530 bp to −1570 containing the two intact but inversely oriented AP1 binding sites. The sequence of the three mutant constructs prepared by PCR containing mutations of the 5′ AP1 site, the 3′ AP1 site, or both AP1 binding sites is demonstrated. (B) To determine the role of one or both AP1 sites in enhancer activity, mutations of either the 5′ AP1 site, the 3′ AP1 site, or both sites were introduced into the shorter construct −1503/−1578 SV40pCAT. This shorter construct includes both AP1 binding sites but eliminates 77 bp of the 5′ region including the GATA and AP2 site and 77 bp of 3′ sequence. The enhancer activity of the original −1426/−1655 SV40pCAT construct was compared with the shorter intact construct −1503/−1578 SV40pCAT and the three mutant constructs, 5′ AP1m −1503/−1578 SV40pCAT, 3′ AP1m −1503/−1578 SV40pCAT, and Dbl AP1m −1503/−1578 SV40pCAT in uninduced and induced K562 cells after transient transfection. Cotransfection with RSV-luciferase was used to control for transfection efficiency. After 48 hours of incubation, cell extracts were assayed after normalization for transfection efficiency. CAT activity of the constructs was determined by thin-layer chromatography and differential extraction. The mean and standard deviation of CAT activity of the mutant constructs from at least three separate electroporations was determined relative to the −1426/−1655 SV40pCAT construct in uninduced K562 cells, which was assigned a value of 1.0.
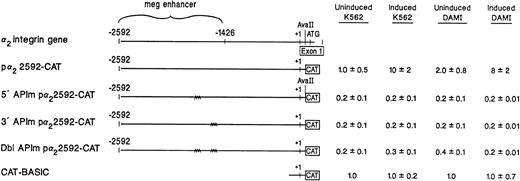
To further demonstrate that the two AP1 sites within the distal 5′ flanking region are required for expression of the α2 integrin gene in cells with megakaryocytic features, either one or both of the AP1 binding sites were mutated in the context of the original intact α2 integrin gene promoter/enhancer construct, pα22592-CAT. Identical mutations of either the 5′ AP1 site, the 3′ AP1 site, or both AP1 sites, as outlined above, were introduced into the original pα22592-CAT construct by site-directed mutagenesis. Mutation of either the 5′ AP1 site, the 3′ AP1 site, or both AP1 sites completely eliminated reporter gene activity mediated by the full-length construct containing the entire 5′ flank of the α2 integrin gene in both induced and uninduced K562 cells (Fig 5), just as it did for the simpler reporter construct (Fig 4B).
Site-directed mutagenesis of the tandem AP1 binding sites markedly reduced the promoter/enhancer activity of the intact p22592-CAT construct. Mutation of the 5′ AP1 site, the 3′ AP1 site, or both AP1 sites was introduced into the original p22592-CAT construct by site-directed mutagenesis. The promoter/enhancer activity of the original p22592-CAT construct was compared with the activity of the construct with mutations in the 5′AP1 site, the 3′ AP1 site, or both AP1 sites after transient transfection into uninduced and induced K562 cells and uninduced and induced DAMI cells. Cotransfection with RSV-luciferase was used to control for transfection efficiency. After 48 hours of incubation, cell extracts were assayed. After normalization for transfection efficiency, CAT activity of the constructs was determined by thin-layer chromatography and differential extraction. The mean and standard deviation of CAT activity of the mutant constructs in either K562 cells or DAMI cells from at least three separate electroporations was determined relative to the pCAT-Basic construct in uninduced K562 or uninduced DAMI cells, respectively, which was assigned a value of 1.0.
Site-directed mutagenesis of the tandem AP1 binding sites markedly reduced the promoter/enhancer activity of the intact p22592-CAT construct. Mutation of the 5′ AP1 site, the 3′ AP1 site, or both AP1 sites was introduced into the original p22592-CAT construct by site-directed mutagenesis. The promoter/enhancer activity of the original p22592-CAT construct was compared with the activity of the construct with mutations in the 5′AP1 site, the 3′ AP1 site, or both AP1 sites after transient transfection into uninduced and induced K562 cells and uninduced and induced DAMI cells. Cotransfection with RSV-luciferase was used to control for transfection efficiency. After 48 hours of incubation, cell extracts were assayed. After normalization for transfection efficiency, CAT activity of the constructs was determined by thin-layer chromatography and differential extraction. The mean and standard deviation of CAT activity of the mutant constructs in either K562 cells or DAMI cells from at least three separate electroporations was determined relative to the pCAT-Basic construct in uninduced K562 or uninduced DAMI cells, respectively, which was assigned a value of 1.0.
The two AP1 sites serve enhancer function in DAMI cells.
The enhancer activity of the two AP1 sites in DAMI cells, another model of megakaryocytic differentiation, was established. Uninduced DAMI cells express low levels of the α2β1integrin. In DAMI cells, α2β1 integrin expression is markedly increased by the addition of PDB.14Reporter gene activity of the pα22592-CAT construct with mutations of the 5′ AP1 site, the 3′ AP1 site, and both AP1 sites was compared with the original pα22592-CAT construct in uninduced and induced DAMI cells. The pα22592-CAT construct exhibited low levels of reporter gene activity in uninduced DAMI cells and high levels of reporter activity in induced DAMI cells. Mutations of either the 5′ AP1 site, the 3′ AP1 site, or both AP1 sites completely eliminated reporter gene activity of the pα22592-CAT construct in uninduced and induced DAMI cells as well (Fig 5). These findings provide additional evidence that the tandem AP1 sites located in the distal 5′ flank of the α2 integrin gene are required for α2 integrin gene activity in hematopoietic cells either poised to become megakaryocytic or induced with phorbol dibutyrate to acquire megakaryocytic features.
DNA-protein complex formation with the tandem AP1 binding sites.
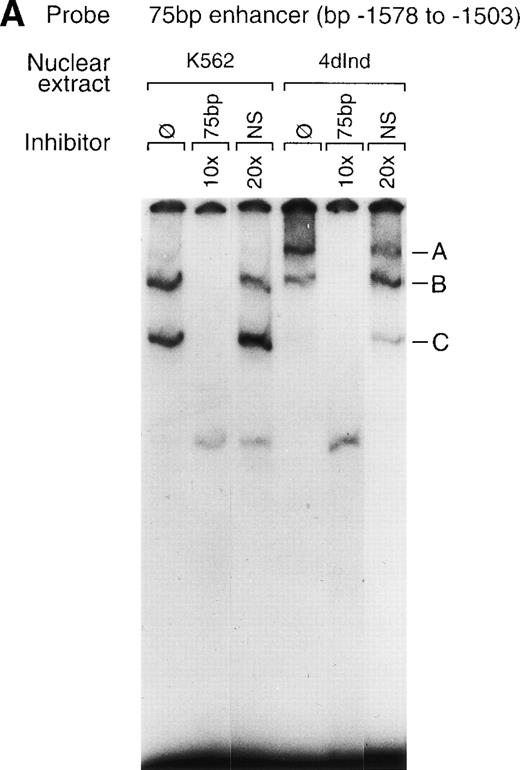
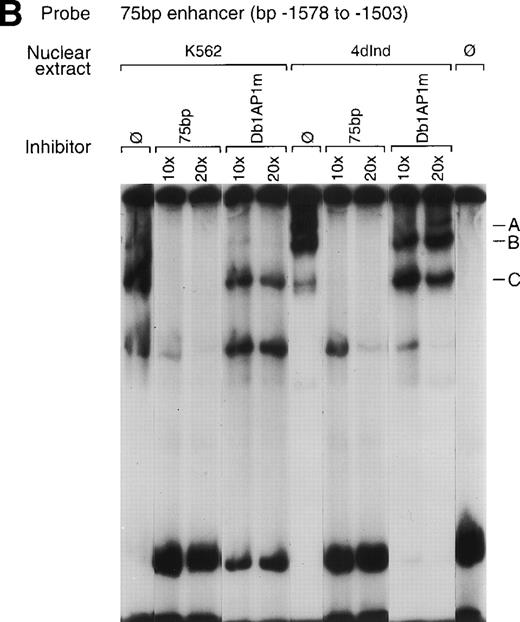
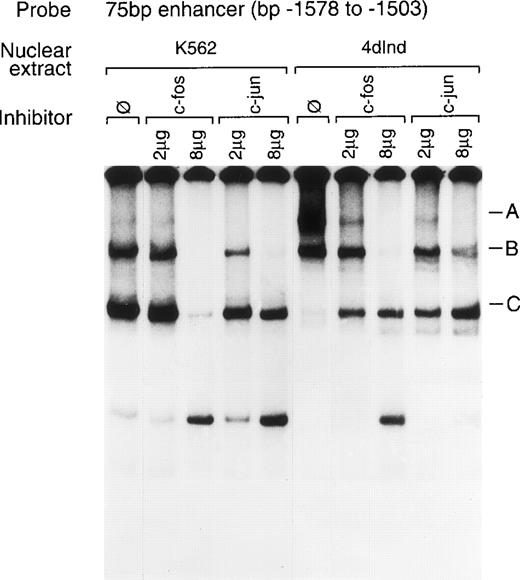
To identify transcription factors that specifically interact with the α2 integrin enhancer, binding of nuclear proteins from uninduced and induced K562 cells to the 75-bp double-stranded DNA fragment (bp −1503 to −1578) containing the two tandem AP1 sites was evaluated by gel mobility shift assays. Nuclear proteins from both uninduced K562 cells and K562 cells induced to differentiate for 4 days with phorbol dibutyrate bound the 75-bp enhancer element (Fig 6A). Nuclear extracts from both uninduced K562 cells and K562 cells induced with phorbol dibutyrate formed a complex of intermediate migration (B). Nuclear extracts from uninduced K562 cells also formed a faster migrating complex (C) that was not as prominent with extracts from induced cells. Furthermore, nuclear extracts from K562 cells induced to develop megakaryocytic features bound the 75-bp DNA fragment to form an additional, slower migrating complex (A) that was not as abundant with extracts from uninduced cells. Formation of all three DNA-protein complexes could be specifically inhibited by the unlabeled 75-bp DNA fragment but not by an irrelevant 119-bp double-stranded DNA fragment. The unlabeled 75-bp fragment containing a mutation of the 5′ AP1 site, the 3′ AP1 site, or both AP1 sites (Dbl AP1m) failed to inhibit complex formation (Fig 6B, data not shown). More rapidly migrating, nonspecific complexes, as judged by the inability of the unlabeled DNA fragment to compete, were present in some experiments.
Gel mobility shift analysis of the megakaryocyte enhancer. (A and B) Gel mobility shift analysis was performed with32P end-labeled double-stranded DNA fragment containing the 75-bp enhancer region (bp −1503/−1578) with two tandem AP1 binding sites and 1 μg of nuclear protein from either uninduced K562 cells (K562) or K562 cells induced for 4 days (4d Ind) with phorbol. Three major DNA protein complexes are indicated as A, B, and C. DNA-protein complexes were formed in the absence of competitive inhibitor (ø) (A and B), in the presence of a non-specific (NS) inhibitor consisting of 119-bp double-stranded DNA fragment from the coding region of the 2 cDNA (A), or in the presence of 75-bp DNA fragment containing mutations to both AP1 binding sites (Dbl AP1m) (B). The presence of an unlabeled specific inhibitor (75 bp) inhibited DNA-protein complex formation (A and B). Gel mobility shift analysis was performed as described in Materials and Methods.
Gel mobility shift analysis of the megakaryocyte enhancer. (A and B) Gel mobility shift analysis was performed with32P end-labeled double-stranded DNA fragment containing the 75-bp enhancer region (bp −1503/−1578) with two tandem AP1 binding sites and 1 μg of nuclear protein from either uninduced K562 cells (K562) or K562 cells induced for 4 days (4d Ind) with phorbol. Three major DNA protein complexes are indicated as A, B, and C. DNA-protein complexes were formed in the absence of competitive inhibitor (ø) (A and B), in the presence of a non-specific (NS) inhibitor consisting of 119-bp double-stranded DNA fragment from the coding region of the 2 cDNA (A), or in the presence of 75-bp DNA fragment containing mutations to both AP1 binding sites (Dbl AP1m) (B). The presence of an unlabeled specific inhibitor (75 bp) inhibited DNA-protein complex formation (A and B). Gel mobility shift analysis was performed as described in Materials and Methods.
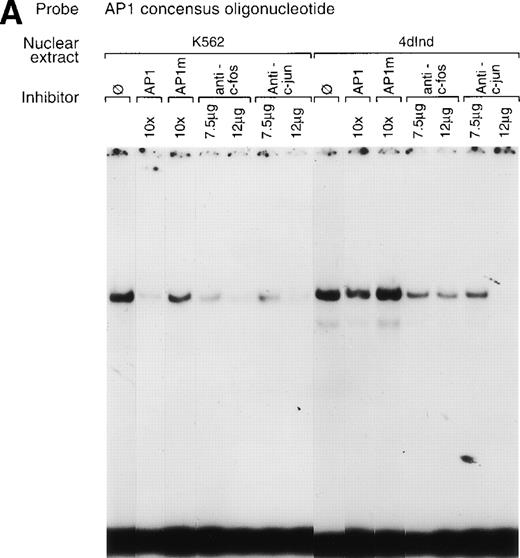
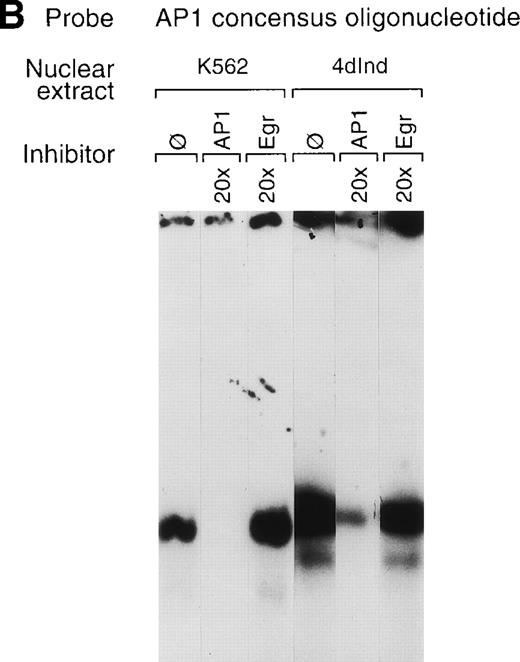
The involvement of two tandem AP1 consensus elements in the enhancer suggests a role for the AP1 family of transcription factors in α2 integrin gene regulation. A double-stranded oligonucleotide containing only a single consensus AP1 site specifically bound nuclear extracts from both uninduced and induced K562 cells (Fig 7A, B, and C). The complex formed between the consensus AP1 oligonucleotide and nuclear extracts from induced K562 cells was more abundant than the complex formed with nuclear extracts from uninduced K562 cells. Complex formation between the consensus AP1 site and nuclear extracts from both uninduced and induced K562 cells was partially inhibited by 10-fold and almost completely inhibited by 20-fold addition of the identical unlabeled oligonucleotide and completely inhibited by 10-fold or 20-fold addition of the 75-bp enhancer fragment but was not inhibited by the AP1 consensus oligonucleotide with a 2-bp mutation (AP1m; Fig 7A and C). In addition, an oligonucleotide containing the consensus binding site for the Egr family of transcription factors failed to inhibit complex formation (Fig 7B).
A consensus AP1 site forms DNA-protein complexes. (A, B, and C) Gel mobility shift experiments were performed with32P end-labeled double-stranded oligonucleotide containing a single AP1 consensus binding site and 1 μg of nuclear extract from uninduced K562 cells (K562) or phorbol ester-induced K562 cells (4d Ind) (A, B, and C). A DNA-protein complex formed in the absence of competitive inhibitor (ø) (A, B, and C), in the presence of an identical oligonucleotide with a 2-bp mutation in the consensus binding site (AP1m) (A), in the presence of the unlabeled specific 75-bp enhancer region (bp −1503/−1578) with two tandem AP1 binding sites (C), or in the presence of an irrelevant double-stranded oligonucleotide containing the consensus binding site for transcription factors of the early growth response family (Egr) (B). The unlabeled AP1 oligonucleotide inhibited DNA-protein complex formation (A, B, and C). Monoclonal or polyclonal antibodies against either c-fos or c-jun (at the indicated concentration) were preincubated with nuclear extracts for 18 hours at 4°C before the addition of 32P end-labeled probe, as described in Materials and Methods. Antisera against both c-fos and anti–c-jun inhibited DNA-protein complex formation in a concentration-dependent manner. The molar excess of unlabeled competitor or the concentration of antibody is indicated. Gel mobility shift analyses were performed as described in Materials and Methods.
A consensus AP1 site forms DNA-protein complexes. (A, B, and C) Gel mobility shift experiments were performed with32P end-labeled double-stranded oligonucleotide containing a single AP1 consensus binding site and 1 μg of nuclear extract from uninduced K562 cells (K562) or phorbol ester-induced K562 cells (4d Ind) (A, B, and C). A DNA-protein complex formed in the absence of competitive inhibitor (ø) (A, B, and C), in the presence of an identical oligonucleotide with a 2-bp mutation in the consensus binding site (AP1m) (A), in the presence of the unlabeled specific 75-bp enhancer region (bp −1503/−1578) with two tandem AP1 binding sites (C), or in the presence of an irrelevant double-stranded oligonucleotide containing the consensus binding site for transcription factors of the early growth response family (Egr) (B). The unlabeled AP1 oligonucleotide inhibited DNA-protein complex formation (A, B, and C). Monoclonal or polyclonal antibodies against either c-fos or c-jun (at the indicated concentration) were preincubated with nuclear extracts for 18 hours at 4°C before the addition of 32P end-labeled probe, as described in Materials and Methods. Antisera against both c-fos and anti–c-jun inhibited DNA-protein complex formation in a concentration-dependent manner. The molar excess of unlabeled competitor or the concentration of antibody is indicated. Gel mobility shift analyses were performed as described in Materials and Methods.
Antibodies against c-fos/c-jun heterodimers prevent DNA-protein complex formation.
The AP1 consensus element of the α2 integrin enhancer is separated by one nucleotide (TGANTCA).36 c-fos/c-jun heterodimers prefer such DNA targets. In contrast, ATF/CREB-containing dimers prefer AP1 consensus elements separated by two nucleotides (TGANNTCA).37 Antibodies against c-fos or c-jun family members inhibited complex formation between K562 nuclear extracts and the oligonucleotide containing the AP1 consensus site, but not oligonucleotides containing the SP1 or NFκB consensus sites in a concentration-dependent manner under our experiment conditions (Fig 7A and data not shown).
Under similar experimental conditions, antibodies against c-fos also inhibited formation of complexes B and C between the 75-bp enhancer fragment and uninduced K562 cell extracts, as shown in Fig 8. Anti–c-fos antibodies inhibited complexes A and B formed between the 75-bp enhancer and nuclear extracts from induced K562 cells. An antibody against c-jun family members diminished formation of complex B formed by the 75-bp enhancer and uninduced nuclear extracts and formation of complexes A and B between the 75-bp fragment and extracts from induced K562 cells. These findings suggest that members of the c-fos and c-jun family bind to the 75-bp enhancer fragment of the α2 integrin gene.
Anti–c-fos and anti–c-jun antibodies inhibit DNA-protein complex formation. Gel mobility shift experiments were performed with 32P end-labeled 75-bp enhancer fragment (bp −1578 to bp −1503) and 1 μg of nuclear extract from either uninduced K562 cells (K562) or K562 cells induced for 4 days with phorbol dibutyrate (4d Ind). Monoclonal or polyclonal antibodies against either c-fos or c-jun (at the indicated concentration) were preincubated with nuclear extracts for 18 hours at 4°C before the addition of 32P end-labeled probe, as described in Materials and Methods. Anti–c-fos and anti–c-jun inhibited DNA-protein complex formation.
Anti–c-fos and anti–c-jun antibodies inhibit DNA-protein complex formation. Gel mobility shift experiments were performed with 32P end-labeled 75-bp enhancer fragment (bp −1578 to bp −1503) and 1 μg of nuclear extract from either uninduced K562 cells (K562) or K562 cells induced for 4 days with phorbol dibutyrate (4d Ind). Monoclonal or polyclonal antibodies against either c-fos or c-jun (at the indicated concentration) were preincubated with nuclear extracts for 18 hours at 4°C before the addition of 32P end-labeled probe, as described in Materials and Methods. Anti–c-fos and anti–c-jun inhibited DNA-protein complex formation.
A role for the MAPK cascade in α2 integrin gene regulation.
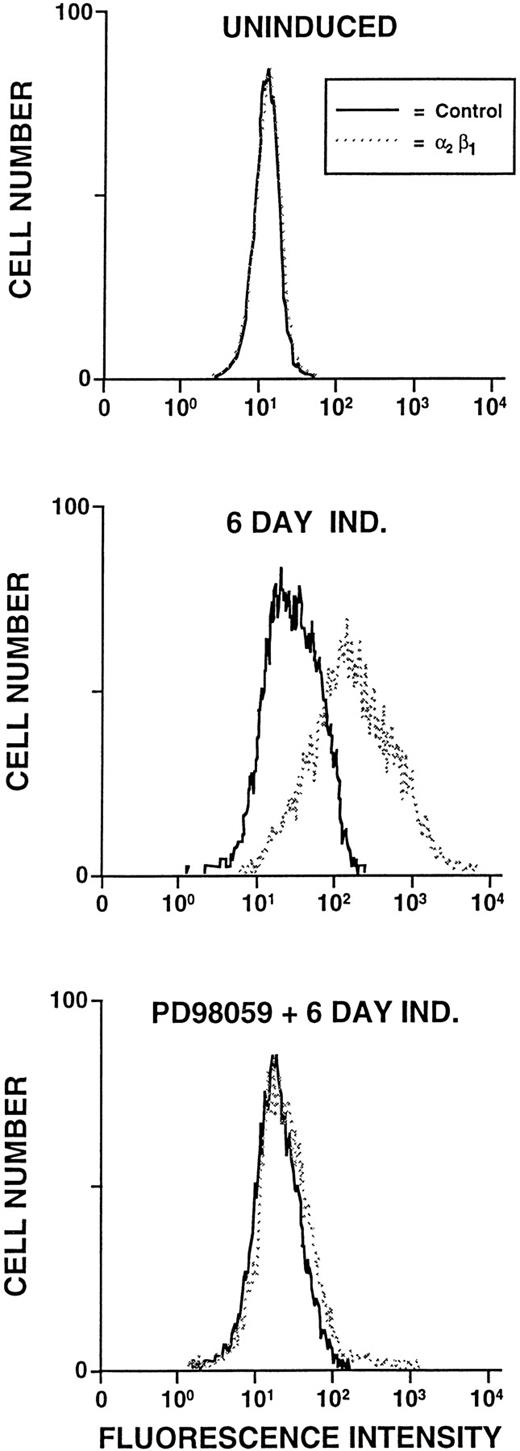
The signaling cascades that lead to AP1 activation include numerous mitogen activated protein kinase intermediates and transcriptional induction of c-fos and c-jun proteins.38,39 Recent observations showed that inhibition of MAPK kinase 1 (MAPKK1) prevented some features of phorbol-induced megakaryocytic differentiation, including expression of the αIIb and β3integrin genes.40 The role of MAPKK1 in regulating α2 integrin gene expression was therefore evaluated. As shown previously, uninduced K562 cells fail to express detectable levels of the α2β1 integrin (Fig 9). After 6 days of PDB induction, expression of the α2β1 integrin is greatly increased. The addition of PD98059, the MAPKK1 specific inhibitor, to K562 cells 15 minutes before the addition of PDB completely eliminated α2β1 integrin subunit expression, as shown in Fig 9.
Inhibition of 2β1 integrin expression by PD98059. Flow cytometric analysis of the 2β1 integrin expression by uninduced K562 cells, K562 cells induced for 6 days with PDB (40 nmol/L) (6d Ind), or K562 cells treated with PD98059 (50 μmol/L) for 15 minutes before the addition of PDB (40 nmol/L) (PD98059 + 6d Ind).
Inhibition of 2β1 integrin expression by PD98059. Flow cytometric analysis of the 2β1 integrin expression by uninduced K562 cells, K562 cells induced for 6 days with PDB (40 nmol/L) (6d Ind), or K562 cells treated with PD98059 (50 μmol/L) for 15 minutes before the addition of PDB (40 nmol/L) (PD98059 + 6d Ind).
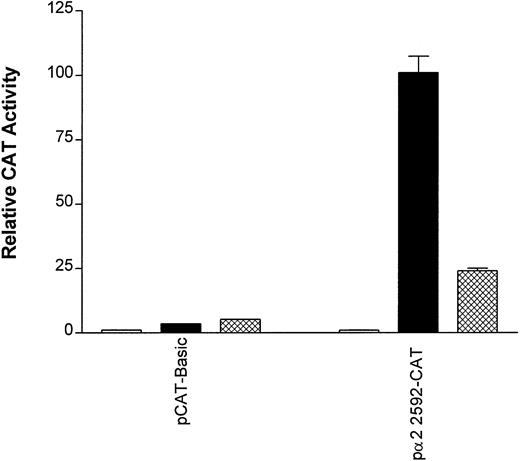
The role of the MAPK cascade in activating the α2promoter/enhancer was assessed. As shown in Figs 2B and10, the reporter gene activity of the original intact α2 integrin promoter/enhancer construct pα22592-CAT was high in induced K562 cells and at background levels in uninduced K562. Reporter gene activity of the pα22592-CAT in K562 cells treated with both PDB and PD98059 was markedly reduced compared with the reporter activity in PDB induced K562 cells (Fig 10). Inhibition of the MAPK kinase cascade diminished the function of the α2 integrin promoter/enhancer. Based on these findings, we propose a model by which activation of the MAPK cascade leads to α2 integrin gene expression in cells induced to become megakaryocytic (Fig 11).
Diminution of 2 promoter/enhancer function by PD98059. The constructs pCAT-Basic and p22592-CAT were transfected into uninduced K562 cells (□), K562 cells treated with PDB (40 nmol/L; ▪), and K562 cells treated for 15 minutes with PD98059 (50 μmol/L) before the addition of PDB (40 nmol/L; ⊠). Cotransfection with RSV-luciferase was used to control for transfection efficiency. After 48 hours of incubation, cell extracts were assayed. After normalization for transfection efficiency, CAT activity of the constructs was determined using thin-layer chromatography. The mean and standard deviation of CAT activity in uninduced K562 cells, PDB-induced K562 cells, or K562 cells treated with PD98059 and PDB was determined. CAT activity relative to the pCAT-Basic construct in uninduced K562, which was assigned a value of 1.0, is shown.
Diminution of 2 promoter/enhancer function by PD98059. The constructs pCAT-Basic and p22592-CAT were transfected into uninduced K562 cells (□), K562 cells treated with PDB (40 nmol/L; ▪), and K562 cells treated for 15 minutes with PD98059 (50 μmol/L) before the addition of PDB (40 nmol/L; ⊠). Cotransfection with RSV-luciferase was used to control for transfection efficiency. After 48 hours of incubation, cell extracts were assayed. After normalization for transfection efficiency, CAT activity of the constructs was determined using thin-layer chromatography. The mean and standard deviation of CAT activity in uninduced K562 cells, PDB-induced K562 cells, or K562 cells treated with PD98059 and PDB was determined. CAT activity relative to the pCAT-Basic construct in uninduced K562, which was assigned a value of 1.0, is shown.
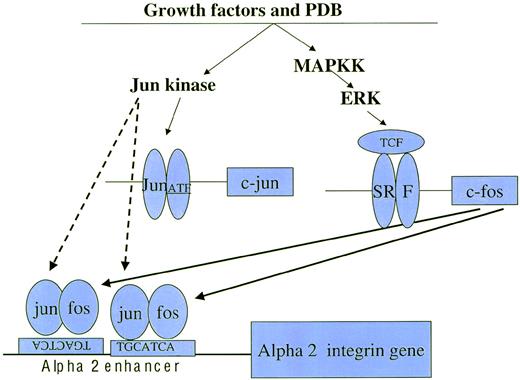
A proposed pathway for increased expression of the 2 integrin subunit in cells with megakaryocytic features. Both growth factors and PDB indirectly activate the MAPK cascade, including the mitogen-activated protein kinase kinase (MAPKK), extracellular receptor activated kinase (Erk), and Jun kinase via intermediate signaling molecules, including protein kinase C (PKC). Activated Erk and Jun kinase increase c-fos and c-jun synthesis by phosphorylation and activation of transcription factors, including complex factors (TCF), serum response factors (SRF), c-jun, and activating transcription factors (ATF).38 In addition, Erk and Jun kinase phosphorylation augment the activity of both c-fos and c-jun. Newly synthesized (→) and phosphorylated (- - →) c-fos and c-jun family members heterodimerize to bind the tandem and inversely oriented AP1 binding sites of the 2 integrin enhancer and upregulate 2integrin gene expression.
A proposed pathway for increased expression of the 2 integrin subunit in cells with megakaryocytic features. Both growth factors and PDB indirectly activate the MAPK cascade, including the mitogen-activated protein kinase kinase (MAPKK), extracellular receptor activated kinase (Erk), and Jun kinase via intermediate signaling molecules, including protein kinase C (PKC). Activated Erk and Jun kinase increase c-fos and c-jun synthesis by phosphorylation and activation of transcription factors, including complex factors (TCF), serum response factors (SRF), c-jun, and activating transcription factors (ATF).38 In addition, Erk and Jun kinase phosphorylation augment the activity of both c-fos and c-jun. Newly synthesized (→) and phosphorylated (- - →) c-fos and c-jun family members heterodimerize to bind the tandem and inversely oriented AP1 binding sites of the 2 integrin enhancer and upregulate 2integrin gene expression.
DISCUSSION
In earlier studies we demonstrated that expression of the α2 integrin gene in cells with megakaryocytic features, both induced K562 cells and DAMI cells, is regulated by three distinct regions of the 5′ flank of the α2 integrin gene containing both positive and negative elements.14 The three regions include a core promoter between bp −30 and −92, a silencer between bp −92 and −351, and megakaryocytic enhancers between bp −1426 and −2592. In this study, using a combination of deletional and mutational analyses, we identified a megakaryocytic enhancer of the α2 integrin gene. The enhancer requires two tandem inversely oriented AP1 consensus binding sites for interaction with c-fos/c-jun family members. These data suggest a model in which signaling via the MAPK cascade leads to activation of the megakaryocytic enhancer of the α2integrin gene (Fig 11). Extracellular stimuli such as phorbol dibutyrate under our experimental conditions or growth factors during thrombopoiesis directly or indirectly activate the MAPK signaling cascade.
The MAP kinase cascade involves at least three distinct signaling pathways, including the extracellular regulated kinases (ERK), jun kinase, and p38 MAPK. The best delineated pathway initiated via growth factor receptors or phorbol esters initiates a cascade from ras, raf, MAPKK1 (MEK1), or MAPKK2 (MEK2) to ERK1 or ERK2.39,41 The activated ERK proteins then translocate to the nucleus to phosphorylate and activate nuclear transcription factors, such as ternary complex factors (TCF).42 ERK activation directly increases AP1 activity via c-jun activation and increased c-fos synthesis, leading to an increase in c-fos/c-jun heterodimerization and DNA binding.43
A role for the MAP kinase pathway in megakaryocytic differentiation was originally suggested by the ability of phorbol esters to initiate megakaryocytic differentiation and induce the expression of megakaryocyte-specific genes.10,11 A direct role for the MAP kinase pathway in megakaryocytic differentiation has been demonstrated by a number of investigators.39-41,44,45Megakaryocytic differentiation of K562 cells induced with phorbol dibutyrate, including the morphologic aspects of cell adhesion and cell spreading and the expression of the megakaryocytic genes encoding the αIIb and β3 integrin subunits, was either partially or completely inhibited by PD98059, the MAPKK1 specific inhibitor. In addition, constitutive activation of the pathway via expression of the constitutively active mutants of MAPKK1 or MAPKK2 induced the expression of the platelet/megakaryocytic-specific genes αIIb and β3. The regulatory elements required for expression of megakaryocyte-specific proteins αIIbβ3, platelet factor 4 (PF4), glycoprotein Ib, and c-mpl have been mapped.23-28 Adjacent GATA and ets consensus binding sites identified in the promoters of the αIIb, PF4, glycoprotein Ib, and c-mpl are essential for transcriptional activation of these genes.27,28 46-48 GATA and ets transcription factors are activated by the MAPK cascade.
The important role that adjacent GATA and ets binding sites play in activation of many megakaryocyte-specific genes led us to evaluate the role of the adjacent GATA and AP2 sites in enhancer activity of the α2 integrin gene. Surprisingly, the GATA and AP2 sites did not serve enhancer function and served to slightly repress the activity of the distal enhancer of the α2 integrin gene. Although the architecture of the α2 promoter/enhancer that consists of a promoter, a silencer, and a distal enhancer resembles other megakaryocytic genes, the enhancer is distinctly different and consists of two adjacent but inversely oriented AP1 binding sites.
Mutation of either or both AP1 sites markedly reduced enhancer activity in the irrelevant promoter construct SV40pCAT, as well as in the intact α2 promoter/enhancer construct extending through the distal 5′ flank of the α2 integrin gene. Consensus AP1 binding sites have not been demonstrated to play an important role in the enhancer activity of other megakaryocytic genes. However, the inverse orientation of the two AP1 sites resembles the dyad symmetry element (DSE) required for c-fos induction after 12-0-tetradecanoyl phorbol-13-acetate (TPA)-induced megakaryocytic differentiation of K562 cells.42 AP1, a sequence-specific transcription factor composed of c-fos and c-jun family members, was initially identified as the factor that mediates TPA induction.43,49,50 Activation and expression of AP1 is induced by multiple stimuli, including growth factors, cytokines, UV irradiation, and the different members of the MAPK cascade.38 50
The NF-E2 binding site consists of either one or tandem, extended AP1 binding sites.51-54 A possible role for NF-E2 in α2 integrin gene regulation was considered.51-55 The NF-E2 binding sites in the hypersensitivity site 2 (HS2) enhancer of the β globin gene resemble the two tandem AP1 binding sites in the α2 integrin enhancer.54 56 However, the additional -G- in theGCTGAGTC sequence that is required for NF-E2 binding is absent from both of the tandem AP1 binding sites in the α2 integrin enhancer. In addition, although nuclear proteins from induced K562 cells bind to an oligonucleotide containing the consensus NF-E2 binding site and the unlabeled oligonucleotide containing the NF-E2 consensus binding site successfully inhibits the binding of nuclear proteins to NF-E2, the unlabeled NF-E2 oligonucleotide fails to compete for binding to the megakaryocyte enhancer region (data not shown). In addition, expression of the hematopoietic restricted component p45 failed to induce megakaryocytic differentiation or expression of the α2β1integrin (data not shown).
Based on the studies presented here, the MAPK pathway is required for α2 integrin gene expression in cells induced to differentiate along the megakaryocytic lineage. Two tandem and inversely oriented AP1 sites in the α2 integrin gene enhancer bind to members of the AP1 family of transcription factors. We propose a model by which either PDB, under experimental conditions, or growth factors such as thrombopoietin (TPO), under physiologic conditions, activate the MAPK cascade and ultimately lead to α2 integrin gene expression in cells induced to be megakaryocytic. The mechanisms by which PDB activates c-fos/c-jun family members is well delineated.38TPO has been shown to directly activate the MAPK cascade after binding to c-Mpl on megakaryocytic progenitors.57 58 This model provides a physiologically relevant series of events that lead to expression of the α2β1 integrin.
ACKNOWLEDGMENT
The authors thank Dr Samuel A. Santoro for encouragement and constructive criticism of the manuscript. We also thank Mary Beth Flynn for excellent secretarial assistance.
Supported by National Institutes of Health Grant No. R01 HL51450-04.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Mary M. Zutter, MD, Washington University School of Medicine, Department of Pathology, St Louis, MO 63110.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal