Abstract
The mechanisms whereby chromosomal translocations are consistently associated with specific tumor types are largely unknown. A generally accepted hypothesis is that the physical proximity of the involved chromosomal regions may be one important factor in the genesis of these phenomena. Accordingly, a likely possibility is that such a proximity may occur in a cell-lineage and cell-differentiation stage-specific manner. In this work, we have addressed this issue using as models the ABL and BCR genes of t(9;22) and the PML and RAR genes of t(15;17). By using in situ hybridization and confocal microscopy, we have measured the distances between these two pairs of genes in three-dimensionally preserved hematopoietic cells belonging to different cell lineages, at various stages of differentiation, and at various stages of the cell cycle, with the following results. (1) Intergenic distances vary periodically during the cell cycle and a significant association of ABL with BCR and of PML with RAR is seen at the transition between S and G2, which persists during G2 and prophase (such a behavior is not observed for distances between ABL or PML and the β-globin genes, used as a control). (2) The proportion of cells in which PML and RAR or ABL and BCR are closely associated is higher in hematopoietic precursors than in B-lymphoid cells (whereas the distances between ABL or PML and the β-globin genes are not affected by cell type). (3) When intergenic distances in unstimulated bone marrow CD34+ cells were compared with those in CD34+ cells treated with interleukin-3 (IL-3), a trend towards a higher proximity of the ABL and BCR genes in the former and of the PML and RAR genes in the latter is observed. (4) Analysis of B-lymphoid cells during mitosis shows that intergenic distances at metaphase are strongly influenced by physical constraints imposed by the chromosomal location of the gene, by the size of the respective chromosome, and by the geometry of the metaphase plate. These findings suggest that intrinsic spatial dynamics, established early in hematopoiesis and perpetuated differentially in distinct cell lineages, may facilitate the collision of individual genes and thus reciprocal recombination between them at subsequent stages of hematopoietic differentiation.
SINCE THE DISCOVERY of the t (9;22) in chronic myeloid leukemia (CML), several types of reciprocal chromosomal translocations have been consistently associated with human cancer.1,2 The molecular characterization of chromosomal breakpoints has shown that the same genomic regions are systematically involved in each specific type of translocation, but, despite this knowledge, the mechanisms underlying interchromosomal recombination phenomena in somatic cells are largely unknown. In those circumstances in which loci of antigen-receptor genes are involved, it has been postulated that the enzymatic systems responsible for the normal somatic VDJ rearrangements of these genes in normal B- or T-cell ontogeny may be subverted to promote the illegitimate recombination with other genomic regions.1,3 Also, it has been suggested that the higher susceptibility of certain chromosomal regions to breakage and rearrangement may be related to local structural features of the chromatin fiber that make them more susceptible to damage than other genomic regions.4 Recently, it has been further proposed that reciprocal rearrangements such as those underlying the formation of complex BCR-ABL genes in cells of CML patients might be mediated by the mutual attraction of Alu sequences located in heterologous chromosomes.5
Irrespective of the exact nature of the biological mechanisms responsible for the occurrence of chromosomal translocations in somatic cells, a widely accepted assumption is that the spatial proximity of the involved chromosomal regions is likely to be an obligatory requirement for the exchange event to occur.6 Recent reports appear to support this view, because the ABL and BCR genes were found to be in close proximity in two-dimensional human lymphocytes7 and total bone marrow cells8 more often than would have been expected by chance. However, a still open question is whether the spatial relationships between these or other loci frequently engaged in reciprocal recombination phenomena in leukemic cells relate to a particular cell lineage, stage of cell differentiation, and/or to specific phases of the cell cycle.
In this work, we sought to investigate these issues using as models the ABL and BCR genes from the t(9;22) of CML and the PML and RARα genes from t(15;17) of acute promyelocytic leukemia. Apart from their consistent involvement in specific types of human neoplasia, a further reason making these genes specially suited for the addressing of these questions is that the stage of hematopoietic differentiation in which each translocation takes place can be predicted with reasonable certainty: the t(9;22) is present in cells of all hematopoietic lineages in patients with CML,9 whereas the t(15;17) has been exclusively observed in malignant promyelocytes.10
Accordingly, the BCR-ABL rearrangement can be envisaged as representing an oncogenic molecular lesion of a very-early hematopoietic progenitor, whereas the PML-RARα rearrangement is likely to occur in a precursor already committed to the myelo-erythroid11 or granulocytic12 differentiation.
We have therefore analyzed the distances between these genes in hematopoietic cells belonging to different cell lineages and at various stages of differentiation and of the cell cycle. The analysis of gene positioning in bidimensional cells may induce important artifacts that make it difficult to interpret the results from in situ hybridization techniques,13 thus requiring the use of mathematical corrections for three-dimensional (3-D) reconstruction of the data.7 8 Therefore, we have measured intergenic distances in 3-D preserved cells using nonisotopic in situ hybridization and confocal microscopy. We show that, irrespective of their behavior in earlier stages of the cell cycle, each of these pairs of genes tends to become closely associated at the transition of late-S to G2 in all cell types analyzed and, in hematopoietic precursors in interphase, the interdistances PML-RARα and ABL-BCR are smaller than those observed on cells of mature B phenotype. Also, when intergenic distances in unstimulated CD34+ cells were compared with those in cells treated with interleukin-3 (IL-3), a tendency towards a higher proximity of the ABL and BCR genes in the former and of the PML and RARα genes in the latter were observed.
MATERIALS AND METHODS
Cells
Hematopoietic precursors.
Hematopoietic progenitors were enriched from 20 to 30 mL heparinized normal bone marrow obtained from donors of the bone marrow transplantation program, after informed consent had been obtained. Mononuclear cells were isolated by density gradient centrifugation (Ficoll-Paque; Pharmacia, Uppsala, Sweden) and CD34+ cells subsequently obtained using the MiniMACS separation column (Miltenyi Biotec Inc, Auburn, CA). Technical procedures were performed according to the instructions included in the CD34 Progenitor Cell Isolation Kit (QBEND/10, Auburn, CA). In preliminary experiments, the phenotypic profile of the selected CD34+ cells was assessed by flow cytometric analysis in five independent bone marrow samples, with the following results (mean ± SEM): CD34+, 90.8% ± 2.1%; CD38+, 97.4 % ± 0.4%; HLA-DR+, 92% ± 1.6%; CD33+, 59.7% ± 4.5%; CD7+, 6.2% ± 0.9%; CD3+, 1.8% ± 0.7%; CD19+, 24.5% ± 5.6%; CD56+, 1.0% ± 0.2 %; and CD71+, 25.8% ± 7.1% (all antibodies from Becton Dickinson, Mountain View, CA). Double-labeling experiments performed in three of the five samples further showed that the CD34+ cells simultaneously expressed HLA-DR in 97.1% ± 0.63%, CD38 in 98.9% ± 0.53%, and CD33 in 66.4% ± 4.85%. Accordingly, unstimulated cells with these characteristics were used in this study as representative of a cell pool enriched in early hematopoietic precursors.
To obtain a cell population preferentially committed to the myeloid differentiation, CD34+ cells were cultured in 96-microwell plaques at 105 cells/mL in Iscove’s Modified Dulbecco’s Medium (IMDM) supplemented with 5% fetal calf serum (FCS; GIBCO-BRL, Gaithersburg, MD), 1% penicillin/streptomycin (GIBCO-BRL), and recombinant IL-3 (10 ng/mL; Sandoz-Pharma, Basel, Switzerland) at 37°C and 5% CO2. Morphological analysis of cytospin preparations obtained from three independent samples of CD34+ cells on 7 consecutive days and stained with May-Grunwald-Giemsa showed the following: first 48 hours—undifferentiated blasts, 70% to 82%; day 3—undifferentiated blasts (45% to 48%) and promyelocytes (52% to 55%); day 4—undifferentiated blasts (30% to 33%), promyelocytes (55% to 65%), and monocytes (2% to 15%); day 5—undifferentiated blasts (12% to 15%), promyelocytes (40% to 45%), myelocytes (20% to 24%), and monocytes (17% to 19%); day 6—undifferentiated blasts (7% to 8%), promyelocytes (18% to 20%), myelocytes (8% to 10%), and monocytes (40% to 55%); day 7—undifferentiated blasts (5% to 7%), promyelocytes (18% to 23%), myelocytes (8% to 10%), and monocytes (46% to 50%). No erythroid differentiation was observed in these conditions. On the basis of these results, CD34+cells treated with IL-3 for 48 hours were subsequently used in the study as a cell population enriched in progenitors already committed to the myeloid differentiation.
Lymphoid cells.
A mature B-lymphoid cell line (IM-9; ECACC, Salisbury, UK) was used as representative of a nonmyeloid cell population. This cell line has a diploid modal chromosomal number and was cultured in suspension in RPMI-1640 (GIBCO-BRL) supplemented with 10% FCS (GIBCO-BRL).
In Situ Hybridization in 3-D Preserved Cells
Cell preparation.
Unstimulated CD34+ cells (CD34+), CD34+ stimulated with IL-3 for 48 hours (CD34+, IL-3), and IM-9 cells were harvested by centrifugation at 50gfor 5 minutes and applied onto poly-L-lysine–coated 10 mm/10 mm cover slips. This was immediately followed by fixation and permeabilization under conditions that preserve the three-dimensionality of the cells. Briefly, the cells were fixed and extracted in 3.7% paraformaldehyde/0.5% Triton X-100/HPEM (65 mmol/L PIPES, 30 mmol/L HEPES, 10 mmol/L EGTA, 2 mmol/L MgCl2, pH 6.9) for 15 minutes at room temperature,14followed by permeabilization in 0.7% Triton X-100/0.1 N HCl/phosphate-buffered saline (PBS) for 10 minutes, on ice, as described.14 Subsequently, cells were incubated with 0.1 mg/mL RNase A in 2× SSC for 30 minutes at 37°C. Before the hybridization, cells were denatured in 50% formamide in 2× SSC for 20 minutes at 80°C.15
Molecular probes and in situ hybridization.
The PML, RARα, ABL, and BCR (Mbcr) genes were visualized with specific probes purchased from Oncor (Gaithersburg, MD) according to the instructions of the manufacturer.
A c-DNA probe for the β-globin gene (a 3.7-kb ClaI/Kpn I fragment; kindly provided by Dr Mike Antoniou, Guy’s Hospital, London, UK) was nick-translated with dinitrophenyl-11-dUTP (DNP; Molecular Probes, Leiden, The Netherlands), as described by Johnson et al.16 The β-globin genes (GLB) were used in this study as a control, representing gene sequences that are not usually involved in recombination phenomena in leukemic cells.
Chromosome painting probes for chromosomes 9 and 11 (digoxigenin-labeled; Oncor) and 15 (biotin-labeled; Cambio, Cambridge, UK) were hybridized and detected according to the instructions of the manufacturer. For double-hybridization experiments, additional probes for chromosomes 9, 17, and 22 were kindly provided by Dr J. Gray (Biomedical Science Division, Lawrence Livermore National Laboratory, San Francisco, CA)17 and nick-translated with biotin-16-UTP (Boehringer Mannheim, Mannheim, Germany; chromosome 9) or digoxigenin-11-dUTP (Boehringer Mannheim; chromosomes 17 and 22). For each sample, 30 ng/μL of each labeled probe (in the case of noncommercial painting probes for chromosomes 15 and 17) or 100 ng/mL (for chromosome 9 biotin-labeled probe) were combined with Human Cot 1 DNA (GIBCO-BRL; 0.560 mg/mL in the case of probe for chromosome 9 and 2 mg/mL for the remaining probes), ethanol-precipitated, air-dried, and dissolved in hybridization buffer (50% formamide, 2× SSC, 10% dextran sulphate, 50 mmol/L phosphate buffer, pH 7.0).
All the procedures for in situ hybridization, signal detection, and simultaneous visualization of the nuclear envelope were performed as previously described.13,18 19 The probe for the GLB genes was denatured for 10 minutes at 75°C, combined with the other gene-specific probes, and, after the hybridization procedure, detected with a rabbit anti-DNP antibody (1:100; Molecular Probes) at 37°C for 30 minutes and washed in 0.05% Tween 20/PBS (3× for 5 minutes), followed by incubation with a Texas-Red–conjugated goat antirabbit Ig (1:50; Jackson Immunoresearch Laboratories, West Grove, PA). For the simultaneous visualization of hybridization sites and the nuclear envelope, the samples were incubated immediately after the detection of the hybridization signals with a rabbit anti–lamin-B antibody (kindly provided by Dr S. Georgatos, Heidelberg, Germany), diluted at 1:100 at 37°C for 30 minutes and washed as described, followed by incubation with goat Texas-Red–conjugated antirabbit Ig (Jackson Immunoresearch Laboratories) at 37°C for 30 minutes.
Identification of the Cell Cycle Phases
Cells were cultivated under the presence of Bromodeoxyuridin (BrdU) as previously described.13 Briefly, BrdU (Boehringer Mannheim) was added at 10 μmol/L to the cell culture for 20 minutes, followed by fixation and permeabilization procedures. The detection of incorporated BrdU was performed after the in situ hybridization step using a Cy5-conjugated anti-BrdU antibody (1:50; Jackson Immunoresearch Laboratories). In these conditions, it is possible to identify cells in the S-phase (those that incorporated BrdU) as well as to discriminate different stages within the S-phase based on the morphological patterns of BrdU incorporation.20
To identify cells in the G2 phase of the cell cycle, the following pulse and chase strategy was used: the duration of the G2 phase was first determined in the IM-9 cell line according to the methods previously described21 22 and found to be approximately 2 hours and 30 minutes. On the basis of this information, the cells were then cultured in the presence of BrdU at 10 μmol/L for 20 minutes, followed by the removal of the culture medium by centrifugation at 50g for 5 minutes and subsequently cultured in fresh medium without BrdU for a period of time inferior to the duration of G2 (2 hours and 15 minutes). At this time point, the cells were harvested, fixed, permeabilized, and hybridized as described above. In these conditions, those cells exhibiting a late-S pattern of BrdU incorporation were considered to be in G2 and were selected for analysis of intergenic distances (assuming that the duration of G2 does not vary markedly in different cell types, the same value was accepted for the duration of G2 in bone marrow cells).
In IM-9 cells, the intergenic distances were also analyzed along the sequential phases of mitosis (prophase, metaphase, anaphase, and telophase) that were directly identified under the microscope by DNA staining with 4′6-diamidino-2-phenylindole (DAPI).
Determination of Intergenic Distances
Criterion for gene proximity.
Intergenic distances ≤2 μm were used as a criterion for gene proximity. The rationale was the following. First, mathematical models emerging from studies of clastogenesis induced by ionizing irradiation indicate that “free ends from double strand breaks initially formed as much as ∼2 μm apart can apparently interact,”6 a possibility that implies a certain degree of freedom as to the movement of chromatin within the nucleoplasm. Second, the latter assumption is supported by recent in vivo observations in which it is shown that particular chromatin segments do indeed undergo free Brownian movements within a constrained subregion, the radius of which is 0.5 μm in yeast nuclei and 0.9 μm in Drosophila embryo nuclei.23Accordingly, an absolute requirement for two loci to interact would be the overlap of their confined regions,23 which could easily happen if two chromatin segments are separated by less than 2 μm in a human hematopoietic cell nucleus with a diameter of 7 to 8 μm. Third, further evidence suggesting that a distance of 2 μm may be sufficient for the establishment of functional (physical?) interactions between two distinct chromatin regions is that provided by the work of LaSalle and Lalande,24 in which it is shown that the imprinted homologous loci of Prader-Willi and Angelman syndromes become transiently associated, at distances ≤2 μm, in the nucleus of T-lymphoid cells during a short period of the S phase. Taken together, the above arguments indicate that what could have been considered, at first sight, a substantial distance for an average nucleus with a diameter of 8 μm may well be compatible with the physical interaction of two distinct chromatin regions.
Confocal microscopy analysis.
The analysis of hybridization signals in 3-D preserved nuclei was performed with the confocal microscope Zeiss LSM-410 (Carl Zeiss, Oberkochen, Germany) as previously described.18 19 The nuclei were selected for analysis according to the following criteria: adequate morphological preservation of chromatin (as assessed by DAPI staining); integrity of the nuclear envelope (visualized with an anti-lamin B antibody); and identification of all alleles by the presence of distinct hybridization signals.
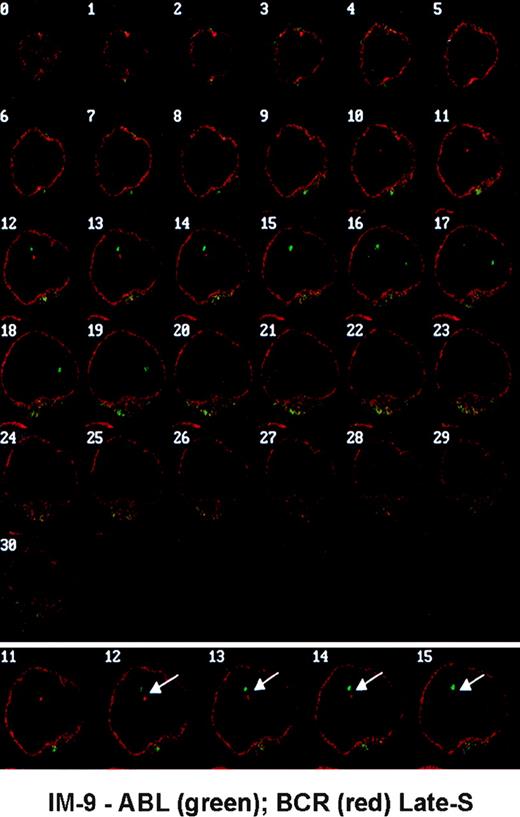
Thirty-one optical sections were obtained for each nucleus. Because in almost all cells the hybridization signal for each gene could be seen in several sequential planes, the section containing the brightest signal was selected and recorded for further analysis with a macro specifically developed for the NIH Image 1.55 software (Wayne Rasbaud, NIH, Springfield, VA). For each gene, the absolute coordinates of the center of each hybridization signal were determined (15 pixels = 1 μm in the x and y axes; the average increment between sections along the z axis was 0.25 μm, varying from 0.2 to 0.3 μm). Each of the distances between homologous genes (dx-x) and the minimum distance between heterologous genes (dx-y) were then calculated. One example of optical series obtained in IM-9 cells with closely associated ABL and BCR genes is shown in Fig 1.
Optical series obtained in IM-9 cells hybridized with gene-specific probes showing a nucleus from a late-S cell with closely associated ABL (green) and BCR (red) genes (arrows). The nuclear envelope is labeled with an anti-lamin B antibody (red).
Optical series obtained in IM-9 cells hybridized with gene-specific probes showing a nucleus from a late-S cell with closely associated ABL (green) and BCR (red) genes (arrows). The nuclear envelope is labeled with an anti-lamin B antibody (red).
Statistical Analysis
The χ2 test was applied to investigate the hypothesis that the proportion of cells with dx-y ≤2 μm was the same throughout the subphases of the cell cycle for each pair of genes studied. When the null hypothesis was rejected, multiple comparisons were made between successive subphases using multiple χ2tests. A similar strategy was used to investigate whether the proportion of cells with dx-y ≤2 μm was the same between each pair of genes and its control. The level of significance chosen for all the tests was α = .05. A total of 2,864 nuclei were analyzed, 50 for each pair of genes and each subphase of the cell cycle, except in G2 (30 to 35 nuclei) and distances between the ABL and PML genes and the control gene GLB in interphase (30 nuclei for each subphase and pair of genes). Because the procedures for BrdU pulse and chase experiments (see above) were consistently accompanied by inadequate 3-D preservation in freshly eluted CD34+ cells, the G2 phase of the cell cycle was not investigated in unstimulated CD34+cells. The data concerning the PML-RARα and ABL-BCR interdistances in unstimulated CD34+ cells in G1 were obtained from two independent bone marrow donors: cells with intergenic distances ≤2 μm for PML and RARα genes were 50% and 52%, respectively (P = .932), and for ABL and BCR genes were 50% and 62%, respectively (P = .536).
RESULTS
Intergenic Distances PML-RARα and ABL-BCR in IM-9 Cells Throughout the Cell Cycle
The analysis of intergenic distances in IM-9 cells grown in the presence of BrdU, in conditions that allow the discrimination of G1, early-S, middle-S, late-S, and G2, shows that (Fig 2) (1) in all phases of the cell cycle there are cells in which the intergenic distances are ≤2 μm and (2) the proportion of cells with closely associated genes vary during the cell cycle. As to the PML and RARα genes in interphase (Fig 2A), this proportion decreases from G1 to early-S (proportion ± standard error; 41% ± 0.074% and 17% ± 0.057%, respectively;P = .013), remaining stable during early-S and middle-S (16% ± 0.056%). At the transition of late-S to G2, a gradual increase in the proportion of cells with gene proximity was observed (21% ± 0.063% to 29% ± 0.071%, respectively) that persists in G2 and prophase (30% ± 0.062% in prophase), reaches a maximum in metaphase (64% ± 0.071%; prophase v metaphase, P< .001), decreases in anaphase (41% ± 0.068%; metaphasev anaphase, P = .023), and returns to values similar to those of G1 in telophase (31% ± 0.076%; Fig 2A).
Distribution of IM-9 cells with intergenic distances ≤2 μm during the cell cycle. (A) PML-RAR interdistances. (B) ABL-BCR interdistances. See text (Results) for statistically significant differences. E-S, early-S; M-S, middle-S; L-S, late-S; Pro, prophase; Met, metaphase; Ana, anaphase; Tel, telophase.
Distribution of IM-9 cells with intergenic distances ≤2 μm during the cell cycle. (A) PML-RAR interdistances. (B) ABL-BCR interdistances. See text (Results) for statistically significant differences. E-S, early-S; M-S, middle-S; L-S, late-S; Pro, prophase; Met, metaphase; Ana, anaphase; Tel, telophase.
The analysis of the distances between the genes ABL and BCR shows that the proportion of cells with intergenic distances ≤2 μm is stable during G1 and all S phase (G1, 22% ± 0.061%; early-S, 23% ± 0.060%; middle-S, 27% ± 0.063%; and late-S, 18% ± 0.056%; Fig 2B). Again, after late-S, a progressive increase of cells with close ABL and BCR genes, which persists in G2 and prophase (late-Sv G2, P = .016), can be observed (36% ± 0.074% and 34% ± 0.072%, respectively). However, in contrast with the PML and RARα genes, metaphase is the phase of the cell cycle in which the proportion of cells with close ABL-BCR genes reaches its minimum (11% ± 0.045%; prophase v metaphase, P = .007). A return to values of G1 is again observed in telophase (14% ± 0.051% in anaphase and 27% ± 0.065% in telophase; Fig 2B).
In summary, the data show that the distances between PML and RARα and between ABL and BCR genes vary along the sequential phases of the cell cycle. Also observed is a common emerging pattern, which consists in an increase of cells with gene proximity beyond the late stages of S phase that persists during G2 and prophase and a return to a G1 pattern of interdistances in telophase.
Topography of Chromosomes 15, 17, 9, and 22 During Mitosis in IM9 Cells
To investigate the reasons for the opposite behavior of PML-RARα and ABL-BCR interdistances in metaphase, and assuming that they might be caused by constraints inherent to the formation of the metaphase plate, we subsequently proceeded to the analysis of the spatial relationships between the chromosomes where these genes are located during the stages of mitosis. Thus, genomic libraries specific to chromosomes 15, 17, 9, and 22 were used in double hybridization experiments in 3-D preserved mitotic cells. The results obtained are shown in Fig 3. A high percentage of cells with close proximity of chromosomes 15 and 17 (Fig 3A) and chromosomes 9 and 22 (Fig 3B) was observed in all stages of mitosis, which differed significantly from distances between chromosomes 15 and 11 and 9 and 11: proximities between chromosomes 15 and 17 versus chromosomes 11 and 17 were P < .05 in metaphase and anaphase; and proximities between chromosomes 9 and 22 versus 9 and 11 were P< .05 in metaphase and telophase. It is noteworthy that, in metaphase, chromosomes 9 and 22 were adjacent in more than 60% of cells, contrasting with the ABL and BCR genes, which were found to be in close proximity in only 11% of metaphase cells (see Figs 2B and3B). The reason for this discrepancy became evident when observing the arrangement of chromosomes in the metaphase plate. In fact, during metaphase, the chromosomes always assume a spatial arrangement in which the centromeres are oriented toward the center of the rosette, whereas the long and short arms are extended towards the periphery (see diagram in Fig 4). Because the ABL gene is a subtelomeric gene in the long arm of chromosome 9, which is substantially larger than chromosome 22, and the BCR gene is located not very far from the centromere of chromosome 22, one of the shortest human chromosomes, the two genes are forced to move apart during metaphase, even if they have been in close proximity in earlier stages of the cell cycle (see illustrating examples in Fig4). Such a behavior is not observed for the PML and RARα genes, because both are located approximately at the middle of two short chromosomes with similar sizes.
Distribution of IM-9 cells with adjacent chromosomes during mitosis. (A) Proximities between chromosomes 15 and 17 versus chromosomes 11 and 17: P < .05 in metaphase and anaphase. (B) Proximities between chromosomes 9 and 22 versus chromosomes 9 and 11:P < .05 in metaphase and telophase. The legends are as in Fig 2.
Distribution of IM-9 cells with adjacent chromosomes during mitosis. (A) Proximities between chromosomes 15 and 17 versus chromosomes 11 and 17: P < .05 in metaphase and anaphase. (B) Proximities between chromosomes 9 and 22 versus chromosomes 9 and 11:P < .05 in metaphase and telophase. The legends are as in Fig 2.
Optical sections from three IM-9 cells in metaphase hybridized with painting probes for chromosomes 9 (red) and 22 (green). Note that the signal corresponding to chromosome 22 is smaller and more centrally located than that of chromosome 9. The diagram on the right represents the arrangement of chromosomes in a metaphase rosette and its potential effect on ABL-BCR and PML-RAR interdistances.
Optical sections from three IM-9 cells in metaphase hybridized with painting probes for chromosomes 9 (red) and 22 (green). Note that the signal corresponding to chromosome 22 is smaller and more centrally located than that of chromosome 9. The diagram on the right represents the arrangement of chromosomes in a metaphase rosette and its potential effect on ABL-BCR and PML-RAR interdistances.
It is thus possible to conclude that intergenic distances during metaphase are strongly influenced by physical and geometrical constraints related to the location of the gene in the chromosome, the size of the chromosome, and the geometry of the metaphase plate.
Comparison of PML-RARα and ABL-BCR Interdistances With Those Between PML and β-Globin and ABL and β-Globin Genes in IM-9 Cells
Having determined the dynamics of intergenic distances during the cell cycle, we subsequently asked whether the observed proximity was specific to these pairs of genes. Therefore, the PML-RARα and ABL-BCR interdistances were compared with those between PML-GLB and ABL-GLB in all stages of the cell cycle. As shown in Fig 5A and B, there are no significant differences during G1, early-S, and middle-S between the two cell populations (all P > .05). However, a clear divergence was observed after late-S, with a progressive reduction in the proportion of cells with close PML-GLB (31% ± 0.076% in late-S and 19% ± 0.065% in G2; Fig 5A) and ABL-GLB (27% ± 0.075% in late-S and 20% ± 0.080% in G2; Fig 5B) as opposed to the increase of cells with close PML-RARα (21% ± 0.063% in late-S and 29% ± 0.071% in G2) and ABL-BCR (18% ± 0.056% in late-S and 36% ± 0.074% in G2). Therefore, it appears that the PML and RARα or ABL and BCR are specifically associated during G2 and prophase in IM-9 cells.
Distribution of IM-9 cells with intergenic distances ≤2 μm during the cell cycle. (A) Comparison of distances PML-RAR with PML-βGLB. (B) Comparison of distances ABL-BCR with ABL-βGLB. (C) Comparison of distances PML-RAR with PML-PML and RAR-RAR. (D) Comparison of distances ABL-BCR with ABL-ABL and BCR-BCR. See text (Results) for statistically significant differences. The legends are as in Fig 2.
Distribution of IM-9 cells with intergenic distances ≤2 μm during the cell cycle. (A) Comparison of distances PML-RAR with PML-βGLB. (B) Comparison of distances ABL-BCR with ABL-βGLB. (C) Comparison of distances PML-RAR with PML-PML and RAR-RAR. (D) Comparison of distances ABL-BCR with ABL-ABL and BCR-BCR. See text (Results) for statistically significant differences. The legends are as in Fig 2.
The comparison of the distances PML-RARα and ABL-BCR with those between homologous genes (Fig 5C and D) also shows that distances between heterologous genes are shorter than those between homologous genes. Statistically significant differences are as follows: for PML-RARα versus PML-PML, P < .05 in G1, G2, metaphase, and anaphase; for PML-RARα versus RARα-RARα, P < .05 in G2, prophase, metaphase, and anaphase; for ABL-BCR versus ABL-ABL,P < .05 in G1, late-S, G2, prophase, and telophase and for ABL-BCR versus BCR-BCR in G1 and middle-S.
PML-RARα and ABL-BCR Interdistances in Immature Normal Hematopoietic Cells
Having studied the intergenic distances in B-lymphoid cells, the analysis has subsequently progressed to normal hematopoietic progenitors in interphase.
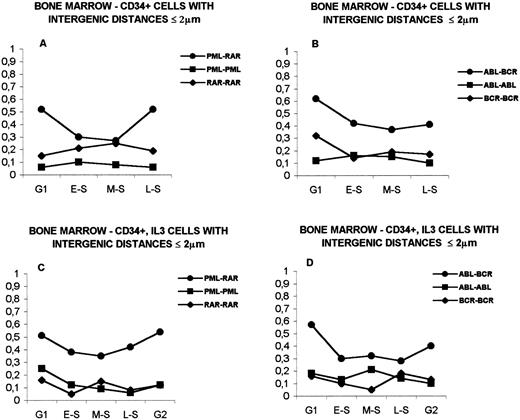
The analysis of PML and RARα genes in unstimulated CD34+in G1 and S phases (G2 not performed) is shown in Fig 6A. As in IM-9 cells, variations along the sequential stages of the cell cycle can be observed. The proportion of cells with gene proximity were 52% ± 0.072% in G1, 30% ± 0.066% in early-S, 27% ± 0.061% in middle-S, and 39% ± 0.089% in late-S (P = .027 at the transitions of G1 to early-S and of middle-S to late-S). However, the proportion of CD34+ cells with close PML and RARα genes is always higher than that observed in IM-9 cells in all subphases analyzed (P = .007 in late-S; see Fig 8A). Furthermore, in contrast with IM-9 cells, the proportion of cells with PML-RARα proximity is always higher than that of cells with PML-GLB proximity (cells with close PML-GLB genes were 38% ± 0.068% in G1, 23% ± 0.075% in early-S, 16% ± 0.066% in middle-S, and 7% ± 0.089% in late-S), a difference that becomes more noticeable after middle-S (P < .0001 in late-S; Fig 6A).
Distribution of bone marrow cells with intergenic distances ≤2 μm during interphase. (A) CD34+ cells. Comparison of distances PML-RAR with PML-βGLB. (B) CD34+ cells. Comparison of ABL-BCR with ABL-βGLB. (C) CD34+ cells stimulated with IL-3. Comparison of distances PML-RAR with PML-βGLB. (D) CD34+ cells stimulated with IL-3. Comparison of distances ABL-BCR with ABL-βGLB. See text (Results) for statistically significant differences. The legends are as in Fig 2.
Distribution of bone marrow cells with intergenic distances ≤2 μm during interphase. (A) CD34+ cells. Comparison of distances PML-RAR with PML-βGLB. (B) CD34+ cells. Comparison of ABL-BCR with ABL-βGLB. (C) CD34+ cells stimulated with IL-3. Comparison of distances PML-RAR with PML-βGLB. (D) CD34+ cells stimulated with IL-3. Comparison of distances ABL-BCR with ABL-βGLB. See text (Results) for statistically significant differences. The legends are as in Fig 2.
The analysis of ABL-BCR distances in unstimulated CD34+cells is shown in Fig 6B. Cells with close ABL and BCR genes were 56% ± 0.068% in G1, 42% ± 0.069% in early-S, 37% ± 0.066% in middle-S, and 41% ± 0.091% in late-S (P = .045 at the transition of G1 to early-S). Similar to what was observed for the PML and RARα genes, the proportion of cells with close ABL and BCR genes is higher than those with ABL-GLB proximity (P < .05 in early-S; cells with close ABL and GLB genes were 41% ± 0.091% in G1, 14% ± 0.066% in early-S, 30% ± 0.083% in middle-S, and 20% ± 0.070% in late-S) and markedly higher than in IM-9 cells, with statistically significant differences in G1 (P < .0001), early-S (P = .044), and late-S (P = .026) (see Fig 8B).
The results obtained in CD34+ stimulated with IL-3 for 48 hours, in interphase, are depicted in Fig 6C and D. There is again a higher proportion of cells with close PML-RARα than with close PML-GLB (respectively: G1, 51% ± 0.070% and 30% ± 0.083%; early-S, 38% ± 0.067% and 19% ± 0.070%; middle-S, 35% ± 0.065% and 27% ± 0.080%; late-S, 42% ± 0.069% and 23% ± 0.077%; G2, 55% ± 0.086% and 24% ± 0.079%;P = .015 in G2; Fig 6C) and with close ABL-BCR genes than with close ABL-GLB (respectively: G1, 57% ± 0.070% and 26% ± 0.078%; early-S, 30% ± 0.072% and 33% ± 0.086%; middle-S, 33% ± 0.071% and 26% ± 0.078%; late-S, 28% ± 0.094% and 13% ± 0.062%; G2, 40% ± 0.086% and 20% ± 0.073%; P = .006 in G1 and P = .019 in late-S; Fig 6D). The distances ABL-BCR differ significantly at the transition G1-early S (P = .01). In IL-3–stimulated cells, the proportions of cells with close PML and RARα genes were higher than in IM-9 cells (P = .020 in early-S, P = .043 in middle-S, and P = .036 in late-S; see Fig 8A), the same happening for interdistances ABL-BCR (P = .001 in G1; see Fig8B).
The proportion of cells with PML-RARα or ABL-BCR proximity is also higher than that with proximity between homologous genes in CD34+ cells with or without stimulation with IL-3, a difference that is more conspicuous than that observed in the lymphoid cell line (Fig 7; see also Fig 5). In CD34+ cells, PML-RARα interdistances differed significantly from PML-PML in all subphases (P < .005) and from RARα-RARα in G1 and late-S (P < .005; Fig 7A); the ABL-BCR distances differed significantly from ABL-ABL and from BCR-BCR in all subphases (P < .0001 in G1 and P < .05 in the remaining phases for ABL-BCR v ABL-ABL; P< .005 in G1 and early-S and P < .05 in middle-S and late-S for ABL-BCR v BCR-BCR; Fig 7B).
Distribution of bone marrow cells with intergenic distances ≤2 μm during interphase. (A) CD34+ cells. Distances PML-RAR versus PML-PML and PML-RAR versus RAR-RAR. (B) CD34+ cells. Distances ABL-BCR versus ABL-ABL and ABL-BCR versus BCR-BCR. (C) CD34+ cells stimulated with IL-3. Distances PML-RAR versus PML-PML and PML-RAR versus RAR-RAR. (D) CD34+ cells stimulated with IL-3. Distances ABL-BCR versus ABL-ABL and ABL-BCR versus BCR-BCR. See text (Results) for statistically significant differences. The legends are as in Fig 2.
Distribution of bone marrow cells with intergenic distances ≤2 μm during interphase. (A) CD34+ cells. Distances PML-RAR versus PML-PML and PML-RAR versus RAR-RAR. (B) CD34+ cells. Distances ABL-BCR versus ABL-ABL and ABL-BCR versus BCR-BCR. (C) CD34+ cells stimulated with IL-3. Distances PML-RAR versus PML-PML and PML-RAR versus RAR-RAR. (D) CD34+ cells stimulated with IL-3. Distances ABL-BCR versus ABL-ABL and ABL-BCR versus BCR-BCR. See text (Results) for statistically significant differences. The legends are as in Fig 2.
In CD34+ cells stimulated with IL-3, interdistances PML-RARα differed significantly from PML-PML and RARα-RARα in all subphases (P < .05 in G1 and P < .0005 in the remaining phases for PML-RARα v PML-PML; P < .0001 in G1, early-S, late-S, and G2 and P < .05 in middle-S for PML-RARα v RARα-RARα; Fig 7C). The distances ABL-BCR differed from ABL-ABL in all subphases except middle-S (P < .0001 in G1, early-S, late-S, and G2) and from BCR-BCR in G1 (P< .0001), early-S (P < .05), and middle-S (P < .0005) (Fig 7D).
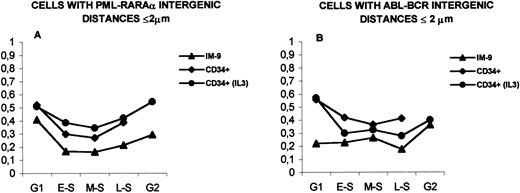
When the distances PML-RARα and ABL-BCR in unstimulated CD34+ cells were compared with those observed in IL-3–stimulated CD34+ cells, no statistically significant differences were found. However, a higher proximity for the ABL and BCR genes was observed in unstimulated CD34+ cells (Fig 8B), whereas the distances between PML and RARα were shorter in cells stimulated with IL-3 (Fig 8A).
Comparison of bone marrow and IM-9 cells with intergenic distances ≤2 μm during interphase. (A) Distances PML-RAR in IM-9 cells, CD34+ cells and CD34+ cells stimulated with IL-3. (B) Distances ABL-BCR in IM-9 cells, CD34+ cells and in CD34+ cells stimulated with IL-3. See text (Results) for statistically significant differences. The legends are as in Fig 2.
Comparison of bone marrow and IM-9 cells with intergenic distances ≤2 μm during interphase. (A) Distances PML-RAR in IM-9 cells, CD34+ cells and CD34+ cells stimulated with IL-3. (B) Distances ABL-BCR in IM-9 cells, CD34+ cells and in CD34+ cells stimulated with IL-3. See text (Results) for statistically significant differences. The legends are as in Fig 2.
In summary, the analysis of intergenic distances in early hematopoietic precursors shows that, similar to what happens in IM-9 cells, there are variations in intergenic distances during interphase. It was also observed that the proportion of bone marrow cells in interphase with close proximity between PML and RARα or ABL and BCR genes is higher than that observed in IM-9 cells. Furthermore, a higher ABL-BCR proximity in unstimulated CD34+ cells and a higher PML-RARα proximity was observed in cells that were stimulated with IL-3 for 48 hours.
DISCUSSION
In this work, we have measured the distances between PML and RARα and between ABL and BCR genes in 3-D preserved hematopoietic cells belonging to different cell lineages at various stages of differentiation and of the cell cycle. We observed that each pair of genes tends to become associated at the transition of late-S to G2 in all cell types analyzed, a behavior not observed for distances between ABL or PML and the β-globin genes. Furthermore, the proportion of cells in which PML and RARα or ABL and BCR are closely associated is higher in hematopoietic precursors than in B-lymphoid cells, in contrast with distances between ABL or PML and the β-globin genes, which are not affected by cell type.
Because the cell cycle phase is known to influence the topography of chromatin within the nucleus,25-27 we first asked whether intergenic distances change during the sequential phases of the cell cycle. The results in IM-9 cells show that distances between heterologous genes vary periodically during the cell cycle as a return to a G1-pattern of gene interdistances was consistently observed at telophase. Furthermore, the observed fluctuations in the percentage of cells with gene proximity indicate that in each phase of the cell cycle there is a proportion of cells in which all the different pairs of loci under analysis are in close proximity. However, when PML-RARα or ABL-BCR interdistances were compared with those between PML, or ABL, and the control gene, the emerging pattern was that, starting at the transition of late-S to G2, there is a trend towards a greater proximity between the PML and RARα, or ABL and BCR genes, as opposed to distances between PML or ABL and the β-globin gene. These findings contrast with those reported by Kozubek et al7 in which distances between the ABL and BCR genes were found to increase during S/G2 in human lymphocytes. This discrepancy might be explained, at least partly, by cells in S and G2 phases having been merged into a single cell population and analyzed as such.7 This could result in a dilution effect of G2 cells, due to the short duration of G2 as compared with S phase28 that, according to our results, would bias the measured distances towards the greater values we consistently observed in S phase cells. Consequently, the trend towards greater gene proximity observed at the transition of late-S to G2 would have been missed had not these subphases been specifically discriminated.
The data obtained in IM-9 cells show that the proximity between the ABL and BCR and between PML and RARα genes persists during G2 and prophase. At metaphase, the pattern of intergenic distances changes abruptly, with cells having PML and RARα genes in close proximity reaching its maximum (64% of metaphase cells had PML-RARα interdistances ≤2 μm), whereas cells with close ABL and BCR genes reached its minimum (10%). The analysis of the spatial relationships between the chromosomes where those genes are located showed that at least one chromosome 9 (containing the ABL gene) and one chromosome 22 (containing the BCR gene) were in adjacent positions in more than 60% of the metaphases analyzed. It was thus clear that the increase in distance between the ABL and BCR genes in metaphase was not due to a separation of the respective chromosomes during this stage of the cell cycle, but rather to physical constraints imposed by the size of the chromosome, the chromosomal location of the gene, and the geometrical arrangement of the metaphase rosette.
Having studied the intergenic distances in B-lymphoid cells, the analysis has subsequently progressed to normal hematopoietic progenitors in distinct stages of differentiation. We observed that, similar to IM-9 cells, intergenic distances change during interphase. A high percentage of cells with closely associated genes was always present in G1 (for each pair of genes), which decreased at early-S to start augmenting again in middle-S or late-S. These findings thus show that a tendency towards a reassociation of these genes in the late stages of interphase is present in hematopoietic cells of different cell lineages and in distinct stages of cell differentiation. However, the most striking aspects of this analysis are the higher gene proximity in bone marrow cells than in IM-9 cells and the trend toward a closer association of ABL and BCR in unstimulated CD34+when compared with IL-3–treated cells, the opposite pattern being observed for the PML and RARα genes. It seems then that, in hematopoietic cells of different lineages and in distinct stages of differentiation, there is always a proportion of cells, varying in different cell types and stages of the cell cycle, in which the ABL and BCR, or PML and RARα genes, are separated by a distance that is theoretically compatible with their physical contact (see criterion for gene proximity in Materials and Methods) and, presumably, with their reciprocal recombination. The higher proximity of these genes in hematopoietic precursors would favor their recombination in those cells. Likewise, the higher proximity of the ABL and BCR genes in CD34+ cells (which correspond to an immature cell population that contains the pool of hematopoietic stem cells), and of the PML and RARα genes in cells treated with IL-3 (mostly committed to myeloid differentiation), shows an interesting correlation with the type of cells in which these gene rearrangements are thought to occur in vivo. However, the fact that in some cells the control gene is also in close proximity to the ABL or PML genes equally reinforces the notion that the proximity between loci, necessary as it may be, cannot account solely for recombination phenomena in somatic cells. In this context, the pattern of variation of intergenic distances during the cell cycle is of special interest. In fact, if the nature of chromosomal arrangements in prometaphase and metaphase explain the variations observed for the PML-RARα and ABL-BCR interdistances at the prophase-metaphase transition, the same cannot be said about their behavior in interphase, where little is known as to the spatial distribution of chromosomes and the rules that underlie that arrangement (see previous reviews29-32). Especially intriguing was the association of these genes at the S-G2 transition and its persistence during G2 and prophase. The specificity of the phenomenon raises the possibility that such an association might obey some, as yet unknown, functional imperative. The already mentioned transient association between homologous imprinted genes, presumably for trans-regulation effects,24 is an example that phenomena of this type may occur and be part of the cell strategies for epigenetic regulation of gene transcription. If this is so, it is then likely that the recombinogenic potential of gene proximity may differ in distinct phases of the cell cycle and/or in different cell types. In other words, the specific association of these loci in G2 and prophase in hematopoietic precursors might reflect an intrinsic dynamical behavior that is established early in hematopoietic development and perpetuated in different cell lineages, but whose putative functional meaning may be restricted to a short window of hematopoietic differentiation. If, for the sake of functionality, the spatial association of these genes is accompanied by local chromatin decondensation, then the conditions that might facilitate their reciprocal rearrangement may well be created. There are, of course, other alternatives. One is that the recombination may occur with similar frequencies in different hematopoietic cell types, but only cause cell immortalization in specific moments of differentiation. This possibility would explain the rare finding of BCR-ABL transcripts in the peripheral blood of healthy individuals.33 Another possibility is that these translocations may occur rarely, being frequently observed due to the selective advantage that the fusion protein may confer in a specific cell type.
The uncovering of the biological role of the proteins coded by these genes in normal hematopoiesis will hopefully help to clarify these issues.
ACKNOWLEDGMENT
The authors are grateful to Dr S. Georgatos for the anti-lamin B antibody, to Dr Michael Antoniou for the β-globin probe, to Dr Joe Gray for the chromosome-specific libraries, to Amgen (Thousand Oaks, CA) for the MiniMACS isolation system, and to Sandoz-Pharma for the gift of recombinant IL-3.
Supported by Program PRAXIS XXI. H.N. is supported by a fellowship from Junta Nacional de Investigação Cientı́fica.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Leonor Parreira, MD, PhD, Instituto de Histologia e Embriologia, Faculdade de Medicina de Lisboa, Av. Prof. Egas Moniz 1699 Lisboa Codex, Portugal; e-mail: hleonor@fml.fm.ul.pt.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal