Abstract
The mi locus of mice encodes a transcription factor of the basic-helix-loop-helix-leucine zipper protein family (MITF). The MITF encoded by the mutant mi allele (mi-MITF) deletes 1 of 4 consecutive arginines in the basic domain. The mice of mi/migenotype express mi-MITF, whereas the mice of tg/tggenotype have a transgene at the 5′ flanking region of themi gene and do not express any MITF. To investigate the function of mi-MITF in cultured mast cells (CMCs), we took two approaches. First, mRNA obtained from mi/mi CMCs ortg/tg CMCs was subtracted from complementary (c) DNA library of normal (+/+) CMCs, and the (+/+-mi/mi) and (+/+-tg/tg) subtraction libraries were obtained. When the number of clones that hybridized more efficiently with +/+ CMC cDNA probe than with mi/mi or tg/tg CMC cDNA probe was compared using Southern analysis, the number was larger in the (+/+-mi/mi) library than in the (+/+-tg/tg) library. Second, we compared mRNA expression of six genes betweenmi/mi and tg/tg CMCs by Northern analysis. The transcription of three genes encoding mouse mast cell proteases was impaired in both mi/mi and tg/tg CMCs. On the other hand, the transcription of three genes encoding c-kit receptor, tryptophan hydroxylase, and granzyme B was markedly reduced inmi/mi CMCs, but the reduction was significantly smaller intg/tg CMCs. These results indicated the inhibitory effect ofmi-MITF on the transactivation of particular genes in CMCs.
THE MUTANT MICE OF mi/mi genotype were found by Hertwig1,2 among the offsprings of an X-irradiated male mouse. The mi/mi mice show microphthalmia, depletion of pigment in both hair and eyes, osteopetrosis, and decrease in number of mast cells.3-7 The mutant gene was introduced into C57BL/6 background and has been kept in the Jackson Laboratory (Bar Harbor, ME). The mi locus was demonstrated to encode a member of the basic-helix-loop-helix-leucine zipper (bHLH-Zip) protein family of transcription factors (hereafter calledmi-transcription factor [MITF]).8,9 The MITF encoded by the mutant mi allele deletes 1 of 4 consecutive arginines in the basic domain (hereaftermi-MITF).8,10,11 The mi-MITF is defective in the DNA binding activity and the nuclear localization potential,12,13 and it does not transactivate target genes.13-17
A VGA-9-tg/tg transgenic mouse possessing the transgene-insertional mutation at the 5′ flanking region of themi gene was produced by Hodgkinson et al8 and Tachibana et al.18 The expression of MITF transcripts was undetectable in various tissues of VGA-9-tg/tgmice except the embryonal retina.8 We introduced thetg transgene into the C57BL/6 background in the Osaka University Medical School (Osaka, Japan). C57BL/6-tg/tg and C57BL/6-mi/mi mice share several phenotypic features, such as microphthalmia, white coat color, and the decrease of mast cells. However, they are clearly distinguishable from each other at least in one respect: the C57BL/6-tg/tg mice do not show osteopetrosis. This fact indicates that the presence of mi-MITF is associated with a more severe impairment than the absence of normal MITF (+-MITF). Although the osteopetrosis of mi/mi mice is attributable to the deficiency of osteoclasts,19,20 the effect of MITF has been analyzed more intensely in mast cells than in osteoclasts. mRNA expression of mi-MITF in mi/mi cultured mast cells (CMCs) was comparable to that of +-MITF in +/+ CMCs,21 but the expression of MITF was not detectable in tg/tg CMCs even with reverse transcriptase modification of polymerase chain reaction (RT-PCR).22 In the present study, we investigated the inhibitory effect of mi-MITF on gene transactivation in CMCs.
Two approaches were taken. First, subtraction libraries of complementary (c) DNAs were produced and analyzed. We have recently elaborated cDNA library from +/+ CMCs and mi/mi CMCs and subtracted the latter from the former.21 The subtraction process was so successful that we could isolate two genes encoding granzyme (Gr) B and tryptophan hydroxylase (TPH) as novel MITF targets from the resulting subtracted cDNA library of (+/+-mi/mi).21 We applied the same subtraction procedures here to tg/tg CMCs. By comparing two subtracted libraries, we found that the (+/+-tg/tg) cDNA library contained +/+ CMC-specific clones at an apparently lower frequency than the (+/+-mi/mi) cDNA library. In addition to the Gr B and TPH genes, we have isolated several genes whose expression was reduced drastically in mi/mi CMCs.14-17 As the second approach, mRNA expression levels of such genes were compared betweenmi/mi and tg/tg CMCs by the Northern analysis. The expression of mouse mast cell protease (MMCP)-4, MMCP-5, and MMCP-6 was impaired in both mi/mi and tg/tg CMCs, but the reduction of expression of three genes encoding Gr B, TPH, and c-kit receptor was apparently smaller in tg/tg CMCs than in mi/mi CMCs. The results obtained by the two experiments consistently indicated that mi-MITF had an inhibitory effect on transcription of particular genes that are expressed in +/+ CMCs.
MATERIALS AND METHODS
Mice and cells.
The original stock of C57BL/6J-mi/+ (mi/+) mice was purchased from the Jackson Laboratory and was maintained in our laboratory by consecutive backcrosses with our own inbred C57BL/6 colony (more than 12 generations at the time of the present experiments). Female mi/+ mice were crossed with malemi/+ mice, and the resulting mi/mi mice were selected on the basis of their white coat color.3,4 The original stock of VGA-9-tg/tg mice,8 in which the mouse vasopressin-Escherichia coli-galactosidase transgene was integrated at the 5′ flanking region of the mi (MITF) gene, was kindly given by Dr H. Arnheiter (National Institutes of Health, Bethesda, MD). The integrated transgene was maintained by repeated backcrosses to our own inbred C57BL/6 colony (more than 12 generations at the time of the present experiments). Female and maletg/+ mice were crossed together, and the resultingtg/tg mice were selected by their white coat color.
Pokeweed mitogen-stimulated spleen cell conditioned medium (PWM-SCM) was prepared according to the method described by Nakahata et al.23 Mice of mi/mi or tg/tg genotype and their normal (+/+) littermates were used to obtain CMCs at an age of 2 to 3 weeks. Mice were killed by decapitation after ether anesthesia and the spleens were removed. Spleen cells derived from +/+, mi/mi,and tg/tg mice were cultured in α-minimal essential medium (α-MEM; ICN Biomedicals, Costa Mesa, CA) supplemented with 10% PWM-SCM and 10% fetal calf serum (FCS; Nippon Biosupp Center, Tokyo, Japan). Half of the medium was replaced every 7 days. Four weeks after initiation of the culture, more than 95% of cells were CMCs.24
The P-815 cell line was originally established from a mastocytoma of a DBA/2 mouse by Dunn and Potter25 and was supplied by the Japanese Cancer Research Bank (Tokyo, Japan). P-815 cells were cultured in α-MEM supplemented with 10% FCS.
Preparation of the subtracted cDNA library.
The detailed process for the preparation of the subtracted cDNA library was described previously.21 To prepare single-stranded plasmid DNA, the plasmid DNA prepared from the +/+ CMC cDNA library was introduced into E coli DH5αF′IQ cells by electroporation. After 1 hour of culture in rich medium (2× YT), transformed cells were infected with R408 helper phages. Single-stranded DNA was then purified from the supernatant of the 8-hour culture. To prepare biotinylated RNA drivers, total RNA was extracted by the guanidine thiocyanate/CsTFA method from mi/miand tg/tg CMCs, and poly (A)+ RNA was purified and labeled by photobiotin (Vector Laboratories, Burlingame, CA). One microgram of single-stranded DNA prepared from the +/+ CMC cDNA library was hybridized with 10 μg of biotinylated RNA at 42°C in 25 μL hybridization buffer containing 40% formamide, 50 mmol/L HEPES (pH 7.5), 1 mmol/L EDTA, 0.1% sodium dodecyl sulfate (SDS), 0.2 mol/L NaCl, and 1 μg of oligo-poly(rA). After hybridization for 42 hours, the mixture was transferred to 400 μL of SB (50 mmol/L HEPES [pH 7.5], 2 mmol/L EDTA, 500 mmol/L NaCl) and 10 μg of streptoavidin was subsequently added. The mixture was incubated at room temperature for 5 minutes and extracted with phenol/chloroform/isoamyl alcohol (25:24:1). The organic phase was back extracted with 100 μL TE (10 mmol/L Tris-HCl [pH 7.5], 1 mmol/L EDTA). The aqueous phases were pooled. Streptoavidin binding and phenol treatment were repeated once more. The recovered single-stranded plasmid DNA was subtracted with biotinylated RNA one more time. After repeating the subtraction process, the recovered single-stranded DNA was converted to double-stranded plasmid DNA by the BcaBEST DNA polymerase (TaKaRa, Otsu, Japan) reaction at 65°C for 30 minutes. After phenol extraction and ethanol precipitation, the DNA was dissolved in 20 μL TE buffer and 3 μL aliquots were introduced into E coliMC1061A cells by electroporation.
Screening of +/+ CMC-specific clones by Southern blot analysis.
Four hundred cDNA clones were prepared from ampicillin-resistant colonies randomly selected from the subtracted cDNA library by an automated plasmid purification machine (PI-100: KURABO, Osaka, Japan). After digestion with both Sma I and Not I to separate the cDNA insert, the plasmid DNA was electrophoresed in an agarose gel and transferred to nylon membranes. For positive controls, the mouse β-actin21 and human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) HindIII-Apa I26 cDNA fragments were also blotted. The cDNA probes were synthesized with reverse transcriptase from poly (A)+ RNA prepared from +/+,mi/mi, or tg/tg CMCs in a reaction mixture containing [α-32P]dCTP. Duplicate membranes were hybridized with the reverse transcribed cDNA probes for 15 hours. The filters were washed several times with a final stringency of 0.1× SSC (1× SSC = 0.15 mol/L NaCl, 15 mmol/L sodium citrate, pH 7.2) and 0.1% SDS at 50°C and autoradiographed at −80°C with an intensifying screen. The numbers of clones that hybridized with higher efficiency to the +/+ cDNA probe compared with the mi/mi ortg/tg cDNA probe were counted.
DNA sequencing.
The plasmid DNA was purified individually from the subtracted cDNA libraries by PI-100 and directly subjected to DNA sequencing. Dideoxy-chain termination sequencing reactions were performed with T7 dye-labeled primers and thermal cycle sequencing kits purchased from LI-COR (Lincoln, NE). The reaction products were analyzed by a Model 4000L Automated DNA Sequencer (LI-COR).
Northern blot analysis.
Five micrograms of total RNA prepared from +/+, mi/mi, ortg/tg CMCs was loaded per lane, fractionated on 1% agarose-formaldehyde gels, and transferred to nylon membranes by capillary action in 20× SSC. Baked membranes were prehybridized for 3 hours at 42°C in a buffer containing 50% formamide, 5× SSC, 5× Denhardt’s solution, and 0.1% SDS. The membranes were hybridized with the [α-32P]dCTP-labeled DNA probes at 42°C for 15 hours in the same buffer. Preparation of the DNA probes was performed according to the random hexamer labeling method. Preparation of MMCP-4, MMCP-5, MMCP-6, c-kit, TPH, Gr B, MITF, and β-actin cDNA probes was described previously.21 27 After hybridization, the membranes were washed to a final stringency of 0.1× SSC and 0.1% SDS at 50°C and autoradiographed at −80°C.
To characterize the subtracted cDNA libraries, 1 μg of theNot I-digested plasmid DNA prepared from the +/+ CMC cDNA library, the (+/+-mi/mi) subtracted library, or the (+/+-tg/tg) subtracted library was used as a template for the T7 RNA polymerase reaction (Stratagene, La Jolla, CA). Two micrograms of synthesized RNA was loaded per lane, fractionated on 1% agarose-formaldehyde gels, and transferred to nylon membranes by capillary action in 20× SSC. The hybridization procedures were the same as described above.
Construction of effector and reporter plasmids and the transient cotransfection assay.
pEF-BOS expression vector28 was kindly provided by Dr S. Nagata (Osaka University). The expression plasmid containing +-MITF ormi-MITF cDNA was constructed as described previously.14,15 The luciferase gene subcloned into pSP72 (pSPLuc) was generously provided by Dr K. Nakajima (Osaka University Medical School). To construct reporter plasmids, the promoter region of the Gr B (nucleotides [nt] −910 to +42, +1 is the transcription start site)29 gene was obtained with PCR and subcloned into the upstream region of the luciferase gene in pSPLuc. Twenty micrograms of a reporter, 5 μg of an effector, and 5 μg of an expression vector containing the β-galactosidase gene were added to cell suspension of P-815 cells (1 × 107) in 0.4 mL of α-MEM. After incubation for 10 minutes on ice, electroporation was performed by a single pulse (975 μF, 320 V) from Gene Pulser II (Bio-Rad Laboratories, Hercules, CA). The cells were suspended in 5 mL α-MEM supplemented with 10% FCS and cultured in a 6-cm dish. The expression vector containing the β-galactosidase gene was used as an internal control. The cells were harvested 24 hours after electroporation and lysed with 0.1 mol/L potassium phosphate buffer (pH 7.4) containing 1% Triton X-100. Soluble extracts were then assayed for luciferase activity with a luminometer LB96P (Berthold GmbH, Wildbad, Germany) and for β-galactosidase activity. The luciferase activity was normalized using the β-galactosidase activity, and the total protein concentration was estimated according to the method described by Yasumoto et al.30 The normalized value was divided by the value obtained after cotransfection with the reporter and pEF-BOS and was expressed as the relative luciferase activity.
Concentration of serotonin.
The concentration of serotonin was measured using high performance liquid chromatography (HPLC) with electrochemical detection.31 Briefly, CMCs were collected, washed with phosphate-buffered saline (PBS), counted, and sonicated for 20 seconds in a sonicator (Tomy, Tokyo, Japan) in 1 mL of ice-cold 3% perchloric acid containing 5 mmol/L EDTA and 1 mmol/L sodium metabissulfite. The homogenate was centrifuged at 10,000g for 15 minutes at 4°C and the supernatant was applied directly to the HPLC column. The concentration of serotonin per 1.0 × 106 cells was calculated.
Cytotoxicity assay.
Mast cell cytotoxicity was measured using a 51Cr release assay according to the procedure described by Bissonnette and Befus.32 As a positive control, spleen cells were freshly prepared from 8-week-old +/+ mice. The target was YAC-1 cells, which were obtained from the American Type Culture Collection (Bethesda, MD). The cells were maintained in α-MEM supplemented with 10% FCS. CMCs derived from +/+, mi/mi, and tg/tg mice, and spleen cells of +/+ mice were washed, suspended in α-MEM supplemented with 10% FCS, and distributed at different concentrations (0.5, 1.0, and 2.5 × 106 cells) in triplicate into 96-well microtiter plates with round bottoms. YAC-1 cells (5 × 106) were labeled with 100 μCi of [51Cr] Na2CrO4 (Amersham, Arlington Heights, IL) for 2 hours, washed three times, and resuspended in α-MEM supplemented with 10% FCS. Labeled YAC-1 cells (1.0 × 104) were mixed with various numbers of CMCs in a total volume of 200 μL/well. Plates were incubated at 37°C for 18 hours in a CO2 incubator and spun at 150g for 10 minutes, and the radioactivity was determined in 100 μL samples of cell-free supernatants. The radioactivity released in the well containing YAC-1 cells alone was designated spontaneous release (SR). Total 51Cr release (TR) was measured by adding 0.01% Triton X-100 to the well containing YAC-1 cells alone. The percentage of specific 51Cr release was calculated using the following formula: (cpm in the presence of CMCs − SR)/(TR − SR) × 100.
RESULTS
Screening of two subtracted cDNA libraries by Southern analysis.
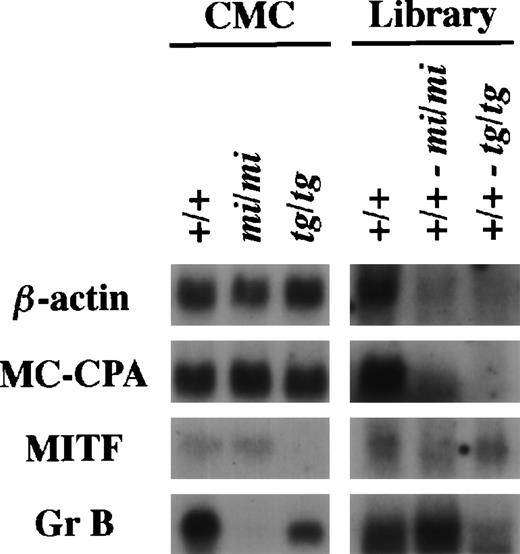
Using the same procedure that was used to establish the subtracted cDNA library of (+/+-mi/mi),21 we elaborated the other subtracted library of (+/+-tg/tg) by the removal of clones carrying inserts complementary to tg/tg CMC mRNAs from the +/+ CMC cDNA library. The (+/+-tg/tg) library was examined by the library-Northern analysis.21 The sense RNAs of the cDNA inserts were synthesized by the T7 RNA polymerase reaction using theNot I-digested plasmid DNA of the +/+, (+/+-mi/mi), and (+/+-tg/tg) cDNA libraries as templates. They were then fixed to nylon membranes and probed with several cDNAs. MC-CPA and β-actin cDNAs were expressed equally in +/+, mi/mi, and tg/tgCMCs, and hybridized bands were rarely detectable in the two subtracted libraries (Fig 1). MITF expression was comparable between +/+ and mi/mi CMCs but not detectable intg/tg CMCs. When probed with MITF cDNA, a faint band was detectable in the RNA obtained from the (+/+-tg/tg) library but not in the RNA obtained from the (+/+-mi/mi) library. When probed with the Gr B cDNA, the intensity of the hybridized band was significantly stronger in the (+/+-mi/mi) library than in the (+/+-tg/tg) library (Fig 1).
Characterization of the subtracted cDNA library by Northern blot analysis. In the left panel, 5 μg of total RNA prepared from +/+, mi/mi, or tg/tg CMCs was loaded in each lane and fixed onto nylon membranes by capillary action. In the right panel, sense RNAs were synthesized by the T7 RNA polymerase reaction using the Not I-digested plasmid DNA of the +/+ CMC cDNA library, the (+/+-mi/mi) subtracted cDNA library, or the (+/+-tg/tg) subtracted cDNA library as a template. Two micrograms of synthesized RNA was loaded per lane and fixed onto nylon membranes. Probes were prepared from the cDNAs for β-actin, MC-CPA, MITF, or Gr B using the random hexamer labeling method.
Characterization of the subtracted cDNA library by Northern blot analysis. In the left panel, 5 μg of total RNA prepared from +/+, mi/mi, or tg/tg CMCs was loaded in each lane and fixed onto nylon membranes by capillary action. In the right panel, sense RNAs were synthesized by the T7 RNA polymerase reaction using the Not I-digested plasmid DNA of the +/+ CMC cDNA library, the (+/+-mi/mi) subtracted cDNA library, or the (+/+-tg/tg) subtracted cDNA library as a template. Two micrograms of synthesized RNA was loaded per lane and fixed onto nylon membranes. Probes were prepared from the cDNAs for β-actin, MC-CPA, MITF, or Gr B using the random hexamer labeling method.
The number of clones that are specifically expressed by +/+ CMCs was compared between two subtracted libraries. Eighty cDNA clones were isolated randomly from the (+/+-mi/mi) subtracted library, and the cDNA insert of each clone was blotted on two membranes. For a positive control, the mouse β-actin and human GAPDH cDNAs were also blotted. One membrane was hybridized with cDNA reverse-transcribed from +/+ CMC poly (A)+ RNA as a probe, and another was hybridized with cDNA reverse-transcribed from mi/mi CMC poly (A)+ RNA (Fig 2, upper panel). According to the same procedure, two blotted membranes were also made using 80 clones from the (+/+-tg/tg) subtracted library and probed with +/+ and tg/tg CMC cDNAs individually (Fig 2, lower panel). After adjustment of the β-actin–specific and GAPDH-specific signals of each membrane to equal intensity, it may be expected that the clones that hybridize more efficiently with the +/+ cDNA probe than with the mi/mi or tg/tg cDNA probe carry cDNA inserts transcribed specifically in +/+ CMCs. Five clones (denoted by arrowheads with numbers in Fig 2, upper panel) corresponded to such ones in the (+/+-mi/mi) library, whereas only one clone (denoted by an arrowhead with a number in Fig 2, lower panel) corresponded in the (+/+-tg/tg) library. By sequencing and a computer-assisted homology search, all six clones turned out to be reported genes. Clone no. 1 of Fig 2 encodes vimentin; clones no. 2 and 5 encode Gr B; clone no. 3 encodes E25 that is a marker for chondro-osteogenic differentiation33; clone no. 4 encodes MMCP-5; and clone no. 6 encodes MMCP-6. Although the expression of vimentin and E25 in mast cells has not been studied intensively, the mRNA expression was detectable in +/+ CMCs and significantly reduced inmi/mi CMCs (data not shown).
Screening of +/+ CMC-specific clones by Southern blot analysis. After digestion with both Sma I and Not I to separate the cDNA insert, plasmid DNAs of 80 clones randomly selected from the subtracted cDNA library [(+/+-mi/mi) or (+/+-tg/tg)] were electrophoresed in 1.0% agarose gel (left panel, stained with ethidium bromide [EdBr]) and bound to nylon membranes. Duplicate membranes were hybridized with32P-labeled cDNAs synthesized from poly(A)+RNA of +/+ (middle panel), mi/mi (right upper panel), ortg/tg (right lower panel) CMCs. In some lanes, an equal amount of the mouse β-actin (A) and human GAPDH (G) cDNA fragments and a λ-BstEII size marker (M) were loaded as a control, and we graphically equalized the intensity of their bands for two filters to normalize the intensity of other sets of the bands. The clones that hybridized to a greater degree with +/+ CMC cDNA than withmi/mi or tg/tg CMC cDNA were identified by arrowheads with numbers.
Screening of +/+ CMC-specific clones by Southern blot analysis. After digestion with both Sma I and Not I to separate the cDNA insert, plasmid DNAs of 80 clones randomly selected from the subtracted cDNA library [(+/+-mi/mi) or (+/+-tg/tg)] were electrophoresed in 1.0% agarose gel (left panel, stained with ethidium bromide [EdBr]) and bound to nylon membranes. Duplicate membranes were hybridized with32P-labeled cDNAs synthesized from poly(A)+RNA of +/+ (middle panel), mi/mi (right upper panel), ortg/tg (right lower panel) CMCs. In some lanes, an equal amount of the mouse β-actin (A) and human GAPDH (G) cDNA fragments and a λ-BstEII size marker (M) were loaded as a control, and we graphically equalized the intensity of their bands for two filters to normalize the intensity of other sets of the bands. The clones that hybridized to a greater degree with +/+ CMC cDNA than withmi/mi or tg/tg CMC cDNA were identified by arrowheads with numbers.
The above-mentioned Southern experiment (experiment no. 1) was repeated four more times (experiments no. 2, 3, 4, and 5). Consequently, we isolated 400 clones from the (+/+-mi/mi) or (+/+-tg/tg) library, subjected them to Southern analysis, and counted the number of clones yielding stronger signals with +/+-probe than with mi/mi- or tg/tg-probe (summarized in Table 1). In every experiment, the number of such clones was larger in the (+/+-mi/mi) subtraction cDNA library than in the (+/+-tg/tg) subtraction library. In total, the proportion of such clones was greater in the (+/+-mi/mi) library than in the (+/+-tg/tg) library. These results suggested that the number of genes transcriptionally downregulated in mi/mi CMCs is larger than the number of genes transcriptionally downregulated intg/tg CMCs.
Screening of the (+/+-mi/mi) and (+/+-tg/tg) Subtracted cDNA Libraries by Southern Analyses
| Subtracted cDNA Libraries . | No. of Clones Fractioned on the Gel . | No. of Clones Yielding Hybridization Signals With +/+-Probe . | No. of Clones Yielding Stronger Signals With +/+-Probe Than With mi/mi- ortg/tg-Probe . | No. of Clones Yielding Stronger Signals With mi/mi- ortg/tg-Probe Than With +/+-Probe . |
|---|---|---|---|---|
| Experiment no. 1 | ||||
| +/+-mi/mi | 80 | 44 | 5 | 0 |
| +/+-tg/tg | 80 | 46 | 1 | 0 |
| Experiment no. 2 | ||||
| +/+-mi/mi | 80 | 35 | 3 | 1 |
| +/+-tg/tg | 80 | 40 | 1 | 0 |
| Experiment no. 3 | ||||
| +/+-mi/mi | 80 | 38 | 4 | 0 |
| +/+-tg/tg | 80 | 39 | 0 | 1 |
| Experiment no. 4 | ||||
| +/+-mi/mi | 80 | 41 | 5 | 0 |
| +/+-tg/tg | 80 | 37 | 2 | 0 |
| Experiment no. 5 | ||||
| +/+-mi/mi | 80 | 40 | 3 | 0 |
| +/+-tg/tg | 80 | 43 | 1 | 0 |
| Average | ||||
| +/+-mi/mi | 80 | 39.6 ± 1.5 | 4.0 ± 0.4 | 0.2 ± 0.2 |
| +/+-tg/tg | 80 | 41.0 ± 1.6 | 1.0 ± 0.3* | 0.2 ± 0.2 |
| Total | ||||
| +/+-mi/mi | 400 | 198 | 20 | 1 |
| +/+-tg/tg | 400 | 205 | 5† | 1 |
| Subtracted cDNA Libraries . | No. of Clones Fractioned on the Gel . | No. of Clones Yielding Hybridization Signals With +/+-Probe . | No. of Clones Yielding Stronger Signals With +/+-Probe Than With mi/mi- ortg/tg-Probe . | No. of Clones Yielding Stronger Signals With mi/mi- ortg/tg-Probe Than With +/+-Probe . |
|---|---|---|---|---|
| Experiment no. 1 | ||||
| +/+-mi/mi | 80 | 44 | 5 | 0 |
| +/+-tg/tg | 80 | 46 | 1 | 0 |
| Experiment no. 2 | ||||
| +/+-mi/mi | 80 | 35 | 3 | 1 |
| +/+-tg/tg | 80 | 40 | 1 | 0 |
| Experiment no. 3 | ||||
| +/+-mi/mi | 80 | 38 | 4 | 0 |
| +/+-tg/tg | 80 | 39 | 0 | 1 |
| Experiment no. 4 | ||||
| +/+-mi/mi | 80 | 41 | 5 | 0 |
| +/+-tg/tg | 80 | 37 | 2 | 0 |
| Experiment no. 5 | ||||
| +/+-mi/mi | 80 | 40 | 3 | 0 |
| +/+-tg/tg | 80 | 43 | 1 | 0 |
| Average | ||||
| +/+-mi/mi | 80 | 39.6 ± 1.5 | 4.0 ± 0.4 | 0.2 ± 0.2 |
| +/+-tg/tg | 80 | 41.0 ± 1.6 | 1.0 ± 0.3* | 0.2 ± 0.2 |
| Total | ||||
| +/+-mi/mi | 400 | 198 | 20 | 1 |
| +/+-tg/tg | 400 | 205 | 5† | 1 |
The result of experiment no. 1 is shown in Fig. 2.
P < .01 by t-test when compared with the value of the (+/+-mi/mi) library.
P < .01 by χ2-test when compared with the value of the (+/+-mi/mi) library.
Difference of transcriptional impairment between mi/miand tg/tg CMCs.
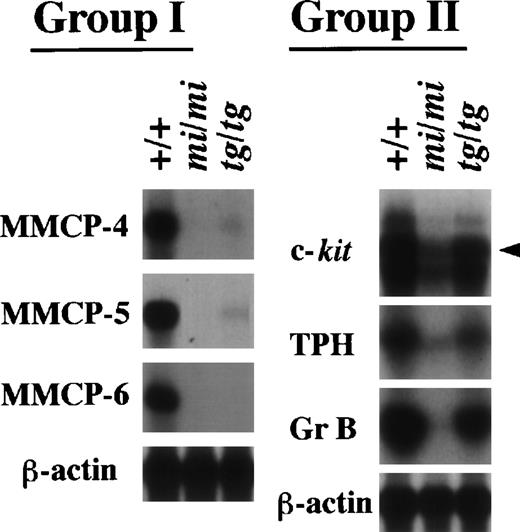
We have demonstrated that several genes indeed show a drastically reduced expression in mi/mi CMCs.14-17 21Therefore, we examined whether the transcription of these genes was also impaired in tg/tg CMCs. We performed Northern analysis with the six cDNA probes of the c-kit, TPH, Gr B, MMCP-4, MMCP-5, and MMCP-6 genes. These genes were divided into two groups according to their expression profile (Fig3). The MMCP-4, MMCP-5, and MMCP-6 genes belonged to the first group. They showed a transcriptional impairment in tg/tg CMCs as severe as in mi/mi CMCs, although just a faint expression of MMCP-4 and MMCP-5 genes was detected in tg/tg CMCs. The c-kit, TPH, and Gr B genes belonged to the second group. Although they showed a reduced expression in tg/tg CMCs, the reduction was much smaller than the reduction observed in mi/miCMCs. By a densitometric analysis, the expression of the three genes was reduced by 90% or more in mi/mi CMCs, whereas the expression was reduced by only 50% in tg/tg CMCs (data not shown). Therefore, the three genes were not included in the (+/+-tg/tg) subtracted library. The presence of genes belonging to the second group was consistent with the result obtained by screening the subtracted libraries.
Expression of the MMCP-4, MMCP-5, MMCP-6, Gr B, TPH, and c-kit genes in +/+, mi/mi, and tg/tg CMCs. Five micrograms of total RNA prepared from +/+, mi/mi,or tg/tg CMCs was loaded in each lane and fixed onto nylon membranes by capillary action. The membranes were hybridized with specific DNA probes. An arrowhead indicates the specific signal for c-kit. Reprobing with the β-actin probe allowed verification that an equal amount of mRNA was loaded per lane. With respect to the expression profiles, the six genes were divided into two groups (groups I and II).
Expression of the MMCP-4, MMCP-5, MMCP-6, Gr B, TPH, and c-kit genes in +/+, mi/mi, and tg/tg CMCs. Five micrograms of total RNA prepared from +/+, mi/mi,or tg/tg CMCs was loaded in each lane and fixed onto nylon membranes by capillary action. The membranes were hybridized with specific DNA probes. An arrowhead indicates the specific signal for c-kit. Reprobing with the β-actin probe allowed verification that an equal amount of mRNA was loaded per lane. With respect to the expression profiles, the six genes were divided into two groups (groups I and II).
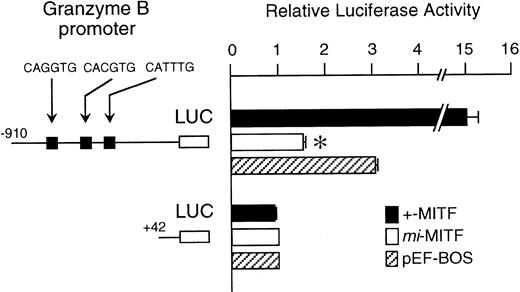
The mi-MITF appeared to function negatively in the transcription of genes belonging to the second group. We examined the effect of mi-MITF on the transactivation of the Gr B gene promoter using the transient cotransfection assay. The 5′-flanking sequence of the Gr B gene (nt −910 to +42)29 was cloned upstream of the luciferase gene. Three CANNTG motifs were present in this region. The Gr B gene appeared to be transactivated through these three CANNTG motifs.21 The luciferase construct was cotransfected into P-815 cells with the expression plasmid pEF-BOS containing no insert, +-MITF cDNA, ormi-MITF cDNA. Neither +-MITF nor mi-MITF showed any transactivation effect on the reporter without the Gr B promoter sequence (Fig 4). When the Gr B promoter-reporter construct was cotransfected with pEF-BOS containing no insert, the luciferase activity increased threefold. The endogenous MITF and other factors appeared to transactivate the luciferase construct in P-815 cells. The cotransfection of pEF-BOS containing +-MITF increased the luciferase activity remarkably. In contrast, that of pEF-BOS containing mi-MITF cDNA significantly reduced the luciferase activity (Fig 4).
The effect of coexpression of +-MITF or mi-MITF cDNA on the luciferase activity under the control of the Gr B gene promoter. Various forms of the reporter and effector constructs were introduced into P-815 cells by electroporation. Three solid squares represent CANNTG motifs between nt −910 and +42; ie, CAGATG (nt −563 to −558), CACGTG (nt −530 to −525), and CATTTG (nt −521 to −516) motifs. The bars represent the mean ± standard error (SE) of the relative luciferase activities obtained by three independent experiments: (▪) +-MITF; (□) mi-MITF; (▨) pEF-BOS. In some cases, the SE was too small to be shown by the bars. *P< .01 by t-test when compared with the luciferase activity, in which pEF-BOS containing no insert was cotransfected.
The effect of coexpression of +-MITF or mi-MITF cDNA on the luciferase activity under the control of the Gr B gene promoter. Various forms of the reporter and effector constructs were introduced into P-815 cells by electroporation. Three solid squares represent CANNTG motifs between nt −910 and +42; ie, CAGATG (nt −563 to −558), CACGTG (nt −530 to −525), and CATTTG (nt −521 to −516) motifs. The bars represent the mean ± standard error (SE) of the relative luciferase activities obtained by three independent experiments: (▪) +-MITF; (□) mi-MITF; (▨) pEF-BOS. In some cases, the SE was too small to be shown by the bars. *P< .01 by t-test when compared with the luciferase activity, in which pEF-BOS containing no insert was cotransfected.
Serotonin concentrations.
Because mRNA expression levels of the TPH and Gr B genes were significantly different between mi/mi and tg/tg CMCs, we examined whether these two CMCs possess different phenotypes in relation to the two genes. Because TPH is the rate-limiting enzyme for serotonin synthesis,34 serotonin concentration was compared between mi/mi and tg/tg CMCs. The serotonin concentration was significantly lower in mi/mi CMCs than in +/+ CMCs, as previously reported.21 The serotonin concentration in tg/tg CMCs was in the middle of both, ie, significantly lower than that of +/+ CMCs and significantly higher than that of mi/mi CMCs (Table 2). These values were well correlated with the mRNA expression levels of the TPH gene.
Concentration of Serotonin in +/+,mi/mi, and tg/tg CMCs
| Genotype of CMCs . | Serotonin Concentration3-150 (nmol/106cells) . |
|---|---|
| +/+ | 7.44 ± 0.29 |
| mi/mi | 1.33 ± 0.083-151,3-152 |
| tg/tg | 4.23 ± 0.213-151 |
| Genotype of CMCs . | Serotonin Concentration3-150 (nmol/106cells) . |
|---|---|
| +/+ | 7.44 ± 0.29 |
| mi/mi | 1.33 ± 0.083-151,3-152 |
| tg/tg | 4.23 ± 0.213-151 |
Means ± SE of three experiments.
P < .05 by t-test when compared with the value of +/+ CMCs.
P < .05 by t-test when compared with the value oftg/tg CMCs.
Cytotoxicity to YAC-1 cells.
Gr B is essential for the induction of DNA fragmentation in target cells caused by natural killer (NK) cells and cytotoxic T lymphocytes.35-37 Because the expression of Gr B gene was drastically reduced in mi/mi CMCs and mildly intg/tg CMCs, it seemed logical to assume thatmi/mi CMCs would show impaired cytotoxic activity but that of tg/tg CMCs would be near the normal level. We examined this possibility using YAC-1 lymphoma cells as a target (Table3). CMCs of +/+, mi/mi, ortg/tg genotype were cultured together with51Cr-labeled YAC-1 cells and 51Cr release from YAC-1 cells was measured after 18 hours. At an effector/target (E/T) ratio of 50, neither +/+, mi/mi, nor tg/tg CMCs showed any cytotoxic activity to YAC-1 cells (Table 3). When the +/+ spleen cells were used as a positive control, they killed YAC-1 cells at the E/T ratio. This was compatible with a previous report36 37 indicating that our assays were reproducible. At an increased E/T ratio of 100 or 250, a significant difference in cytotoxic activity between +/+ and mi/mi CMCs was observed; +/+ CMCs displayed approximately 20% cytotoxicity to YAC-1 cells, whereas no cytotoxicity was observed with mi/mi CMCs. In contrast,tg/tg CMCs showed an apparent cytotoxicity comparable to that of +/+ CMCs.
Cytotoxic Activity of +/+, mi/mi, and tg/tg CMCs to YAC-1 Cells
| E/T Ratio . | Specific51Cr Release (%)* . | |||
|---|---|---|---|---|
| +/+ CMCs . | mi/mi CMCs . | tg/tg CMCs . | +/+ Spleen Cells . | |
| 50 | <0.1 | <0.1 | <0.1 | 50.8 ± 2.4 |
| 100 | 13.9 ± 0.1† | <0.1 | 12.0 ± 0.6† | 57.3 ± 3.8 |
| 250 | 21.1 ± 2.8† | <0.1 | 19.0 ± 1.9† | NE |
| E/T Ratio . | Specific51Cr Release (%)* . | |||
|---|---|---|---|---|
| +/+ CMCs . | mi/mi CMCs . | tg/tg CMCs . | +/+ Spleen Cells . | |
| 50 | <0.1 | <0.1 | <0.1 | 50.8 ± 2.4 |
| 100 | 13.9 ± 0.1† | <0.1 | 12.0 ± 0.6† | 57.3 ± 3.8 |
| 250 | 21.1 ± 2.8† | <0.1 | 19.0 ± 1.9† | NE |
E/T ratio is the ratio of CMCs or spleen cells to YAC-1 cells.
Abbreviation: NE, not examined.
Means ± SE of three experiments.
P < .01 by t-test when compared with the value ofmi/mi CMCs.
DISCUSSION
We compared mi/mi and tg/tg CMCs to examine the effect of mi-MITF on the transcription. The former possesses the mutated MITF (mi-MITF) that is deficient in DNA-binding and nuclear localization potential.12,13 The latter practically lacks MITF.8,22 To study the difference of the gene expression between these two types of CMCs, we evaluated subtracted cDNA libraries: a subtraction of mi/mi CMC mRNA from +/+ CMC cDNA library (+/+-mi/mi) and another subtraction oftg/tg CMC mRNA from +/+ CMC cDNA library (+/+-tg/tg). As previously reported,21 we found a number of new genes whose expression was downregulated in mi/mi CMCs when the (+/+-mi/mi) library was screened. In contrast, the (+/+-tg/tg) library that was newly established in the present study contained a significantly smaller number of clones whose expression might be downregulated in tg/tg CMCs. We have isolated several genes whose expression severely decreases inmi/mi CMCs.14-17 21 When we examined the expression levels of such genes in tg/tg CMCs, we found that they were divided into two groups. The first group showed a transcriptional impairment in tg/tg CMCs as severe as in mi/mi CMCs. The MMCP-4, MMCP-5, and MMCP-6 genes are included in this group. The presence of +-MITF appeared to be essential for the transactivation of the genes belonging to this group. The second group also showed a reduced expression in tg/tg CMCs, but the magnitude of reduction was much smaller when compared with that of mi/miCMCs. The c-kit, TPH, and Gr B genes belonged to the second group. The result obtained by Southern analysis (Fig 2 and Table 1) and that of Northern analysis (Fig 3) were consistent and indicated that the number of genes whose expression was downregulated in tg/tgCMCs is significantly smaller than the number of genes downregulated inmi/mi CMCs. These results also suggested that mi-MITF functioned negatively in the transactivation of genes belonging to the second group.
We have reported that MITF is involved in transcriptional activation of the c-kit,14 TPH,21 and Gr B21 genes through the recognition of the CANNTG motif. The transactivation effect of +-MITF on the c-kit, TPH, and Gr B promoter-reporter construct was threefold or fourfold as large as the control level.14,21 The effect was comparable with the induction of the expression of those genes observed in +/+ CMCs compared with the expression in tg/tg CMCs (Fig 3). On the other hand, the expression level of the three genes in +/+ CMCs appeared to be greater than 10-fold of the expression level observed inmi/mi CMCs. The difference between mi/mi andtg/tg CMCs may be explained by the involvement of other transcription factor(s) in transactivation of the three genes. In fact, Babichuk et al38,39 recently reported that AP-1 and PEBP2 binding sites are primarily responsible for the high level expression of Gr B in lymphocytes. The two sites were located about 350 bases downstream of three MITF binding sites that were reported by us.21 Interaction of AP-1 or PEBP2 with mi-MITF may occur in the cytoplasm, and the interaction with mi-MITF may have an inhibitory effect on nuclear localization potential of AP-1 and PEBP2.13 There is another explanation, because an appreciable amount of mi-MITF appeared to enter the nucleus.13 In the nucleus, the mi-MITF might prevent AP-1 or PEBP2 from binding their recognition sites.
We also examined the phenotypic differences in relation to the TPH and Gr B genes. Serotonin concentrations of +/+, mi/mi, andtg/tg CMCs were well correlated with the expression levels of the TPH gene. The cytotoxic activity to YAC-1 cells showed a big difference between mi/mi and tg/tgCMCs. CMCs of tg/tg genotype killed YAC-1 cells as effectively as +/+ CMCs, whereas mi/mi CMCs were defective in the cytotoxicity. The results were consistent with those obtained by the Northern analysis that the Gr B gene expression reduced to nondetectable levels in mi/mi CMCs and that the Gr B expression level of tg/tg CMCs was approximately 50% that of +/+ CMCs. The difference among these three types of CMCs with respect to their cytotoxic activity suggests that Gr B is essential for cytotoxicity in CMCs as well as in NK cells.
In conclusion, we examined the differences of transcriptional impairment and the related phenotypes between mi/mi andtg/tg CMCs. We found that mi/mi CMCs suffered from transcriptional impairment more severely than tg/tg CMCs. Two phenotypic features examined correlated with the mRNA expression. The results reinforced our previous suggestion that mi-MITF possessed an inhibitory effect on transactivation of particular target genes.13 Further investigation by comparing mi/miand tg/tg CMCs will lead to the better understanding of the transcriptional events mediated by MITF.
ACKNOWLEDGMENT
The authors thank Kirin Brewery Co Ltd (Tokyo, Japan) for supplying with rmSCF.
Supported by grants from the Ministry of Education, Science and Culture, the Ministry of Health and Welfare, the Osaka Cancer Society, the Naito Foundation, the Mochida Memorial Foundation, the Ryoichi Naito Foundation, and Japan Foundation for Health Sciences.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Yukihiko Kitamura, MD, Department of Pathology, Osaka University Medical School, Yamada-oka 2-2, Suita 565-0871, Japan.

![Fig. 2. Screening of +/+ CMC-specific clones by Southern blot analysis. After digestion with both Sma I and Not I to separate the cDNA insert, plasmid DNAs of 80 clones randomly selected from the subtracted cDNA library [(+/+-mi/mi) or (+/+-tg/tg)] were electrophoresed in 1.0% agarose gel (left panel, stained with ethidium bromide [EdBr]) and bound to nylon membranes. Duplicate membranes were hybridized with32P-labeled cDNAs synthesized from poly(A)+RNA of +/+ (middle panel), mi/mi (right upper panel), ortg/tg (right lower panel) CMCs. In some lanes, an equal amount of the mouse β-actin (A) and human GAPDH (G) cDNA fragments and a λ-BstEII size marker (M) were loaded as a control, and we graphically equalized the intensity of their bands for two filters to normalize the intensity of other sets of the bands. The clones that hybridized to a greater degree with +/+ CMC cDNA than withmi/mi or tg/tg CMC cDNA were identified by arrowheads with numbers.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/4/10.1182_blood.v93.4.1189/5/m_blod40432002w.jpeg?Expires=1767708981&Signature=avdNCLcQ3A1KPg~J8lWDrXywN3yThTuejzVRUCnjL6~oayX8U-V-1LPnAfZ6gCs4TfP0rsnTYe14D-0gUqwzt85JVubjnSoWCjWyK~p-tJJlL8PooxEPy0vV9q06l6iUlsIVJ0zGAzFL5GngrhZ1uMYkfB79xti2GHdIDKDKTX3sERM8IsZ7ab9XFYVD~Cl412XI7kUDr5mgfnkoxl6peo6cxW7k9kT45z5Yf1f62I1K95X7w526lC0WBhhMDx44aTh9rIogDdNNhe7bM2kYdifqzPTyNj7EP49tLZiSxHSITEr2NmL~5nDLtoHC4sEXZv0I7RlBZcS8XViSAFWnJQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal