Abstract
The recent discovery of chemokine receptors as coreceptors for human immunodeficiency virus–type 1 (HIV-1) entry offers new avenues for investigating the pathogenesis of acquired immunodeficiency syndrome (AIDS)-related cytopenias. To this end, we sought to (1) phenotype human hematopoietic cells for CD4 and the HIV-1 coreceptors CXCR4, CCR5, CCR3, and CCR2b; (2) correlate CD4 and chemokine receptor expression with their susceptibility to HIV-1 infection; and (3) examine any potential interplay between inflammatory cytokines released during HIV-1 infection and regulation of chemokine receptor expression. Fluorescence-activated cell sorting (FACS) analysis of bone marrow mononuclear cells (BMMNC), cells derived from serum-free expanded hematopoietic lineages (colony-forming unit–granulocyte-macrophage [CFU-GM], colony-forming unit-megakaryocyte [CFU-Meg], and burst-forming unit-erythroid [BFU-E]), and CD34+ cells showed differential expression of chemokine receptors and CD4 with some lineage specificity. Significantly, FACS-sorted CXCR4+/CD34+ cells had the same clonogeneic potential as CXCR4−/CD34+ cells. Reverse transcriptase-polymerase chain reaction (RT-PCR) analysis of FACS-sorted human candidate stem cells (HSC; CD34+, c-kit+, Rho123low) showed the presence of CXCR4 mRNA but not CD4 mRNA. Infection studies with HIV-1 Env-pseudotyped luciferase reporter viruses indicated that X4 Env (CXCR4-using) pseudotypes infected megakaryocytic cells, whereas R5 Env (CCR5-using) pseudotypes did not. Similarly, R5 but not X4 Env-pseudotyped viruses infected granulocyte-macrophage cells in a CD4/CCR5-dependent manner. Erythroid cells were resistant to R5 or X4 viral infection. Finally, we found that γ-interferon treatment upregulated CXCR4 expression on primary hematopoietic cells. In summary, the delineation of chemokine receptor expression on primary hematopoietic cells is a first step towards dissecting the chemokine-chemokine receptor axes that may play a role in hematopoietic cell proliferation and homing. Furthermore, susceptibility of hematopoietic cells to HIV-1 infection is likely to be more complicated than the mere physical presence of CD4 and the cognate chemokine receptor. Lastly, our results suggest a potential interplay between γ-interferon secretion and CXCR4 expression.
PATIENTS INFECTED BY human immunodeficiency virus–type 1 (HIV-1) frequently exhibit a variety of different hematological abnormalities, including anemia, neutropenia, and thrombocytopenia, in addition to the invariable loss of CD4+ lymphocytes.1,2 The discovery of chemokine receptors as coreceptors for HIV-1 entry offers new avenues for increasing our understanding of the mechanisms underlying HIV-1–associated bone marrow dysfunction.3 At this point, there are 11 reported chemokine or orphan receptors that function as HIV-1 coreceptors: CXCR-4, CCR5, CCR2b, CCR3, CCR8, STRL33, GPR1, V28, ChemR23, GPR15, and APJ (reviewed previously4,5). All HIV-1 strains studied to date use CCR5 (R5 strains), CXCR4 (X4), or both receptors (R5X4) to enter cells, and individuals who lack CCR5 are highly resistant to virus infection (reviewed in McNicholl et al6). The in vivo relevance of coreceptors other than CCR5 and CXCR4 has yet to be determined, although their ability to support infection by more limited numbers of virus strains raises the possibility that their use may be involved in the myriad pathologies associated with HIV-1 infection, including the hematologic abnormalities. As such, exploring the chemokine receptor expression pattern on subsets of hematopoietic progenitors may shed light on the susceptibility of various subsets to either direct infection by HIV-1 or other forms of modulation such as chemokine-induced inhibition/proliferation or perhaps envelope (Env)-mediated toxicity. With regard to the latter point, recent studies have shown that soluble HIV-1 and SIV Env can induce G-protein–mediated signal transduction through their cognate coreceptors.7,8 Therefore, intracellular signaling cascades mediated through chemokine receptors by HIV-1 Env may lead to hematopoietic derangements even in the absence of productive infection of hematopoietic progenitor populations. This is supported by studies showing an inhibitory effect of recombinant viral envelope glycoprotein on CD34+ progenitor cells.9-11
Studies to date have looked at HIV-1 coreceptor expression in bone marrow progenitor cells only at the mRNA level.12,13 The use of in vitro serum-free cultures for expanding relatively pure, lineage-committed hematopoietic progenitors along with recently developed monoclonal antibodies (MoAbs) against the major HIV-1 coreceptors has allowed us to define coreceptor/chemokine receptor expression on erythroid, megakaryocytic, and granulo-macrophage lineages. Although the pathogenesis of acquired immunodeficiency syndrome (AIDS)-related cytopenias is likely to be multifactorial (reviewed in Moses et al14), the delineation of coreceptor and CD4 antigen expression will allow a preliminary determination of hematopoietic subsets that may be susceptible to either direct infection by HIV-1 or to HIV-1 Env-mediated cytotoxicity. In addition, it will now be possible to determine whether the many proinflammatory cytokines (tumor necrosis factor-α [TNF-α], γ-interferon [γ-IFN], etc) secreted in excess during chronic HIV-1 infection15,16 have any influence on cognate coreceptor expression. Because the chemokine-chemokine receptor axes may be involved in hematopoietic proliferation and homing,17 any pertubation of chemokine receptor expression may not only result in the expansion or restriction of HIV-1 tropism, but also contribute to the pathogenesis of the many cytopenias observed in HIV-1 disease.
We report here that HIV-1 coreceptor expression exhibited some lineage specificity and that megakaryocytic cells were infectable by X4 viruses, whereas granulo-macrophage lineage cells were infectable by R5 viruses. Furthermore, we determined that CXCR4 was expressed even on the earliest candidate human stem cells (HSC), although only about half of clonogeneic hematopoietic progenitor cells (HPC) were CXCR4+/CD34+ cells. We also found that γ-IFN could upregulate the expression of CXCR4 on BMMNC, suggesting that proinflammatory cytokines released during chronic HIV infection may influence the dynamics of HIV-1 replication by altering chemokine receptor expression levels.
MATERIALS AND METHODS
Selection of HSC candidates by fluorescence-activated cell sorting (FACS).
Light-density bone marrow mononuclear cells (BMMNC) were obtained from 12 consenting healthy donors and depleted of adherent cells and T lymphocytes (A−T−MNC) as described.18 MNC (∼3 to 6 × 107) were simultaneously labeled with phycoerythrin (PE)-conjugated anti-CD34 MoAb (anti-HPCA-2PE; Becton Dickinson, Mountain View, CA), an antihuman Kit receptor MoAb (SR-1; kind gift of Dr V. Broudy, University of Washington, Seattle, WA) detected with a Cy 5-labeled conjugate, and Rh123 at concentrations previously shown to be nontoxic to hematopoietic cells. CD34+, Kit+, Rh123low (defined as the dimmest 5% to 10% of Rh123-labeled cells) were isolated by FACS as described previously.19 We have also isolated by FACS a fraction of CD34+, Kit+, Rh123bright cells (defined as the brightest 50% of Rh123 labeled cells) that is enriched in HPC.19
Isolation of CXCR4+ cells.
BMMNC were stained with CXCR4 MoAb and subsequently isolated by using immunomagnetic beads (Dynal, Oslo, Norway) according to the manufacturer’s protocol and as described.20 In some experiments, FACS-sorted CD34+/CXCR4+ and CD34+/CXCR4− cells were isolated from total bone marrow. Briefly, BMMNC were stained with CXCR4 MoAb (R&D Systems, Minneapolis, MN) and detected with fluorescein isothiocyanate (FITC)-conjugated goat antimouse polyclonal Abs (Sigma, St Louis, MO), followed by staining with PE-conjugated CD34+ MoAb. Subsequently, cells were washed twice (1× phosphate-buffered saline [PBS] with 2% calf serum) and FACS sorted for both CD34+/CXCR4+ and CD34+/CXCR− cells using FACStarPlus (Becton Dickinson).
In vitro clonogeneic assays for hematopoietic progenitors.
Immunomagnetically isolated CXCR4+ cells (as described above) or FACS-sorted CXCR+/CD34+ or CXCR4−/CD34+ cells were plated in HCC-17 methylcellulose medium (StemCell Technologies, Vancouver, British Columbia, Canada) as described.19 Colony-forming unit-mix (CFU-Mix) colonies were stimulated with a cocktail of recombinant human (rH) growth factors: kit ligand (KL; 10 ng/mL), interleukin-3 (IL-3; 20 U/mL), granulocyte-macrophage colony-stimulating factor (GM-CSF; 5 ng/mL), erythropoietin (Epo; 2 U/mL), and IL-6 (40 U/mL). Burst-forming unit-erythroid (BFU-E) growth was stimulated with Epo (2 U/mL) and KL (10 ng/mL) and colony-forming unit–granulocyte-macrophage (CFU-GM) growth was stimulated with IL-3 (20 U/mL) and GM-CSF (5 ng/mL), whereas colony-forming unit-megakaryocyte (CFU-Meg) growth was stimulated with thrombopoietin (TPO; 50 ng/mL) and IL-3 (20 U/mL). Cytokines were from R&D Systems. Cultures were incubated at 37°C in a fully humidified atmosphere supplemented with 5% CO2. Colonies were scored at day 15 (CFU-Mix) and day 11 (BFU-E, CFU-GM, and CFU-Meg), respectively.
Ex vivo expansion of normal human hematopoietic cells.
CD34+ cells were expanded in serum-free liquid system as described.19-22 Briefly, CD34+A−T− BMNC were resuspended in Iscove Dulbecco’s modified Eagle’s medium (DMEM; GIBCO BRL, Grand Island, NJ; 104/mL) supplemented with 25% of artificial serum containing 1% delipidated, deionized, and charcoal-treated bovine serum albumin (BSA), 270 μg/mL iron-saturated transferrin, insulin (20 μg/mL), and 2 mmol/l L-glutamine (all from Sigma). BFU-E growth was stimulated with rH Epo (2 U/mL) and rH KL (10 ng/mL) and CFU-GM growth was stimulated with rH IL-3 (20 U/mL) and rH GM-CSF (5 ng/mL), whereas CFU-Meg growth was stimulated with rH TPO (50 ng/mL) and IL-3 (20 U/mL). Cytokines were from R&D Systems. Cultures were incubated at 37°C in a fully humidified atmosphere supplemented with 5% CO2. Under these conditions, approximately 100% of BFU-E–derived cells were glycophorin A positive, 65% to 80% of CFU-Meg cells were gpIIa/IIIb positive, and 100% of CFU-GM–derived cells were glycophorin A and gpIIb/IIIa negative and expressed CD33.21-23
Flow cytometry analysis.
The expression of CXCR4, CCR5, CCR2, CCR3, and CD4 on normal human hematopoietic cells was evaluated by FACS. The following MoAbs were used: 12G5 (J.A. Hoxie, University of Pennsylvania, Philadelphia, PA) and clone #701 (R&D Systems) for CXCR4; clones #529, #531, and #549 for CCR5 (R&D Systems); biotinylated clone #RO2 and #R05 for CCR2 (a generous gift from Carlos Martinez-A., Universidad Autonoma de Madrid, Madrid, Spain); 7B11 for CCR3 (NIH AIDS Reference Reagent Program); and Leu3A for CD4 (Becton Dickinson). Flow cytometric staining and analysis of the receptors were performed as described.23 Briefly, the cells were stained in PBS (Ca and Mg free) supplemented with 5% bovine calf serum (BCS). Primary MoAbs were detected with secondary PE- or FITC-conjugated goat antimouse MoAbs (Sigma; 1:100) or PE-conjugated streptavidin (Pharmingen, San Diego, CA) at 0.25 mg/mL for biotinylated primary antibodies. After the final washes, cells were fixed in 1% paraformaldehyde before FACS analysis using FACScan (Becton Dickinson, San Jose, CA). BMMNC or cells isolated from in vitro expanded liquid cultures of BFU-E, CFU-GM, and CFU-Meg cells were also assayed for the binding of biotinylated macrophage inflammatory protein-1α and monocyte chemotactic protein-1 (R&D Systems) according to the manufacturer’s protocols. Data analysis was performed using the Cell Quest (Becton Dickinson, San Jose, CA).
Reverse transcription-polymerase chain reaction (RT-PCR) studies.
RNA was extracted from FACS-sorted CD34+, Kit+, Rh123dull and CD34+, Kit+, Rh123bright cells using a poly A-mRNA purification kit (Pharmacia, Piscataway, NJ) according to the manufacturer’s protocol. The isolated RNA was dissolved in triple-distilled and autoclaved water and stored at −20°C until used. For RT-PCR, mRNA (0.5 μg) was reverse-transcribed with 500 U of Moloney murine leukemia virus reverse transcriptase (MoMLV-RT) and 50 pmol of an ODN primer complementary to the 3′ end of the following sequence of CXCR4 (CAA GGA AGC TGT TGG CTG AAA) or CD4 (5′-TTGGCGCCTTCGGTGCCGGCA-3′),24 according to reported cDNA sequences. The resulting cDNA fragments were amplified using 5 U of Thermus aquaticus (Taq) polymerase with the addition of primers specific for the 5′ end of CXCR4 (5′-CGA GGC AAG TGA CGC CGA GGG CCT G-3′) and CD4 (5′-GTGTGG GGACCCACCTCCCCTAAG-3′).24 Amplified products (10 μL) were electrophoresed on a 2% agarose gel and documented photographically. Specificity of the amplified products was further confirmed by Southern blotting. Electrophoresed gel fragments were transferred to a nylon filter and filters were prehybridized and probed with a 32P end-labeled ODN specific for the cDNA of CXCR4 or CD4. Hybridization was detected by autoradiography as described.18
Viral infection assay.
Luciferase reporter viruses were prepared as previously described25 26 by cotransfecting 293T cells with the indicated Envs and the NL4-3 luciferase virus backbone (pNL-luc-E−R−) plasmids. Full-length gp160 env genes from R5 (ADA, JRFL) and X4 (HXB2, NL4-3) viruses were cloned into pSV7d, where expression is driven off a constitutive SV40 promoter. These plasmids were generously provided by John Moore (Aaron Diamond AIDS Research Center, New York, NY). The NL4-3 luciferase virus backbone (pNL-luc-E−R−) was provided by Ned Landau (Aaron Diamond AIDS Research Center). This backbone was constructed with a frame-shift mutation in its env gene and a luciferase gene inserted into the nef coding region. Forty-eight hours after CaPO4 transfection, the supernatant was collected, filtered through a 0.2-μm filter, and stored at −80°C until further use. Infections were performed on the indicated target cells in the presence of 8 μg/mL of diethyl aminoethyl (DEAE)-dextran. Four days postinfection, cells were lysed with 0.5% TX-100 in PBS and an appropriate aliquot was analyzed for luciferase activity. Chemiluminescence from substrate conversion by luciferase was measured in a Wallac Microbeta Trilux luminometer and data were presented in relative light units (RLU). For inhibition assays, the appropriate inhibitor (chemokine or antibody) at the indicated concentrations was added 30 minutes before the addition of the reporter virus.
Statistics.
Arithmetic means and standard deviations were calculated on a MacIntosh computer using Instat 1.14 (GraphPad, San Diego, CA) software. Data were analyzed using the Student’s t-test for unpaired samples. Statistical significance was defined as P < .01.
RESULTS
Expression of CXCR4, CCR5, CCR2, and CCR3 on normal human BMMNC.
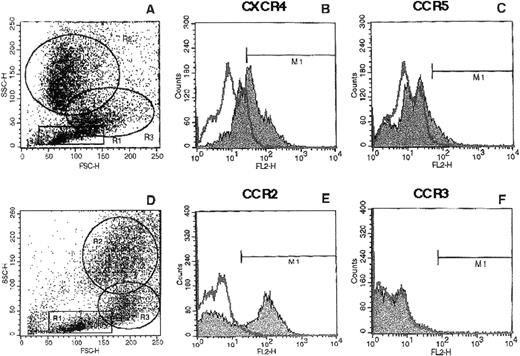
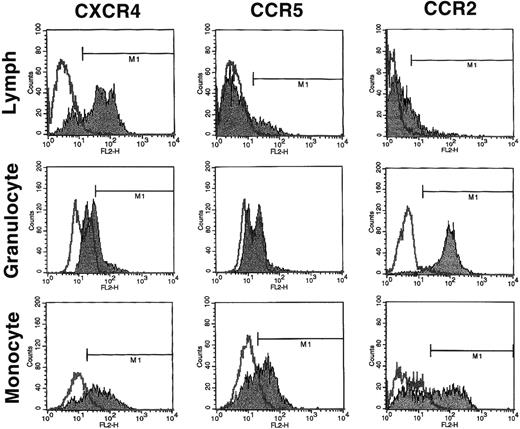
Because the expression of the major HIV-1 coreceptors in the various bone marrow hematopoietic populations has not been systematically examined, we first evaluated the expression of chemokine receptors on normal human BMMNC isolated by Ficoll-gradient centrifugation. As can be seen in Fig 1, CXCR4 (58% ± 6% positive), CCR5 (13% ± 2% positive), and CCR2 (51% ± 6% positive) but not CCR3 were variously present in total BMMNC. Because no detectable CCR3 was expressed on BMMNC, no further analysis of CCR3 was performed. We next determined the expression of these chemokine receptors in different subpopulations of BMMNC (lymphocyte-R1, monocyte-R2, and granulocyte-R3 gates; Fig2) based on their forward versus side-scatter properties (Fig 1). As summarized in Table 1, we found that CXCR4 was expressed predominantly on cells from the lymphocyte and monocyte gates, CCR5 predominantly in the monocyte gate, and CCR2 mostly in the monocyte and granulocyte progenitor gates. These results indicate that chemokine receptor expression exhibits some degree of lineage specificity.
Expression of chemokine receptors on total BMMNC. BMMNC were isolated from bone marrow aspirates of healthy donors by Ficoll-gradient centrifugation; stained with MoAbs to CXCR4 (B), CCR5 (C), CCR2 (E), and CCR3 (F); and subjected to FACS analysis as described in Materials and Methods. The histogram represents analysis of 10,000 events acquired in the total ungated population (A and D). The isotyped matched negative control is shown in the overlay. M1 gate represents the positive populations. Data from at least 3 different donors were analyzed with similar results. Data from a representative donor are presented.
Expression of chemokine receptors on total BMMNC. BMMNC were isolated from bone marrow aspirates of healthy donors by Ficoll-gradient centrifugation; stained with MoAbs to CXCR4 (B), CCR5 (C), CCR2 (E), and CCR3 (F); and subjected to FACS analysis as described in Materials and Methods. The histogram represents analysis of 10,000 events acquired in the total ungated population (A and D). The isotyped matched negative control is shown in the overlay. M1 gate represents the positive populations. Data from at least 3 different donors were analyzed with similar results. Data from a representative donor are presented.
FACS analysis of CXCR4, CCR5, and CCR2 on subpopulations of BMMNC. Total BMMNC were stained with anti-CXCR4, anti-CCR5, and anti-CCR2 antibodies as described and FACS analysis was performed on the gated populations as indicated in Fig 1A and D. R1, R2, and R3 represent the lymphocyte, granulocyte precursor, and monocyte gates, respectively. The isotype negative controls are overlaid (bold line), and M1 represents the positive populations. Data from at least 3 different donors were analyzed. The mean percentage of positive cells for each chemokine receptor plus or minus the standard deviation is summarized in Table 1. Histograms from a representative donor are presented.
FACS analysis of CXCR4, CCR5, and CCR2 on subpopulations of BMMNC. Total BMMNC were stained with anti-CXCR4, anti-CCR5, and anti-CCR2 antibodies as described and FACS analysis was performed on the gated populations as indicated in Fig 1A and D. R1, R2, and R3 represent the lymphocyte, granulocyte precursor, and monocyte gates, respectively. The isotype negative controls are overlaid (bold line), and M1 represents the positive populations. Data from at least 3 different donors were analyzed. The mean percentage of positive cells for each chemokine receptor plus or minus the standard deviation is summarized in Table 1. Histograms from a representative donor are presented.
FACS Analysis of Normal Human BMMNC
| Chemokine Receptor . | Lymph . | Granulocyte . | Monocyte . |
|---|---|---|---|
| CXCR4 | 62 ± 20 | 41 ± 27 | 78 ± 4 |
| CCR5 | 24 ± 9 | 7 ± 4 | 55 ± 8 |
| CCR2 | 23 ± 5 | 84 ± 14 | 55 ± 4 |
| Chemokine Receptor . | Lymph . | Granulocyte . | Monocyte . |
|---|---|---|---|
| CXCR4 | 62 ± 20 | 41 ± 27 | 78 ± 4 |
| CCR5 | 24 ± 9 | 7 ± 4 | 55 ± 8 |
| CCR2 | 23 ± 5 | 84 ± 14 | 55 ± 4 |
Freshly isolated BMMNC from at least 3 different donors were stained and FACS analyzed as described in the text. Gates were set so that less than 5% of cells in the negative isotype control were in the positive gate. Data are presented as the mean percentage of positive cells plus or minus the standard deviation.
Chemokine receptor expression in in vitro expanded hematopoietic lineages.
To further examine the apparent lineage specificity of chemokine receptor expression in hematopoietic subsets and to control for uncharacterized factors in serum that might unduly affect chemokine receptor expression, we sought to determine chemokine receptor expression in erythroid, megakaryocyte, and granulo-macrophage cells expanded under serum-free conditions21 22 in liquid culture. Table 2 summarizes the expression of CCR5, CXCR4, and CCR2 on liquid cultured ex vivo expanded BFU-E–, CFU-Meg–, and CFU-GM–derived cells, as well as on mature erythrocytes and platelets. CCR5 and CXCR4 were both present on CFU-Meg– and CFU-GM–derived cells but were absent on BFU-E–derived cells. In contrast, CCR2 was predominantly present on erythroid cells. Thus, the pattern of chemokine receptor expression reflects some lineage specificity, with CCR2 restricted to the erythroid lineage cells and CCR5 and CXCR4 restricted to megakaryocytic and granulo-macrophage lineage cells.
FACS Analysis of Chemokine Receptor Expression on Ex Vivo Expanded Hematopoietic Cells
| Cell Type . | CXCR4 . | CCR5 . | CCR2b . |
|---|---|---|---|
| BFU-E derived | — | — | 67 ± 7 |
| CFU-Meg derived | 80 ± 12 | 31 ± 4 | — |
| CFU-GM derived | 64 ± 13 | 44 ± 17 | — |
| Erythrocytes* | — | — | — |
| Platelets* | 87 ± 6 | — | — |
| Cell Type . | CXCR4 . | CCR5 . | CCR2b . |
|---|---|---|---|
| BFU-E derived | — | — | 67 ± 7 |
| CFU-Meg derived | 80 ± 12 | 31 ± 4 | — |
| CFU-GM derived | 64 ± 13 | 44 ± 17 | — |
| Erythrocytes* | — | — | — |
| Platelets* | 87 ± 6 | — | — |
CD34+ BMMNC were obtained from at least 3 different healthy donors and serum-free expanded in liquid cultures as described in Materials and Methods. Gates were set so that less than 5% of cells in the negative isotype control were in the positive gate. Data are presented as the mean percentage of positive cells plus or minus the standard deviation.
Abbreviation: —, means less than or equal to isotype-matched negative control.
Primary hematopoietic material, not ex vivo expanded.
Chemokine receptor expression on CD34+ BMMNC.
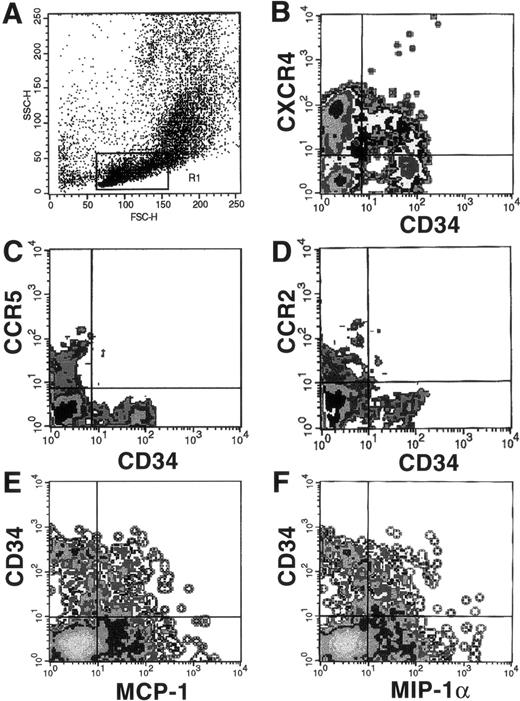
Because ligands to CXCR4, CCR5, and CCR2 have been reported to have effects on hematopoiesis, we next tried to determine if they were expressed on the surface of CD34+ BMMNC. Dual-color flow cytometric analysis of CD34+ BMMNC showed that, whereas greater than 50% of CD34+ cells were positive for CXCR4 (Fig 3B), less than 5% of CD34+ cells were positive for CCR2 (Fig 3D). At the same time, CCR5 was not present on CD34+ cells (Fig 3C).
FACS analysis of chemokine receptor expression on CD34+ cells using MoAbs or biotinylated ligands. BMMNC were costained with an FITC-conjugated MoAb to CD34 and MoAbs to CXCR4 (B), CCR5 (C), and CCR2 (D), followed by PE-conjugated antimouse IgG or streptavidin-PE as described. Alternatively, cells stained with PE-conjugated anti-CD34 MoAb were also costained with biotinylated MIP-1 (E) or MCP-1 (F), followed by avidin-FITC. The forward versus side-scatter characteristics of the gated population, R1, is shown (A). Negative gates were drawn according to the threshold seen with either the isotype-matched negative controls or in the case of the biotinylated ligands, after the addition of neutralizing antichemokine antibodies provided by the manufacturer (R&D Systems).
FACS analysis of chemokine receptor expression on CD34+ cells using MoAbs or biotinylated ligands. BMMNC were costained with an FITC-conjugated MoAb to CD34 and MoAbs to CXCR4 (B), CCR5 (C), and CCR2 (D), followed by PE-conjugated antimouse IgG or streptavidin-PE as described. Alternatively, cells stained with PE-conjugated anti-CD34 MoAb were also costained with biotinylated MIP-1 (E) or MCP-1 (F), followed by avidin-FITC. The forward versus side-scatter characteristics of the gated population, R1, is shown (A). Negative gates were drawn according to the threshold seen with either the isotype-matched negative controls or in the case of the biotinylated ligands, after the addition of neutralizing antichemokine antibodies provided by the manufacturer (R&D Systems).
Although MoAbs to CCR2, CCR3, CCR5, and CXCR4 are available, antibodies to many other chemokine receptors have not yet been developed. Therefore, we used the biotinylated chemokines MIP-1α and MCP-1 as probes to determine if additional chemokine receptors are expressed on CD34+ cells. MIP-1α binds to CCR1 and CCR4 in addition to CCR5, whereas MCP-1 also binds to CCR4 and CCR1 in addition to CCR2.27 28 Because CCR5 was not detectable on CD34+ cells and CCR2 was only detectable on less than 5% of the cells, binding to MIP-1α or MCP-1 would indicate the presence of either CCR1 or CCR4. Therefore, BMMNC were bound to biotinylated MIP-1α and MCP-1 and costained with an anti-CD34 MoAb. Dual-color FACS analysis showed that close to 50% of CD34+ cells were also positive for MIP-1α and MCP-1 receptors (Fig 3E and F). This binding was specific, because coincubation with neutralizing antichemokine antibodies abolished all specific binding activity (data not shown). There was also a distinct CD34−population of cells positive for MIP-1α and MCP-1 receptors, consistent with the CCR5+/CD34− and CCR2+/CD34− populations seen in Fig 3C and D. These binding data suggest that CCR1 and/or CCR4 must be present in significant amounts on CD34+ cells, although it is possible that as yet uncharacterized receptors to MIP-1α and MCP-1 may account for these data.
CXCR4+ cells are enriched in clonogeneic hematopoietic progenitors.
Because greater than 50% of human CD34+ cells coexpress CXCR4, we were interested if CXCR4 is expressed not only on CD34+ cells, but also on the clonogeneic human HPC. This issue is particularly germane, because mice lacking the SDF-1 gene, the natural ligand for CXCR4, appear to have severe defects in B-cell lymphopoiesis and bone marrow myelopoiesis.29 To address this issue, the CXCR4+ cells were isolated by using immunomagnetic beads as described in Materials and Methods. Immediately after isolation, CXCR4+ cells were plated in serum-free methylcellulose cloning medium and stimulated to grow CFU-Mix, BFU-E, CFU-GM, and CFU-Meg colonies by adding the appropriate cytokine cocktail. We found that human bone marrow CXCR4+ cells were clonogeneic and contain hematopoietic progenitors belonging to all major hematopoietic lineages (data not shown).
To further evaluate the distribution of clonogeneic HPC between CD34+/CXCR4+ and CD34+/CXCR4− cells, we FACS-sorted CD34+/CXCR4+ and CD34+/CXCR4− cells from nonadherent T-cell–depleted BMMNC (Fig 3B). Both fractions of cells were subsequently plated serum-free in methylcellulose cultures and stimulated to grow CFU-Mix, BFU-E, CFU-GM, and CFU-Meg colonies. We found that both fractions of CD34+ cells, positive or negative for CXCR4, contained hematopoietic progenitors belonging to the mixed, erythroid, myeloid, and megakaryocytic lineage (Table 3). Therefore, we conclude that human HPC were distributed equally in both CXCR4+CD34+ and CXCR4−CD34+ cells and that lack of CXCR4 in amounts that will allow for their isolation does not restrict the clonogeneic potential of HPC, at least not in the in vitro assays used.
Clonogeneic Potential of FACS-Sorted CXCR4+/CD34+ and CXCR4−/CD34+ BMMNC
| Colony Type . | CXCR4+/CD34+ . | CXCR4−/CD34+ . | P Value . |
|---|---|---|---|
| CFU-Mix | 6 ± 3 | 10 ± 3 | .03 |
| BFU-E | 56 ± 24 | 82 ± 29 | .07 |
| CFU-GM | 136 ± 39 | 171 ± 32 | .07 |
| CFU-Meg | 32 ± 20 | 30 ± 15 | .75 |
| Colony Type . | CXCR4+/CD34+ . | CXCR4−/CD34+ . | P Value . |
|---|---|---|---|
| CFU-Mix | 6 ± 3 | 10 ± 3 | .03 |
| BFU-E | 56 ± 24 | 82 ± 29 | .07 |
| CFU-GM | 136 ± 39 | 171 ± 32 | .07 |
| CFU-Meg | 32 ± 20 | 30 ± 15 | .75 |
Each data entry constitutes four independent clonogeneic assays from 2 different donors. Given the inherent variability between donors,P values greater than .01 are not considered significant.
Expression of CXCR4 mRNA in early human hematopoietic cells.
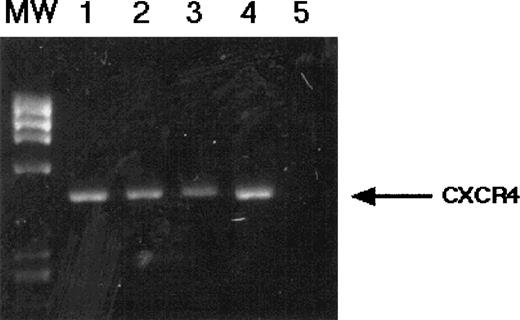
Our results clearly demonstrate that CXCR4 is expressed on the surface of CD34+ cells from human BMMNC. Moreover, CXCR4+ cells isolated from BMMNC have clonogeneic potential as they grow in vitro colonies belonging to all hematopoietic lineages. Therefore, we tried to determine if CXCR4 is expressed on the earliest human HSC. To address this issue, we isolated human CD34+c-kitR+Rh123low cells, which we have previously demonstrated to be highly enriched in HSC19 and CD34+, c-kitR+, Rh123bright cells that are enriched in HPC.19The mRNA was extracted from both populations of cells, and CXCR4 mRNA expression was analyzed by RT-PCR. As shown in Fig 4, both populations of cells enriched in either HSC (CD34+c-kitR+Rh123low) or in HPC (CD34+c-kitR+Rh123bright) expressed mRNA encoding for CXCR4. To determine if HSC also harbor CD4, the primary receptor for HIV-1 entry, we also performed RT-PCR analysis for CD4 mRNA. However, we were unable to demonstrate the expression of CD4 mRNA in HSC (data not shown).
RT-PCR analysis of CXCR4 mRNA expression in candidate HSC. CD34+, c-kit+, Rh123dull(lanes 1 and 2) and CD34+, c-kit+, Rh123bright (lanes 3 and 4) cells were FACS-sorted as described and subjected to RT-PCR analysis for CXCR4 mRNA. Negative control reaction (no template) is shown in lane 5. Specificity of the PCR products shown was confirmed by Southern blotting (data not shown) .
RT-PCR analysis of CXCR4 mRNA expression in candidate HSC. CD34+, c-kit+, Rh123dull(lanes 1 and 2) and CD34+, c-kit+, Rh123bright (lanes 3 and 4) cells were FACS-sorted as described and subjected to RT-PCR analysis for CXCR4 mRNA. Negative control reaction (no template) is shown in lane 5. Specificity of the PCR products shown was confirmed by Southern blotting (data not shown) .
Megakaryocytic cells are infectable by X4 and myeloid cells by R5 viruses.
Whereas BFU-E–, CFU-Meg–, and CFU-GM–derived cells all expressed one or more HIV-1 coreceptors, virus infection would be expected to occur only if CD4 were also expressed. Therefore, we also phenotyped these cells for CD4 antigen expression. CD4 was barely present on BFU-E–derived cells (Fig 5F) but was substantively present on CFU-Meg–derived (64% ± 9%) and CFU-GM–derived (47% ± 10%) cells (Fig 5D and B), respectively. Because megakaryocytic cells and granulo-macrophage cells cloned under serum-free conditions appear to have CD4 and both of the major HIV-1 coreceptors, we next tried to determine if these cells were indeed infectable by either R5 (M-tropic) or X4 (T-tropic) viruses. Classical viral infection assays rely on culturing virus-innoculated cells for up to 2 weeks and measuring levels of viral p24 or RT activity in the culture supernatant as evidence for a productive viral infection. However, culturing in vitro expanded hematopoietic colony cells even under serum-free conditions for such a long period may lead to changes in cellular phenotype unaccounted for their initial characterization, particularly if HIV-1 infection itself can lead to the secretion of proinflammatory and hematopoietic cytokines.14 Therefore, to determine if these megakaryocytic cells were permissive for viral replication at the time of our characterization of its cellular phenotype, we infected CFU-Meg–derived cells with pseudotyped luciferase reporter viruses. The luciferase reporter virus consists of the NL4-3 provirus with a frame-shift mutation, its env gene rendering it replication incompetent, and a luciferase gene inserted into its nef coding region.25,26 Because this provirus does not have a functional Env of its own, it can be pseudotyped by cotransfecting the proviral backbone with a plasmid coding for any viral Env of interest into the appropriate packaging cells. Viruses thus produced will be capable of a single-cycle infection and if infection proceeds to the point of viral integration and LTR-transcription, luciferase will be produced and productively infected cells can be assayed for luciferase activity. This reporter virus system has been widely used to measure the ability of various cell types to support virus entry and integration.26 30
Expression of CD4 on serum-free expanded hematopoietic progenitor-derived cells. CFU-GM (A and B), CFU-Meg (C and D), and BFU-E (E and F) progenitors were serum-free expanded from CD34+ BMMNC as described and FACS analyzed for CD4 antigen expression. Histograms (B, D, and F) represent the gated populations as indicated (A, C, and E). The isotype negative controls are overlaid (bold line), and M1 represents the positive populations. Data from at least 3 different donors were analyzed. The mean percentage of positive cells for each chemokine receptor plus or minus the standard deviation is summarized in the test. Histograms from a representative donor are presented.
Expression of CD4 on serum-free expanded hematopoietic progenitor-derived cells. CFU-GM (A and B), CFU-Meg (C and D), and BFU-E (E and F) progenitors were serum-free expanded from CD34+ BMMNC as described and FACS analyzed for CD4 antigen expression. Histograms (B, D, and F) represent the gated populations as indicated (A, C, and E). The isotype negative controls are overlaid (bold line), and M1 represents the positive populations. Data from at least 3 different donors were analyzed. The mean percentage of positive cells for each chemokine receptor plus or minus the standard deviation is summarized in the test. Histograms from a representative donor are presented.
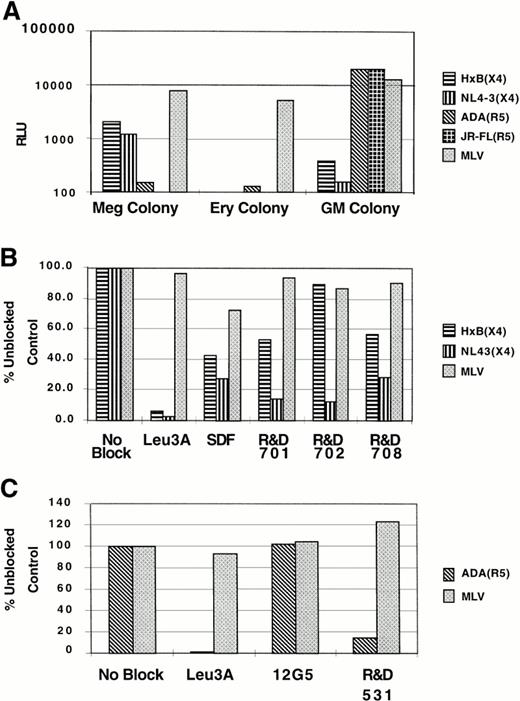
Somewhat surprisingly, we found that megakaryocytic cells were infectable by X4 (HxB, NL4-3) and not R5 (ADA, JRFL) viruses, whereas granulo-macrophage cells were infectable by R5 (ADA, JRFL) but not X4 (HxB, NL4-3) viruses, despite the fact that cells from both lineages express both of the HIV-1 coreceptors (Fig6A). To show that viral entry into these cells was indeed mediated by CD4 and the cognate coreceptor, reporter virus infection was performed in the presence of Leu3A (an anti-CD4 antibody) or antibodies to either CXCR4 (12G5, R&D701, 702,708) or CCR5 (R&D 531). As can be seen in Fig6B and C, Leu3A was highly effective in neutralizing viral entry (as measured by luciferase production), and both anti-CXCR4 and anti-CCR5 antibodies were variously effective in blocking cognate viral entry. The differential susceptibility of the NL4-3 or HxB Env to CXCR4-specific MoAb or SDF-1 inhibition is consistent with reports in the literature showing that inhibition of CXCR4-mediated entry by either anti-CXCR4 MoAb or SDF-1 is highly strain specific.31 32 This indicates that viral entry into megakaryocytic cells was CD4 and CXCR4-dependent and that entry into granulo-macrophage cells was CD4 and CCR5-dependent. By contrast, BFU-E–derived cells were not infectable by either R5 or X4 viruses, consistent with our failure to detect CXCR4 or CCR5 in this cell population. However, these erythroid cells were readily infectable by viruses bearing the amphotorpic MLV Env protein, indicating that the block to infection by R5 and X4 viruses was at the level of viral entry (Fig 6A). Infection with pseudotyped GFP reporter viruses confirmed that only CD41+ cells in CFU-Meg–derived cells are infeactable by X4-env pseudotyped viruses (data not shown). These results in toto indicate that megakaryocytic cells were infectable by X4 viruses and that infection of these cells was mediated through CD4 and CXCR4.
Infection of lineage-specific hematopoietic cells with pseudotyped reporter viruses. (A) Approximately 2 × 105megakaryocytic (Meg colony), erythroid (Ery colony), and granulocyte-macrophage (GM colony) cells were infected with X4 or R5 Env pseudotyped viruses as indicated. Four days after infection, cells were lysed and analyzed for luciferase activity (RLU). The amphotropic MLV Env pseudotyped virus was used to control for cell viability. Megakaryocytic (B) and granulo-macrophage cells (C) were infected with either two different X4 Env pseudotyped viruses or a prototypic R5 Env pseudotyped virus, respectively, in the presence or absence of blocking agents. Leu3A is an MoAb against CD4 that recognizes the HIV Env binding epitope on CD4; SDF-1 is the natural ligand for CXCR4; R&D 701, 702, 708, and 12G5 are MoAbs against CXCR4; and R&D 531 is an MoAb against CCR5. The RLU obtained in the presence of blocking agents is normalized to the RLU obtained without any blocking agents, and the data for infection efficiency are presented as the percentage of unblocked control. Note that none of the blocking agents had any effect on the entry of the MLV pseudotyped virus, indicating the specificity of any blocking effects. All infection and blocking experiments were repeated 2 to 3 independent times with different donors with similar results. Representative experiments are shown.
Infection of lineage-specific hematopoietic cells with pseudotyped reporter viruses. (A) Approximately 2 × 105megakaryocytic (Meg colony), erythroid (Ery colony), and granulocyte-macrophage (GM colony) cells were infected with X4 or R5 Env pseudotyped viruses as indicated. Four days after infection, cells were lysed and analyzed for luciferase activity (RLU). The amphotropic MLV Env pseudotyped virus was used to control for cell viability. Megakaryocytic (B) and granulo-macrophage cells (C) were infected with either two different X4 Env pseudotyped viruses or a prototypic R5 Env pseudotyped virus, respectively, in the presence or absence of blocking agents. Leu3A is an MoAb against CD4 that recognizes the HIV Env binding epitope on CD4; SDF-1 is the natural ligand for CXCR4; R&D 701, 702, 708, and 12G5 are MoAbs against CXCR4; and R&D 531 is an MoAb against CCR5. The RLU obtained in the presence of blocking agents is normalized to the RLU obtained without any blocking agents, and the data for infection efficiency are presented as the percentage of unblocked control. Note that none of the blocking agents had any effect on the entry of the MLV pseudotyped virus, indicating the specificity of any blocking effects. All infection and blocking experiments were repeated 2 to 3 independent times with different donors with similar results. Representative experiments are shown.
Upregulation of CXCR4 expression in human BMMNC after γ-IFN treatment.
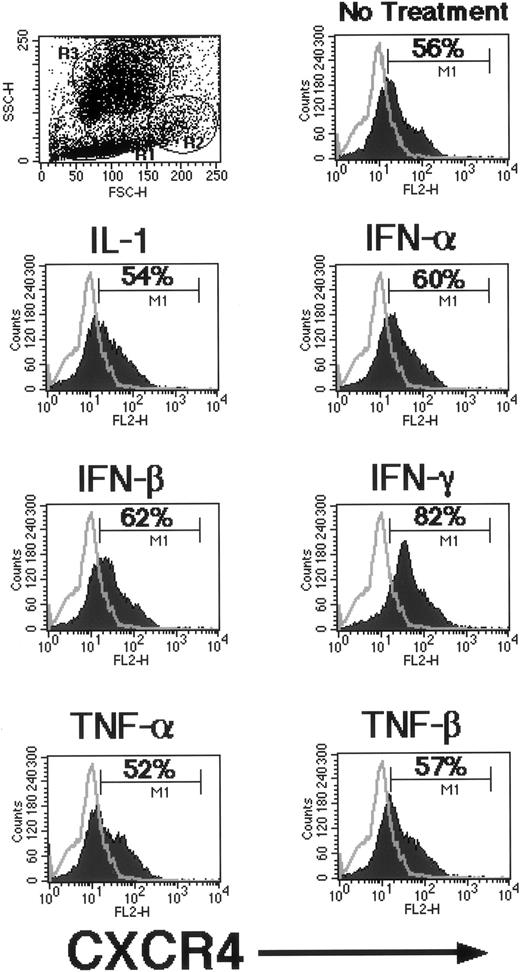
Because different proinflammatory cytokines (IL-1, TNF-α, TNF-β, α-IFN, β-IFN, and γ-IFN) secreted during chronic infections have been reported to either induce or suppress HIV infection in various cell types, we evaluated if these cytokines were able to modulate the expression of HIV-1 coreceptors (CXCR4, CCR5, and CD4) on human BMMNC. To address this issue, BMMNC were resuspended in serum-free medium and stimulated for 36 hours with different proinflammatory cytokines. Subsequently, we evaluated changes in CXCR4, CCR5, and CD4 expression by FACS. As can be seen in Fig 7, of all the proinflammatory cytokines tested, only γ-IFN increased expression of CXCR4. In three independent experiments, γ-IFN increased the number of CXCR4-expressing cells by approximately 20% to 30% of total BMMNC. However, this upregulation was not a global effect on all CXCR4 expressing cells. When the BMMNC were gated based on their forward versus side scatter characteristics, most of the CXCR4 upregulation occurred in the granulocyte precursor and monocyte gates (data not shown). In the parallel experiments, none of the proinflammatory cytokines (IL-1, TNF-α, TNF-β, α-IFN, β-IFN, and γ-IFN) evaluated had any effect on the expression of CCR5 or CD4 (not shown). These results suggest that γ-IFN released during the course of a chronic infection may affect the susceptibility of certain BMMNC to X4 virus infection.
Regulation of CXCR4 expression by γ-IFN. Freshly isolated BMMNC in serum-free media were either left alone or treated with a variety of proinflammatory cytokines as indicated for 36 hours. Expression of CXCR4 was monitored by FACS analysis after the treatment period. The negative isotype control is overlaid on each histogram. A representative experiment is shown of three independent repeats with similar results. M1 represents the positive population; the percentage of positive cells is indicated within each histogram.
Regulation of CXCR4 expression by γ-IFN. Freshly isolated BMMNC in serum-free media were either left alone or treated with a variety of proinflammatory cytokines as indicated for 36 hours. Expression of CXCR4 was monitored by FACS analysis after the treatment period. The negative isotype control is overlaid on each histogram. A representative experiment is shown of three independent repeats with similar results. M1 represents the positive population; the percentage of positive cells is indicated within each histogram.
DISCUSSION
The pathogenesis of HIV-1–associated hematopoietic dysfunction has been a subject of intense investigation and considerable debate. It is likely that no one mechanism can account for the spectrum of hematological abnormalities seen in AIDS. The confluence of experimental results thus far seem to implicate the ability of virus infection or viral gene products to disrupt the hematoregulatory function of bone marrow auxiliary cells (reviewed in Moses et al14). However, the recent discovery of certain chemokine receptors such as HIV-1 coreceptors coupled with the reported ability of cognate chemokine receptor ligands such as MIP-1α and SDF-1 to modulate hematopoietic development29,33 34 has opened a new arena of investigative opportunities regarding the pathogenesis of AIDS-related cytopenias.
Cellular infection by HIV-1 requires the presence of CD4 and at least one additional coreceptor. Accordingly, R5 viruses require CCR5 and X4 viruses require CXCR4 in addition to CD4 for cellular entry.35 Because chemokine receptors may mediate some of the negative influences of the chemokines on the clonogeneic growth of early hematopoietic cells3 and both HIV-1 Env and the ligands to HIV-1 coreceptors can be secreted in excess during HIV-1 infection, deciphering the chemokine/chemokine receptor axes in hematopoietic cells will allow a first approximation as to which hematopoietic subsets might be susceptible to detrimental effects of direct viral infection or Env-mediated cytotoxicity as well as chemokine-mediated hematodysregulation.
In this report, we have examined cell surface expression of the major HIV-1 coreceptors, CCR5 and CXCR4, on various subsets of hematopoietic cells. Although there was pleiotropic expression of these receptors as well as CCR2b to varying degrees on the cells from the lymphocyte, monocyte, and granulocyte gates in total BMMNC, chemokine receptor expression appeared more lineage restricted when examined on cells from serum-free expanded hematopoietic progenitors. CCR5 and CXCR4 were both expressed on cells expanded from CFU-GM and CFU-Meg, whereas CCR2 was predominantly expressed on BFU-E–derived cells. Because we found that CD4 is also expressed on both myeloid and megakaryocytic cells, it was surprising that myeloid cells were only infectable by R5 Env pseudotyped viruses and megakaryocytic cells were only infectable by X4 Env pseudotyped viruses. This finding implies that the physical presence of the appropriate receptors and coreceptors on the cell surface does not necessarily ensure a productive infection. The infectability of megakaryocytic cells by X4 but not R5 viruses has been reported recently.36 Our results confirm and extend these findings by characterizing the coreceptors responsible for infection of both CFU-Meg and CFU-GM derived cells. The apparent discrepancy between the expression of CD4 and the appropriate coreceptor and the restrictive tropism of certain primary cells has precedence in the HIV-1 infection of macrophages. It is becoming increasingly clear that, although both CCR5 and CXCR4 are expressed on macrophages, only R5 viruses can productively replicate in these cells.37,38However, certain R5/X4 viruses can productively infect CCR5-deficient macrophages via CXCR4.37 Whether this restriction of tropism is due to the affinity of the particular Env for the coreceptor in question, the CD4/coreceptor ratio required for productive membrane fusion,39,40 or postentry determinants in the cellular milieu of the target cell remains to be determined.7,8 41However, the restrictive tropism of CFU-Meg and CFU-GM cells offers an additional model in which to sort out the effects that determine viral entry and replication. Furthermore, the susceptibility of CFU-Meg–derived cells to X4 virus infection supports the notion that HIV-1–related thrombocytopenia may be partially explained by the cytopathic effects resulting from direct infection of megakaryocytic precursors. To our knowledge, this is also the first demonstration that erythroid cells are resistant to infection with R5 and X4 viruses. This could be explained by our findings that, although erythroid precursor cells express low levels of CD4, they did not express CXCR4 or CCR5. Therefore, direct infection of erythroid precursor cells probably does not play a major role in the pathogenesis of HIV-related anemia.
It has also been reported recently that CD34+ cells express mRNA for CXCR4 and, to a lesser degree, CCR5.13 In this report, we characterized the expression of chemokine receptors on CD34+ cells at the protein level and the expression of a variety of other chemokine receptors on CD34+ BMMNC. We found that CD34+ BMMNC express CXCR4 but not CCR5, CCR3, or CCR2 proteins. It is also significant that we not only demonstrated cell surface expression of CXCR4 on hematopoietic progenitor cells (CD34+), but also that FACS-sorted CD34+/CXCR4+ cells were clonogeneic and capable of giving rise to all major hematopoietic lineages (CFU-mix, CFU-GM, CFU-Meg, and BFU-E). Interestingly, the clonogeneic potential of hematopoietic precursor cells did not appear to be limited to CXCR4+ cells, because CD34+/CXCR4− cells were also capable of giving rise to multilineage colony formation. This finding implies that CXCR4 may not be a sensitive selection marker for all hematopoietic progenitors. However, in vitro colony-forming assays are only a surrogate for true stem-like regenerative capacity. It remains to be seen if CD34+/CXCR4+ and CD34+/CXCR4− cells possess true stem-like clonogeneic potential by a more stringent test such as SCID-mice repopulation. The potential presence of CXCR4 on human HSC is supported by the fact that we could detect CXCR4 mRNA by RT-PCR in CD34+ Kit+ Rh123low cells that are highly enriched in human hematopoietic stem cells.19Interestingly, our failure to detect expression of CD4 mRNA in the same population of cells could explain why human HSC are resistant to infection by HIV.14,42,43 Nevertheless, the expresssion of CXCR4 on candidate human stem cells as well as on a variety of clonogeneic human progenitor cells has implications for lentiviral gene therapy, because there are HIV-1 and HIV-2 viruses that can use CXCR4 for entry independent of CD4.44-46 Therefore, pseudotyping lentiviral vectors with these CXCR4-dependent, CD4-independent Envs may provide a way of specifically targeting therapeutic genes to hematopoietic stem and progenitor cells.
We also found that, although CD34+ cells were negative for CCR5 and CCR2 by MoAb staining, they were clearly positive for other MIP-1α and MCP-1 receptors as shown by FACS analysis with biotinylated ligands. Because MIP-1α and MCP-1 are also known ligands for CCR1 and CCR4,28 these results imply that CCR1 and/or CCR4 are also present on CD34+ cells. CCR1 and CCR4 mRNA have recently been reported to be expressed in CD34+ cells and the inhibitory effects of MIP-1α on erythropoiesis has been shown to be mediated through CCR1.47 This study shows that CCR1 and/or CCR4 on human CD34+ cells can indeed bind to their respective ligands. Considering that cognate ligands to many of the chemokine receptors examined are secreted in excess during chronic HIV infection,48-50 the delineation of chemokine receptor expression on various subsets of hematopoietic progenitors represents a first step towards teasing apart the intricate network of relationships between chemokine receptors, HIV infection, and hematopoiesis.3
We also tested the hypothesis that some of the proinflammatory cytokines released during chronic infections may modulate the course of HIV infection by augmenting the expression of particular chemokine coreceptors on the surface of hematopoietic cells. Our finding that γ-IFN can upregulate the expression of CXCR4 underscores the interplay between cytokine release during chronic HIV infection and the chemokine/chemokine receptor axes. γ-IFN is greatly increased in lymphoid tissues during HIV-1 infection,51 and other cytokines such as GM-CSF and IL-10 have been shown to decrease or increase the expression of CCR5, respectively.52 53 Thus, cytokine-mediated modulation of coreceptor expression may play a role in the dynamics of HIV replication in vivo.
In conclusion, we have determined the pattern of CD4 and major HIV-1 coreceptor expression on a variety of HPC and correlated this with their susceptibility to HIV-1 infection. We found that productive infection of cells is likely more complicated than the mere physical presence of CD4 and coreceptor on the cell surface. Finally, we also determined that γ-IFN can upregulate the expression of CXCR4 on BMMNC. The results presented represent a guide towards future investigations into the biological consequences of chemokine receptor expression on hematopoietic cells and offer an initial framework in which to sort out the web-like complexity between the myriad cytokine and chemokine networks that may impinge upon the dynamics of HIV-1 replication.
ACKNOWLEDGMENT
The authors thank Monica Tsang (R&D Systems) and Carlos Martines-A. (Universidad Autonoma de Madrid) for generously providing the chemokine receptor antibodies.
B.L. was supported by the Measey Foundation Fellowship for Clinicians (Wistar Institue). R.W.D. was supported by National Institutes of Health (NIH) Grant No. AI-40880. A.M.G. and M.Z.R. were supported by a program project grant in stem cell biology NIH PO1 DK52558-01A1, NIH R01 HL 61796-01.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Mariusz Z. Ratajczak, MD, PhD, Department of Medicine, Hospital of the University of Pennsylvania, 1007 Stellar-Chance Laboratories, 422 Curie Blvd, Philadelphia, PA 19104.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal