Abstract
Hodgkin–Reed-Sternberg (H-RS) cells are clonal B cells carrying Ig gene rearrangements. However, in situ hybridization methods failed to demonstrate Ig gene expression in H-RS cells of classical Hodgkin’s disease (HD). Because somatic mutations rendering potentially functional Ig gene rearrangements nonfunctional were detected in some cases of the disease, it was speculated that H-RS cells in classical HD may have lost the ability to express antigen receptor as a rule. Recently, we established a novel cell line (L1236) from H-RS cells of a patient with mixed cellularity subtype of HD. L1236 cells harbor a potentially functional VH1 and a potentially functional Vκ3 gene rearrangement. However, no antibody expression was detected. To show potential reasons for this lack of Ig expression, we analyzed the genomic organization of the Ig genes and their transcription in the primary and cultivated H-RS cells of this patient. The H-RS cells were found to have switched their isotype to IgG4, confirming their mature B-cell nature. By amplifying cDNA from L1236 cells as well as from frozen biopsy material transcripts of the Vκ3 and the VH1 gene rearrangement were detected for both sources of cDNA. However, Northern blot hybridization of L1236 RNA failed to demonstrate VH1 and Vκ3 transcripts, indicating only a low level of transcription. Sequence analysis of the promoter and leader regions of the VH1 gene rearrangement from L1236 cells as well as from lymphoma-affected tissue showed a somatic mutation in the conserved octamer motif of the promoter region. Somatic mutations were also detected within the 3′ splice site of the leader intron and adjacent nucleotides in the rearranged Vκ light chain gene, leading to aberrant splicing. These mutations might prevent the generation of adequate amounts of functional Ig gene transcripts as template for translation into protein. Thus, mutations in H-RS cells that prevent Ig gene expression might also be located outside the coding region of the Ig genes.
IN HODGKIN’S DISEASE (HD), the malignant Hodgkin–Reed-Sternberg (H-RS) cells represent only a minority of 0.1% to 1% of all cells in affected tissue. They are surrounded by T lymphocytes, histiocytes, fibroblasts, eosinophils, and other nonmalignant cells. Because of the scarcity of H-RS cells in tumor tissue, their genetic characterization has been a major problem in the past. Until recently, clonality and cellular origin of these cells has remained an open question.
The isolation of single H-RS cells from frozen tissue sections1 or cytospins2 by micromanipulation followed by polymerase chain reaction (PCR) analysis now allows molecular analysis of single H-RS cells. Using these methods, detection of Ig gene rearrangements in single H-RS cells of classical HD (ie, nodular sclerosis, mixed cellularity [mc], lymphocyte depleted) has shown their B-cell origin.1,3-8 In most of the cases, Ig gene rearrangements amplified from multiple H-RS cells showed a clonal relationship. Evidence for clonality of H-RS cells in classical HD has also been provided by detection of clonal chromosomal aberrations using fluorescence in situ hybridization on HD-derived cell suspensions with9 and without10 simultaneous immunophenotyping. Thus, H-RS cells arise from a single B-cell clone in most if not all cases. The presence of somatic mutations within the rearranged Ig genes of H-RS cells implied their derivation from germinal center (GC) B cells or their descendants.1,7 8
In 4 of 13 cases of classical HD analyzed by us,1,7,8 stop codons were introduced by somatic mutation into originally functional V gene rearrangements, rendering them nonfunctional. Such crippling mutations physiologically occur in mutating GC B cells. However, GC B cells harboring crippling mutations are usually eliminated by apoptosis and do not leave the GC microenvironment. Only GC B cells that acquired favorable mutations resulting in increased affinity to the respective antigen can leave the GC as memory B cells or plasma cell precursors.11 Thus, the finding of crippling mutations in some cases of classical HD identified GC B cells that escaped apoptosis as the precursors of the tumor clone, at least in these cases. It has been speculated that H-RS cells in classical HD might be derived from crippled GC B cells as a rule.8
In 1 of the 13 cases, potentially functional Ig gene rearrangements for both heavy and κ light chain genes were amplified from single H-RS cells.7 Thus, H-RS cells of this case might have retained the ability to express antigen receptor. Fortunately, a cell line (L1236) could be established from the peripheral blood of this patient suffering from relapse of mc HD.12 Using molecular analysis of Ig gene rearrangements, the derivation of L1236 cells from the H-RS cells in the patient could be demonstrated.7 The in vitro cultivation of the L1236 cells enabled us to perform a detailed analysis of the genomic organization of the Ig genes and their transcription and translation. Class switch recombination to Cγ4 was demonstrated in the rearranged Ig genes in L1236 and primary H-RS cells. Transcription of Ig genes was detected at a very low level, and no protein was found at all. Somatic mutations within their promoter and leader regions were identified as the most likely mechanism for downregulation of Ig gene expression.
MATERIALS AND METHODS
Tissue samples and cell lines.
L1236 is an Epstein-Barr virus (EBV)-negative H-RS cell line established from the peripheral blood of a patient suffering from a relapse of mc HD with extensive bone marrow involvement.12Sections from fresh frozen tissue of the patient’s bone marrow were used to analyze Ig gene expression and structure in primary H-RS cells. L1309 is an EBV-immortalized lymphoblastoid cell line (LCL) that has been established in our laboratory from the peripheral blood of a healthy donor by superinfection of B lymphocytes with the EBV strain B95-8. L1309 was used as positive control for analyzing expression of rearranged Ig genes. CO is an HD-derived cell line with T-cell receptor gene rearrangements13 and was used as a negative control. In addition, U937, a cell line of myeloid origin14 was used as a negative control for the detection of Ig gene transcripts by Northern blot analysis. All cell lines were grown in suspension at 37°C in RPMI 1640 medium (Life Technologies, Eggenstein, Germany) supplemented with 10% fetal calf serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mmol/L glutamine in an atmosphere containing 5% CO2.
DNA extraction and Southern blot analysis.
High molecular weight DNA was extracted from cell lines using standard protocols.15 DNA was extracted from frozen tissue sections using the trizol reagent according to the manufacturer’s instructions (Life Technologies). Ten micrograms of cellular DNA extracted from cell lines was cleaved with the restriction enzyme BamHI, separated by electrophoresis for 36 hours at 1 V/cm on a 0.8% agarose gel, and transferred onto a nylon filter (Hybond N; Schleicher und Schuell, Dassel, Germany). For detection of the Cγ genes, a Cγ4 DNA probe covering all Cγ4 exons and hybridizing to all Cγ genes16 was used. The probes were 32P-labeled and hybridized using standard conditions.15
RNA extraction and Northern blot hybridization.
Total cellular RNA was extracted from cell lines by the guanidinium isothiocyanate method.17 From frozen tissue sections RNA was extracted using the trizol reagent according to the manufacturer’s instructions (Life Technologies). Poly A+ RNA was separated from total RNA according to the protocol for the Oligotex mRNA midi kit (Qiagen, Hilden, Germany). Ten micrograms of total RNA or 5 μg of poly A+ RNA was separated on a 1% agarose gel, transferred onto a nylon filter (Biotechnology Systems, NEN, Research Products, Boston, MA), and hybridized with a 32P-labeled DNA probe under standard conditions.18 For detection of Ig γ heavy chain transcripts, the Cγ4 probe was used16(see also Southern blot analysis). For the detection of Ig κ light chain transcripts, a 2.1-kb Sac I/EcoRI fragment of the human Ig κ light chain constant region gene was used.19
Reverse transcription-PCR (RT-PCR) for amplification of Ig gene transcripts.
cDNA was generated using an adapter oligonucleotide (AP2) AAG GAT CCG TCG ACA TC (T)17 and Moloney murine leukemia virus (M-MLV) reverse transcriptase (Pharmacia, Freiburg, Germany). For amplification of Ig gene transcripts and Ig constant region genes, several oligonucleotides were used (see below). PCR was performed in a 50 μL reaction mixture containing 50 mmol/L KCl, 2.5 or 1.5 mmol/L MgCl2, 200 μmol/L of each dNTP, and 25 μmol/L of each oligonucleotide. PCR was performed according to different protocols (see below). Denaturation was always performed for 1 minute at 95°C. One unit of Taq DNA polymerase (Promega, Mannheim, Germany) was added during the first denaturation step. All amplifications were performed in a Trio Thermoblock (Biometra, Göttingen, Germany). When a two-step PCR was performed, 1 μL of the first round was used as template in the second round.
PCR-oligonucleotides.
Sequences of the VH1 oligonucleotides were as follows: 21-2UIS: gaggtatgaaataatctgtc; VH1LAs: gaatgacagtcttctaaactgg; 5-1CDR2: ccggtgttggctcgacaatg; 3H1: gtggccattgtagttcccac; GIn5: aggaccctgcccctgacctaag; and GC23: ggggtcttcgtggctcacgtc. Sequences of the Vκ3 oligonucleotides were as follows: VK3P: tggtctttgcagctgaaagct; VK3L: atggaagccccagcgcagctt; VK3As: gctaagttggtgccaatgctgtc; 3K3: aacgtcaccggaggccactta; and CΚ: ttcaactgctcatcagatggaggg.
Amplification of DNA and cDNA fragments.
Amplification of the 5′ untranslated region of the VH1 gene rearrangement of L1236 cells was performed by single-step PCR (5′ oligonucleotide: 21-2UIS; 3′ oligonucleotide: 3H1; 2.5 mmol/L MgCl2; annealing at 58°C for 1 minute and extension at 72°C for 3 minutes for 35 cycles; length of the product: 616 bp). Amplification of the 5′ untranslated region of the VH1 gene rearrangement of primary H-RS cells from the bone marrow specimen was performed by PCR (5′ oligonucleotide: 21-2UIS; 3′ oligonucleotide: 3H1; 2.5 mmol/L MgCl2; annealing at 58°C for 1 minute and extension at 72°C for 3 minutes for 40 cycles; length: 616 bp; sequence oligonucleotide VH1LAS). Amplification of the 5′ untranslated region of the Vκ gene rearrangement of L1236 cells was performed by single-step PCR (5′ oligonucleotide: VK3P; 3′ oligonucleotide: 3K3; 2.5 mmol/L MgCl2; annealing at 61°C for 1 minute and extension at 72°C for 90 seconds for 35 cycles; length: 665 bp). Amplification of the 5′ untranslated region of the Vκ gene rearrangement of primary H-RS cells from the bone marrow specimen was performed by a two-step PCR (first round: 5′ oligonucleotide: VK3P; 3′ oligonucleotide: 3K3; 2.5 mmol/L MgCl2; annealing at 61°C for 1 minute and extension at 72°C for 90 seconds for 35 cycles; length: 665 bp; second round: 5′ oligonucleotide: VK3P; 3′ oligonucleotide: VK3As; 2.5 mmol/L MgCl2; annealing at 58°C for 1 minute and extension at 72°C for 90 seconds for 35 cycles; length: 470 bp). A single-step RT-PCR was performed to detect VH1 transcripts in L1236 cells and primary H-RS cells (5′ oligonucleotide: 5-1CDR2; 3′ oligonucleotide: GC23; 1.5 mmol/L MgCl2; annealing at 59°C for 1 minute and extension at 72°C for 90 seconds for 40 cycles; length: 661 bp). Detection of Vκ transcripts in L1236 and primary H-RS cells was performed by single-step RT-PCR (5′ oligonucleotide: VK3L; 3′ oligonucleotide: 3K3; 1.5 mmol/L MgCl2; annealing at 60°C for 1 minute and extension at 72°C for 90 seconds for 40 cycles; length: 356 bp). Vκ/Cκ transcripts in L1236 cells were detected by RT-PCR (5′ oligonucleotide: VK3L; 3′ oligonucleotide: CK; 2.5 mmol/L MgCl2; annealing at 58°C for 1 minute and extension at 72°C for 90 seconds for 40 cycles; length: 440 bp). The Cγ4 gene in L1236 cells was amplified by a single-step PCR (5′ oligonucleotide: GIn5; 3′ oligonucleotide: GC23; 2.5 mmol/L MgCl2; annealing at 58°C for 1 minute and extension at 72°C for 90 seconds for 35 cycles; length: Cγ4: 234 bp).
Sequence analysis.
PCR products were separated on a 1% agarose gel and extracted from the gel using a gel extraction kit (Qiagen, Hilden, Germany). PCR products were directly sequenced using either the Ready Reaction DyeDeoxyTerminator cycle sequencing kit (Applied Biosystems, Weiterstadt, Germany) or using a cycle sequencing kit (Life Technologies). Sequencing reactions were performed according to the manufacturers’ protocols. Sequence reaction products were separated on an 8% polyacrylamide gel under denaturing conditions. Results were obtained by either autoradiography or using an automated sequencer (ABI377; Applied Biosystems).
RESULTS
L1236 cells have performed class switching.
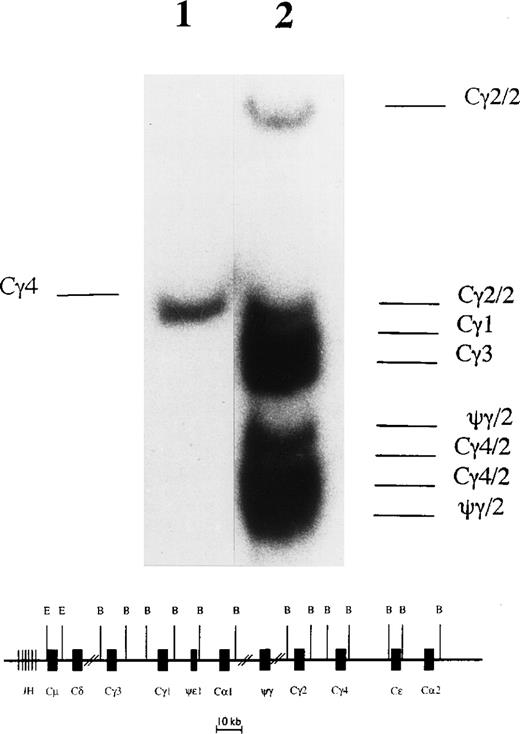
Southern blot hybridization of BamHI-digested L1236 DNA with a Cγ probe showed only one rearranged fragment and none of the germline Cγ fragments (Fig 1). This suggests that all Cγ genes on one chromosome were deleted and that on the other chromosome class switch recombination was targeted to the most 3′ Cγ gene, ie, Cγ4. To confirm this, PCR was performed with genomic DNA of L1236 cells using oligonucleotides suitable to amplify all Cγ genes. Sequence analysis of the resulting PCR product confirmed that only Cγ4 was retained in L1236 cells (data not shown). Further studies showed that class switch recombination to Cγ4 had occurred on the allele harboring the potentially functional VH1 gene rearrangement (see below and Fig 5).
Southern blot analysis to detect class switch recombination in L1236 cells. (Upper part) After digestion of genomic DNA with BamHI, the organization of the Cγ genes was analyzed in L1236 cells (lane 1) in comparison with the HD-derived cell line Co (lane 2) known to have germline configuration of the Cγ genes.20 The Cγ probe used in the analysis recognizes all Cγ genes and the ψγ gene. As expected, the DNA of Co shows all germline fragments for the IgG subclasses. The two IgH loci located on the two chromosomes 14 show a polymorphism for the pseudo Cγ (ψγ), the Cγ2, and the Cγ4 genes resulting in two fragments for these (indicated as /2), whereas only one fragment appears for Cγ1 and Cγ3 each. This pattern is in accordance with the results described by Linsley et al.21 For L1236 DNA, only one fragment is detectable, indicating that on one allele class switch recombination was targeted to the most 3′ γ gene (ie, Cγ4), whereas on the other allele all Cγ genes were deleted. (Lower part) Map of the human IgH locus. This map does not take into account the different haplotypes for the ψγ, the Cγ2, and the Cγ4 genes and is not to scale. B, BamHI; E, EcoRI.
Southern blot analysis to detect class switch recombination in L1236 cells. (Upper part) After digestion of genomic DNA with BamHI, the organization of the Cγ genes was analyzed in L1236 cells (lane 1) in comparison with the HD-derived cell line Co (lane 2) known to have germline configuration of the Cγ genes.20 The Cγ probe used in the analysis recognizes all Cγ genes and the ψγ gene. As expected, the DNA of Co shows all germline fragments for the IgG subclasses. The two IgH loci located on the two chromosomes 14 show a polymorphism for the pseudo Cγ (ψγ), the Cγ2, and the Cγ4 genes resulting in two fragments for these (indicated as /2), whereas only one fragment appears for Cγ1 and Cγ3 each. This pattern is in accordance with the results described by Linsley et al.21 For L1236 DNA, only one fragment is detectable, indicating that on one allele class switch recombination was targeted to the most 3′ γ gene (ie, Cγ4), whereas on the other allele all Cγ genes were deleted. (Lower part) Map of the human IgH locus. This map does not take into account the different haplotypes for the ψγ, the Cγ2, and the Cγ4 genes and is not to scale. B, BamHI; E, EcoRI.
Ig transcripts in L1236 cells are undetectable by Northern blot analysis.
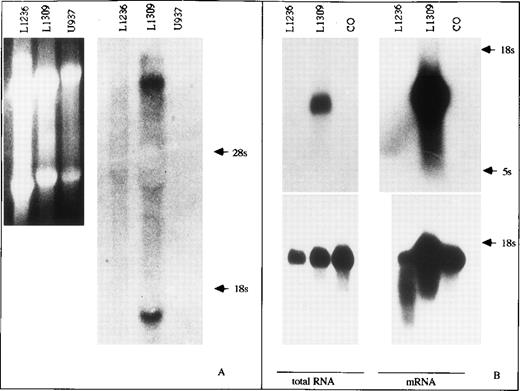
Using various antibodies against human γ heavy as well as κ light chain protein, no staining was observed in the L1236 cell line when flow cytometry (Igκ light chain12) or immunocytochemistry (IgG heavy chain, Igκ light chain; data not shown) was performed. We thus wanted to know if the failure to detect Ig in L1236 cells was caused by a lack of or downregulation of either translation or transcription of the Ig genes. For the detection and quantification of Ig gene transcription in L1236 cells, Northern blot analysis was performed. Hybridization of total cellular L1236 RNA with specific probes for the Cγ or the Cκ genes showed neither Cγ nor Cκ transcripts (Fig 2). By comparison, hybridization of RNA extracted from the B-cell–derived cell line L1309 showed the expected transcripts. The T-cell–derived cell line CO and the myeloid cell line U937, both used as controls, gave the expected negative results. To increase the sensitivity of the method, poly A+ RNA was extracted to detect Cκ transcripts. Again, no transcripts were detected in L1236 by hybridization of the Cκ probe (Fig2). Hybridization with a glyceraldehyde phosphate dehydrogenase (GAPDH) probe to check the quality and amount of mRNA showed that approximately equal amounts of intact RNA were loaded on the gel for the various cell lines. The Northern blot hybridization experiments thus showed that Ig gene transcription in L1236 was below the sensitivity of the method.
Northern blot analysis for IgH and Igκ transcription in L1236 cells. Total cellular RNA of the cell lines was separated on an agarose gel, transferred to a nylon filter, and probed with cDNA clones for Cγ (A, right) and Cκ (B, top, left), respectively. Although the LCL L1309 does express the membrane bound Cγ heavy chain transcript and the secreted Cγ heavy chain transcript (A) as well as the κ light chain transcript (B), neither IgG nor Igκ transcripts are detected in L1236 cells (A, right: ethidium bromide staining of the gel before blotting; B, bottom: hybridization for GAPDH transcripts to control quality of total RNA). The cell lines U937 and CO serve as negative controls for Ig gene transcripts. To increase the sensitivity of Northern blot analysis for Cκ, mRNA was separated on an agarose gel, transferred to a nylon filter, and probed with a cDNA clone (B, left, top). Again, no hybridization signal was obtained in L1236 mRNA. L1309 cells serve as positive control. CO cells serve as negative control. Hybridization with a GAPDH probe to check quality and amount of mRNA showed that approximately equal amounts of intact RNA were loaded on the gel for the various cell lines (B, bottom).
Northern blot analysis for IgH and Igκ transcription in L1236 cells. Total cellular RNA of the cell lines was separated on an agarose gel, transferred to a nylon filter, and probed with cDNA clones for Cγ (A, right) and Cκ (B, top, left), respectively. Although the LCL L1309 does express the membrane bound Cγ heavy chain transcript and the secreted Cγ heavy chain transcript (A) as well as the κ light chain transcript (B), neither IgG nor Igκ transcripts are detected in L1236 cells (A, right: ethidium bromide staining of the gel before blotting; B, bottom: hybridization for GAPDH transcripts to control quality of total RNA). The cell lines U937 and CO serve as negative controls for Ig gene transcripts. To increase the sensitivity of Northern blot analysis for Cκ, mRNA was separated on an agarose gel, transferred to a nylon filter, and probed with a cDNA clone (B, left, top). Again, no hybridization signal was obtained in L1236 mRNA. L1309 cells serve as positive control. CO cells serve as negative control. Hybridization with a GAPDH probe to check quality and amount of mRNA showed that approximately equal amounts of intact RNA were loaded on the gel for the various cell lines (B, bottom).
IgH and Igκ transcripts can be detected by RT-PCR in cell line L1236 and primary H-RS cells.
To increase the sensitivity for the detection of Ig gene transcripts in L1236, RT-PCR was performed using two different oligonucleotide sets. To amplify transcripts from the IgH gene, an oligonucleotide chosen from the CDRII was combined with a consensus oligonucleotide hybridizing to the second exon of all cγ genes. Because of the large intron between the VH region genes and the first exon of the constant region gene, a product can be only amplified from spliced RNA. Thus, the selected oligonucleotides were suitable for specific amplification of transcripts. A product of the expected length was generated by amplification of L1236 cDNA (Fig 3). Sequence analysis showed an identical sequence of the VHDHJHregion gene compared with the sequence previously amplified from genomic DNA of L1236 cells,7 providing evidence that the rearranged VH1 gene is transcribed. Sequence analysis of the constant region confirmed that class switching to Cγ4 had occurred on the respective allele harboring the potentially functional rearrangement and that the JH segment was correctly spliced to the first exon of Cγ4. Igκ transcripts were amplified using oligonucleotides hybridizing to a sequence in the leader region of the rearranged Vκ3 gene and to the Cκ gene or to the CDRIII, respectively (Fig 4). Again, sequencing of the resulting PCR product obtained from L1236 cDNA showed an identical sequence compared with the Vκ3 sequence amplified from genomic DNA of L1236 cells.7 The Jκ gene is correctly spliced to the Cκ gene (see Fig 7).
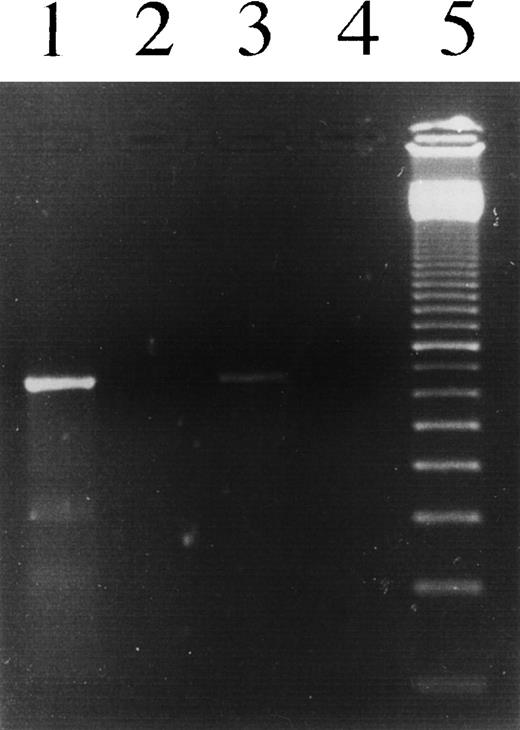
PCR analysis to detect IgH transcripts in L1236 cells as well as in H-RS cells of the patient’s affected bone marrow. Lane 1, L1236 cDNA; lane 2, L1309 cDNA; lane 3, section of the HD-affected bone marrow, cDNA; lane 4, no template; lane 5, 100-bp ladder. Using an oligonucleotide hybridizing to the CDRII of the VH1 gene rearrangement in L1236 cells in combination with an oligonucleotide hybridizing to the second exon of all the 4 γ genes, a specific product was amplified from cDNA obtained from L1236 cells as well as from cDNA obtained from the patient’s bone marrow. No product was amplified from the cDNA obtained from L1309 cells, indicating the specificity of the product. Sequence analysis of the PCR product showed the sequence of the VH1 gene rearrangement switched to Cγ4.
PCR analysis to detect IgH transcripts in L1236 cells as well as in H-RS cells of the patient’s affected bone marrow. Lane 1, L1236 cDNA; lane 2, L1309 cDNA; lane 3, section of the HD-affected bone marrow, cDNA; lane 4, no template; lane 5, 100-bp ladder. Using an oligonucleotide hybridizing to the CDRII of the VH1 gene rearrangement in L1236 cells in combination with an oligonucleotide hybridizing to the second exon of all the 4 γ genes, a specific product was amplified from cDNA obtained from L1236 cells as well as from cDNA obtained from the patient’s bone marrow. No product was amplified from the cDNA obtained from L1309 cells, indicating the specificity of the product. Sequence analysis of the PCR product showed the sequence of the VH1 gene rearrangement switched to Cγ4.
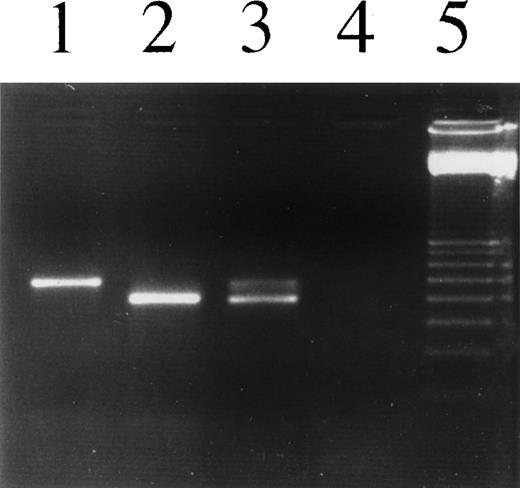
PCR analysis to detect Igκ transcripts in L1236 cells as well as in H-RS cells of the patient’s affected bone marrow. Lane 1, L1236, DNA; lane 2, L1236 cDNA; lane 3, section of the patient’s affected bone marrow, cDNA; lane 4, no template; lane 5, 100-bp ladder. Using an oligonucleotide hybridizing to the Vκ leader sequence and an oligonucleotide hybridizing to the CDRIII of the rearranged Vκ genes, a specific product was amplified from DNA as well as cDNA. The products differ in length due to splicing of the leader intron. Vκ transcripts were amplified and sequenced from L1236 cells as well as from the bone marrow section. The cDNA of the bone marrow section showed a slight contamination with DNA, resulting in an additional band.
PCR analysis to detect Igκ transcripts in L1236 cells as well as in H-RS cells of the patient’s affected bone marrow. Lane 1, L1236, DNA; lane 2, L1236 cDNA; lane 3, section of the patient’s affected bone marrow, cDNA; lane 4, no template; lane 5, 100-bp ladder. Using an oligonucleotide hybridizing to the Vκ leader sequence and an oligonucleotide hybridizing to the CDRIII of the rearranged Vκ genes, a specific product was amplified from DNA as well as cDNA. The products differ in length due to splicing of the leader intron. Vκ transcripts were amplified and sequenced from L1236 cells as well as from the bone marrow section. The cDNA of the bone marrow section showed a slight contamination with DNA, resulting in an additional band.
The transcription of the IgH and Igκ chain genes in L1236 cells as demonstrated by PCR could reflect an in vitro effect observed after culturing of L1236 cells. The VH1 gene rearrangement obtained from L1236 cells was already used as an H-RS cell-specific marker gene to detect these cells during the course of the disease in various affected tissues obtained from the patient from whom the cell line was established.25 26 Therefore, H-RS cell-specific oligonucleotides could be generated for specific amplification of Ig gene transcripts from the H-RS cells in the affected bone marrow specimen of this patient. Because of the low amount of H-RS cells in the biopsy specimen, the sensitivity of PCR was increased by performing 40 cycles of PCR in one round of amplification. H-RS cell-specific γ transcripts (Fig 3) as well as κ transcripts (Fig 4) were amplified from the bone marrow specimen, indicating that Ig genes were also transcribed in vivo. Furthermore, this result confirmed that class switching to Cγ4 had occurred in the H-RS cells in the lymphoma tissue.
The upstream octamer element in the VH1 promoter region is affected by somatic mutation.
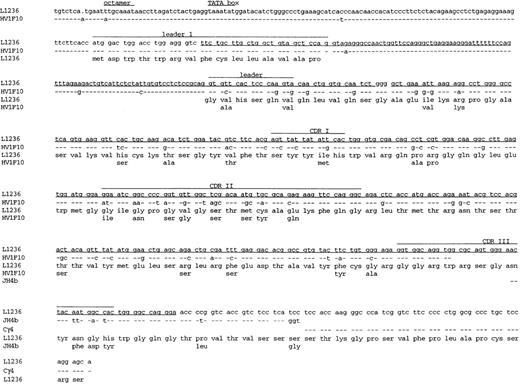
Because it had recently been shown in the mouse that somatic hypermutation can extend to the promoter region of V genes,27 we wondered whether the low amount of Ig gene transcripts in L1236 cells might be due to somatic mutations in this region. Three regulatory elements within the VH promoter region are known to be involved in the regulation of Ig gene transcription.28 Of these, the conserved octanucleotide 5′-ATGCAAAT-3′29,30 is considered to exhibit a dominant effect, both with regard to transcription efficiency and B-cell specificity of Ig gene transcription. This sequence represents a binding site for specific transcription factors31 and is present within all known VH promoter sequences. In vitro mutation of the octamer causes transcriptionally silent IgH constructs.30,32 To analyze the promoter region of the VH1-rearrangement, a 5′ oligonucleotide was selected from the promoter region of the germline sequence HV1F1033to which the rearranged VH1 gene shows the highest homology and combined with oligonucleotides hybridizing specifically to the mutated rearranged VH1 gene. A total of 128 bases 5′ of the leader sequence were amplified from L1236 DNA. In comparison with the germline sequence, two mutations and one nucleotide deletion were detected upstream of the leader exon (Fig 5). One of the mutations affects the first nucleotide of the octamer sequence that probably leads to loss of octamer function (Fig 6 and see below). This mutation was also found in the octamer of the rearranged H-RS cell-specific VH1 gene amplified from the patients’ bone marrow.
Primary sequence data obtained from the 5′ untranslated region of the VH1 gene rearrangement of L1236 cells. The VH1 gene rearrangement was amplified using the oligonucleotide 21-2UIS hybridizing to the 5′ region of the VH1 gene rearrangement and the oligonucleotide 3H1 hybridizing to the CDRIII of the VH1 gene rearrangement. Sequencing was performed using the VH1LAS oligonucleotide hybridizing to the leader region resulting in the antisense sequence. The sequence is given above. Numbers indicate the nucleotide positions on the gel. Note that the first position (number 182) of the octamer (number 175 until number 182) is mutated (A→T; for comparison with germline sequence, see also Fig 6).
Primary sequence data obtained from the 5′ untranslated region of the VH1 gene rearrangement of L1236 cells. The VH1 gene rearrangement was amplified using the oligonucleotide 21-2UIS hybridizing to the 5′ region of the VH1 gene rearrangement and the oligonucleotide 3H1 hybridizing to the CDRIII of the VH1 gene rearrangement. Sequencing was performed using the VH1LAS oligonucleotide hybridizing to the leader region resulting in the antisense sequence. The sequence is given above. Numbers indicate the nucleotide positions on the gel. Note that the first position (number 182) of the octamer (number 175 until number 182) is mutated (A→T; for comparison with germline sequence, see also Fig 6).
Sequence of the VH1 gene rearrangement of L1236 cells including part of the promoter region, the leader intron, and part of the constant region. The sequence obtained from L1236 cells is compared with germline sequences for the corresponding VH, JH, and Cγ4 genes (HV1F10,33JH4b,34 and Cγ4; Chai et al, unpublished data; Genbank Accession No. L23566) -, sequence identity; ., nucleotide deletion. Aminoacid changes are indicated by the germline amino acids given below the amino acid sequence of L1236. Codons are numbered according to Kabat et al.35 Corresponding amino acids are shown below each codon. The leader region, CDRI, CDRII, and CDRIII are indicated. Sequence already published7 is underlined. The sequence of Cγ4 and JH4B was obtained from cDNA. Therefore, no sequence is provided for the intron between JH4B and Cγ4. The RNA is correctly spliced. This sequence is available from the EMBL database under accession no. AJ005570.
Sequence of the VH1 gene rearrangement of L1236 cells including part of the promoter region, the leader intron, and part of the constant region. The sequence obtained from L1236 cells is compared with germline sequences for the corresponding VH, JH, and Cγ4 genes (HV1F10,33JH4b,34 and Cγ4; Chai et al, unpublished data; Genbank Accession No. L23566) -, sequence identity; ., nucleotide deletion. Aminoacid changes are indicated by the germline amino acids given below the amino acid sequence of L1236. Codons are numbered according to Kabat et al.35 Corresponding amino acids are shown below each codon. The leader region, CDRI, CDRII, and CDRIII are indicated. Sequence already published7 is underlined. The sequence of Cγ4 and JH4B was obtained from cDNA. Therefore, no sequence is provided for the intron between JH4B and Cγ4. The RNA is correctly spliced. This sequence is available from the EMBL database under accession no. AJ005570.
The 3′ splice site of the leader intron of the potentially functional Vκ3 gene rearrangement is mutated in L1236 cells and in primary H-RS cells of the same donor.
In search for somatic mutations that might impair the transcription and translation of the rearranged Vκ3 gene, a fragment spanning part of the Vκ promoter, the leader region and part of the framework region I (FRI) were amplified by PCR and sequenced. Oligonucleotides were selected from the germline sequence.20 In the amplified fragment, a total of 32 somatic mutations was detected, 2 within the 5′ untranslated region, 1 within the first leader exon, 22 within the leader intron, 6 within the second leader exon, and 1 within the first 27 bp of the FRI. The two mutations within the 5′ untranslated region did not affect any of the conserved promoter elements. The seven mutations within the leader exons result in five amino acid replacements, four of which are located within the five C-terminal amino acids of the leader protein (Fig 7). One of the mutations within the leader intron affects the 3′ splice site. In eukaryotic cells the sequence at the 3′ end of introns has a common structural motif consisting of 4 bp 5′(A/T/G)CAG3′. The germline sequence of the leader intron of the Vκ3 gene ends with TCAG. In L1236 cells, the G is mutated, destroying the splice site. However, due to somatic mutations 6 bp upstream, a new potential splice site consisting of the consensus sequence TCAG was created by two point mutations changing CCAA into TCAG (Fig 7). Aberrant splicing at this new splicing site leading to the insertion of two amino acids into the leader peptide was indeed demonstrated by amplifying and sequencing a cDNA fragment encompassing the leader and FR1 sequence (Fig 7). The aberrant spliced transcript was also amplified and sequenced from the affected bone marrow tissue, thus excluding an in vitro artifact.
Sequence of the Vκ3 gene rearrangement, including part of the promoter region, the leader intron, and part of the constant region. Sequence format is the same as in Fig 6. The sequence obtained from L1236 cells is compared with germline sequences for the corresponding Vκ, Jκ, and Cκ genes (L2,22Jκ1,23 and Cκ24). The aberrant splicing of the leader intron is indicated. The sequence of Jκ1 and Cκ was obtained from cDNA. Therefore, the intron sequence between Jκ1 and Cκ is not given. The Jκ gene is correctly spliced to the Cκ gene. This sequence is available from the EMBL database under accession no.AJ005571.
Sequence of the Vκ3 gene rearrangement, including part of the promoter region, the leader intron, and part of the constant region. Sequence format is the same as in Fig 6. The sequence obtained from L1236 cells is compared with germline sequences for the corresponding Vκ, Jκ, and Cκ genes (L2,22Jκ1,23 and Cκ24). The aberrant splicing of the leader intron is indicated. The sequence of Jκ1 and Cκ was obtained from cDNA. Therefore, the intron sequence between Jκ1 and Cκ is not given. The Jκ gene is correctly spliced to the Cκ gene. This sequence is available from the EMBL database under accession no.AJ005571.
DISCUSSION
H-RS cells in classical HD represent clonal populations of B-cell–derived lymphoma cells.36 Sequence analysis of rearranged Ig genes obtained from micromanipulated H-RS cells showed the occurrence of somatic mutations within the rearranged Ig genes of H-RS cells in all informative cases, indicating their derivation from GC or memory B cells.1,7,8 Based on the finding of crippling mutations in some of the cases, it was suggested that H-RS cells in classical HD as a rule are derived from crippled GC B cells that lost the capacity to express (high-affinity) antigen receptor due to somatic mutations.8 36
One of the HD cases analyzed in these studies appeared to be an exception to this rule, because in this case both a potentially functional VH and VL chain gene rearrangement were detected in H-RS cells.7 The establishment of a cell line (L1236) permitted further detailed analysis of Ig gene structure and expression in the H-RS cells of this case. Ig gene expression, measured by fluorescence-activated cell sorting (FACS) analysis (Igκ light chain12) or immunostaining on cytospins (IgG heavy chain, Igκ light chain) was undetectable in L1236 cells. An analysis of Ig gene expression in these cells at the RNA level was performed to determine at which stage (transcription and/or translation) antibody expression was blocked. In addition, we wanted to find out whether the potentially functional V gene rearrangements in L1236 cells carry crippling mutations outside the regions of the V genes sequenced previously.7 Moreover, the availability of the cell line L1236 offered the opportunity to investigate whether the Ig genes in these H-RS cells not only accumulated somatic mutations, but also performed class switch recombination.
Class switch recombination in L1236 cells was studied by Southern blot analysis. Hybridization of L1236 DNA with probes detecting the various constant region genes showed that class switch recombination to Cγ4 had occurred on one allele. Whether class switch recombination to Cγ4 leads to the expression of IgG transcripts in L1236 cells was analyzed by RT-PCR using a Cγ oligonucleotide and a VH1 oligonucleotide. A PCR product encoding the potentially functional VH1 gene rearrangement correctly spliced to Cγ4 was amplified, showing that class switch recombination to Cγ4 had taken place on the allele carrying the potentially functional heavy chain gene and had led to the expression of the respective RNA. The VH1-Cγ4 transcript was also amplified from the bone marrow biopsy specimen, showing that this class switch recombination had already occurred in the primary H-RS cells. This is the first demonstration of isotype switching in primary H-RS cells. Interestingly, class switch recombination to Cγ4, which is rarely used by normal B cells37 and can be induced by cytokines preferentially released from TH2 cells, was also described for the HD-derived cell line L428.20 Whether class switch recombination to Cγ4 is a frequent event in H-RS cells has to be established.
Neither Ig protein nor mRNA was detectable in L1236 cells when immunostaining or Northern blot analysis was performed. However, Ig heavy and light chain gene transcripts could be amplified from L1236 cells by RT-PCR. These results indicate that L1236 cells produce only a very low amount of Ig gene transcripts. These transcripts were also detected by RT-PCR in RNA extracted from a section of the affected bone marrow tissue, showing that Ig gene transcription likewise took place in the H-RS cells in vivo. Ig gene transcription in H-RS cells was previously analyzed by several groups using in situ hybridization.38-41 In only 1 of a total number of 95 cases of classical HD analyzed for heavy and/or light chain transcription were Igκ transcripts detected in H-RS cells. Taking these studies and the present work together, it appears that H-RS cells in at least some cases of classical HD can express Ig transcripts, albeit at a low level and below the sensitivity of current in situ hybridization methods. Diffuse staining for both κ and λ light chains was frequently observed within H-RS cells using immunohistochemistry.30,38,42 43 However, this finding was likely caused by artificial uptake of Ig, particularly because it is now known that H-RS cells represent clonal B-cell populations.
Because V gene sequence analysis previously performed in L1236 cells did not show any obvious crippling mutations,7 we wondered whether the low level of Ig gene transcripts might be due to mutations within upstream regulatory elements of the V region genes. Indeed, a point mutation affecting the first nucleotide of the octamer motif (ATGCAAAT) of the heavy chain promoter was detected in L1236 cells as well as in the primary H-RS cells of the bone marrow specimen. An in vitro expression study has shown that a mutation at this position (A into C) drastically reduces Ig gene transcription.30Although a polymorphism at position 2, 4, or 832,44 is sometimes observed within the octamer sequence of human and murine heavy chain promoter regions, sequence variation at the first position was never observed in functional VH genes.32,44 When comparing the inverted octamer sequence (ATTTGCAT) of human and murine Vκ genes, a polymorphism was sometimes observed for positions 1 and 7 of the converted octamer sequence.45Again, a polymorphism at the last position (corresponding to the first position of the octamer of VH genes) was never observed in any of the mouse or human Vκ octamer sequences analyzed so far.45 46 This indicates that sequence variations at distinct positions in the octamer motif occurred during evolution and are at these positions compatible with functionality. However, the absence of polymorphisms at position one of the heavy chain octamer motif further supports the critical role of this position for the functionality of the octamer. Thus, the low level of IgH chain gene transcription in L1236 cells (and presumably also in primary H-RS cells) of this particular case is most likely caused by the somatic mutation affecting the first position of the octamer motif within the heavy chain promoter.
No mutations were found within the promoter elements of the potentially functional Vκ3 gene. Therefore, the low amount of Vκ transcripts could not be attributed to an alteration of the Vκ promoter. By analyzing the Vκ leader intron, several somatic mutations were detected leading to the destruction of the 3′ splice site and to the establishment of a new potential splice site 6 bp upstream. Sequence analysis of Vκ transcripts obtained from L1236 cells as well as from the affected bone marrow tissue indeed showed aberrant splicing of κ pre-mRNA without alteration of the reading frame by using the newly introduced 3′ leader intron splicing site. However, alterations of splice sites have been described that cause a retarded intron removal due to a prolonged binding of spliceosomes.47 This delayed splicing results in nuclear retention of pre-mRNA. Such a mechanism might account for the low amount of Vκ mRNA transcripts in the H-RS cells analyzed.
The present study shows that the lack of Ig expression by H-RS cells in classical HD can not only be caused by crippling mutations within the coding sequence of rearranged Ig genes (like those generating stop codons36), but also by mutations affecting regulatory elements. However, in other cases of HD, H-RS cells may harbor crippling mutations that a priori do not prevent Ig gene expression, eg, mutations that result in a reduced affinity of the antibody to the respective antigen. GC B cells having acquired such mutations also represent crippled GC B cells, because they are destined to die by apoptosis under physiological conditions. H-RS cells that are derived from this type of crippled GC B cells could potentially express antigen receptor. However, expression of antibody in H-RS cells has never been observed (see above). It may be that, in these cases, the aberrant B-cell differentiation status of H-RS cells (exemplified by the absence of B-lineage markers in most cases of classical HD; see Drexler48 for review) leads to the downregulation of Ig gene expression by a disregulation of B-cell–specific transcription factors regulating Ig gene expression.
Taken together, in the H-RS cell line L1236 and the corresponding primary H-RS cells of the patient, transcription of rearranged Ig heavy and light chain genes in H-RS cells was detected at a minimal level. Sequence analysis of DNA strongly suggested that downregulation of Ig gene transcription is due to a mutation in the octamer promoter element of the in-frame heavy chain gene rearrangement. Moreover, the 3′ splice site of the leader intron of the Igκ light chain gene was mutated, resulting in aberrant splicing. These observations, together with the detection of class switch recombination, do not only confirm the mature GC B-cell phenotype of H-RS cells, but also show that in H-RS cell mutations that prevent expression of Ig genes can be located outside the VDJ region.
A.J. and T.Z. contributed equally to this work.
Supported by a grant of the Deutsche Forschungsgemeinschaft through SFB 502.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Jürgen Wolf, MD, Department of Internal Medicine I, LFI E5 R310, Joseph Stelzmann Str. 9, 50931 Cologne, Germany; e-mail: Juergen.Wolf@medizin.uni-koeln.de.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal