Abstract
In this study we have raised the question of whether DNA can be transferred from one cell to another by phagocytosis of apoptotic bodies. We have used integrated copies of the Epstein-Barr virus (EBV) as a marker to follow the fate and expression pattern of apoptotic DNA in the phagocytotic host. Apoptosis was induced in EBV-carrying cell lines by irradiation before cultivation with either human fibroblasts, macrophages, or bovine aortic endothelial cells. Analysis of the expression pattern of EBV-encoded genes was performed by immunofluorescent staining as well as in situ hybridization. Cocultivation of apoptotic bodies from lymphoid cell lines containing integrated but not episomal copies of EBV resulted in expression of the EBV-encoded genes EBER and EBNA1 in the recipient cells at a high frequency. Fluorescence in situ hybridization analysis showed uptake of human chromatin as well as integrated EBV-DNA into the nuclei of bovine aortic endothelial cells. These data show that DNA may be rescued and reused from apoptotic bodies by somatic cells. In addition, our findings suggest that apoptotic bodies derived from EBV-carrying B lymphocytes may serve as the source of viral transfer to cells that lack receptors for the EBV virus in vivo.
THE CONTROL OF cell number in multicellular organisms depends on the fine balance between cell proliferation and cell death. Cell death by apoptosis plays a key role in the elimination of cells during embryonic development as well as in adults. It is, for example, involved in the negative selection of neural cells, removing redundant cells during organ formation.1 In adults, apoptosis plays an important role in the maintenance of tissue homeostasis; in the human small intestine alone, approximately 109 cells apoptose every hour and are sloughed off into the lumen of the gut.2,3 Apoptosis is also of importance in tumor development. It may serve as an anticancer mechanism by restricting the expansion of cells with unleashed proliferative potential. Tumors remain constant in size or dormant when proliferation is balanced by an equal level of cell death.4This state of dormancy may be controlled at the genetic level such as induction of apoptosis by the p53 tumor-suppressor gene or by the inability of the tumor cells to recruit vessels from the microenvironment.5 6 Loss of p53 or the induction of angiogenesis results in lowered incidence of apoptosis and escape from dormancy. Thus, there exists stages during tumor development in which there is a high turnover of genetic material that is fragmented during apoptosis and subsequently taken up by neighboring cells.
The recognition of apoptotic bodies by macrophages or dendritic or neighboring cells is mediated by several different pathways. Surface receptors such as the vitronectin receptor (αvβ3) and thrombospondin receptor (CD36) located on the macrophages may recognize apoptotic cells via the binding of thrombospondin.7Translocation of phosphatidylserine from the inner to outer side of the cell membrane serves as a recognition signal for phagocytosis. Not only macrophages are capable of phagocytosing apoptotic bodies. Injection of apoptotic liver cells in the portal vein in the mouse liver results in uptake of apoptotic bodies in the liver endothelial cells.8In vitro, apoptotic macrophages are rapidly phagocytosed by human fibroblast cells within 1 hour.9
Horizontal transfer of genes has been reported in bacteria and fungi and plays an important role in the generation of resistance to antibiotic drugs as well as adaptation to new environments (reviewed in Porter10 and Mishra11). Transfer of DNA from bacteria to somatic cells may also occur, as shown by in vitro experiments that demonstrate efficient uptake of a β-gal reporter plasmid from attenuated bacteria into the nuclei of the phagocytic cell.12 However, horizontal transfer of genetic information between somatic cells has not yet been described. In this study, we raised the question of whether DNA from apoptotic cells can be salvaged and reused after phagocytosis. We have followed the fate of DNA from apoptotic bodies of Epstein-Barr virus (EBV)-carrying transformed lymphoid cell lines after phagocytosis of normal adherent fibroblasts, macrophages, and endothelial cells. Integrated, non–virus-producing copies of EBV-DNA were used as markers to track the incoming DNA after ingestion as well as the expression of EBV-encoded genes by the phagocytosing cell.
MATERIALS AND METHODS
Cocultivation experiments.
Human fetal lung fibroblasts, bovine aortic endothelial cells (established in the laboratory of Dr Judah Folkman, Children’s Hospital, Boston, MA), human monocytes (purified from normal human blood as previously described13), or human vascular smooth muscle cells (kindly provided by Britt Dahlgren, Karolinska Hospital, Stockholm, Sweden) were trypsinized and transferred to 8-well Lab Tek slides (Nalgene; Nunc, Copenhagen, Denmark) for immunostaining (50,000 cells/well) or 10-cm Petri dishes for fluorescence in situ hybridization (FISH) analysis. The following day, EBV-carrying lymphoid cell lines were irradiated with 150 Gy and added to the cultures at a ratio of 5:1. Alternatively, cells were treated with 16 μg/mL etoposide (Sigma, St Louis, MO) for 24 hours, washed three times in phosphate-buffered saline (PBS), and then added to cultures of human fetal fibroblasts. The tissue culture medium was first changed after 48 hours and then changed every 3 days. Cell lines negative for EBV were also included as negative controls. For immunostaining, cells were washed once with PBS and then fixed in −20°C methanol:acetone, 2:1 (vol:vol) for 5 minutes and air-dried. Cells were rehydrated in balanced salt solution (BSS) and then incubated with human serum antibodies against EBNA1-6 or with serum from an EBV-negative donor for 15 minutes at 37°C and washed in 3× BSS.14 EBNA1 was detected with human serum preadsorbed with an EBNA1-negative, EBNA2-6–positive cell line E95-A-BL28.14 Positive staining was visualized by incubation sequentially with complement and fluorescein isothiocyanate (FITC)-labeled anticomplement antibodies as previously described.15 The mouse monoclonal antibody PE2 was also used to detect EBNA2 expression by immunofluorescence.16The presence of cells in viral lytic cycle was detected by immunostaining against viral capsid antigen (VCA) or early antigen (EA) using FITC-conjugated sera from Burkitt’s lymphoma (EA) or nasopharyngeal carcinoma (VCA) patients according to established protocols.17 18 For detection of EBER1 and 2 nuclear RNAs, cells were fixed in 4% paraformaldehyde for 4 hours, pretreated with 5 μg/mL proteinase K at 37°C for 30 minutes, and hybridized according to the protocol of the manufacturer (DAKO, Glostrup, Denmark).
Apoptosis assays.
For DNA fragmentation assays, 5 × 106 cells were resuspended in 10 mmol/L EDTA, pH 8, 0.5% Triton X-100, and lysed with 10 strokes with a Dounce homogenizer. Cells were pelleted and the resulting supernatant was incubated with 100 μg/mL RNase A at 37°C for 1 hour. Sodium dodecyl sulfate (SDS) and proteinase K were added to concentrations of 1% (wt/vol) and 200 μg/mL, respectively, and incubated at 50°C for 2 hours. The DNA was precipitated overnight at −20°C by adding 1/10 of total volume 5 mol/L NaCl and 2.5 vol of 99% EtOH. Precipitated DNA was collected by centrifugation at 13,000 rpm at 4°C in a microcentrifuge. Fragmentation was analyzed by electrophoresis in 1.5% agarose gels.
FISH.
Total human genomic DNA from a normal donor was labeled with tetramethyl-rhodamine-dUTP using a nick translation kit (GIBCO-BRL, Grand Island, NY). The BamHI W fragment from the EBV genome was labeled with biotin-11-dUTP with the same method. FISH analysis was performed according to the protocol of Pinkel et al.19 Slides were denatured for 5 minutes in 70% formamide, 2× SSC at 75°C and subsequently dehydrated sequentially in cold 70%, 95%, and 100% ethanol and air-dried. Probes were denatured for 10 minutes at 80°C in the hybridization mix (50% formamide, 2× SSC, and 10% dextran sulphate). Slides were hybridized overnight at 37°C and then washed in 50% formamide, 2× SSC for 4 × 3 minutes, and for 3 × 3 minutes in 2× SSC at 46°C. Positive hybridization with the biotinylated BamHI W probe was detected using avidin-conjugated to fluorescein (Vector, Burlingame, CA). The tetra-methyl-rhodamine-dUTP–labeled human probe was viewed directly by fluorescence microscopy without signal amplification. Cells were counterstained with the DNA fluorochrome 4,6-diamidino-2-phenyl indole (DAPI) and mounted in antibleach medium. Results were analyzed using a fluorescence microscope (Leitz-DMRB; Leica, Heidelberg, Germany) equipped with a Hamamatsu 4800 CCD camera (Hamamatsu, Herrsching, Germany). Digital images were processed in Adobe Photoshop (Adobe, Mountain View, CA). For three-dimensional imaging, pictures were sampled in the z-axis and deconvoluted using digital confocal imaging (Openlab, Improvision, Coventry, UK). The resulting images were processed using the 3-D rendering software (Openlab).
RESULTS
Expression of EBV-encoded genes in fibroblasts after uptake of apoptotic bodies.
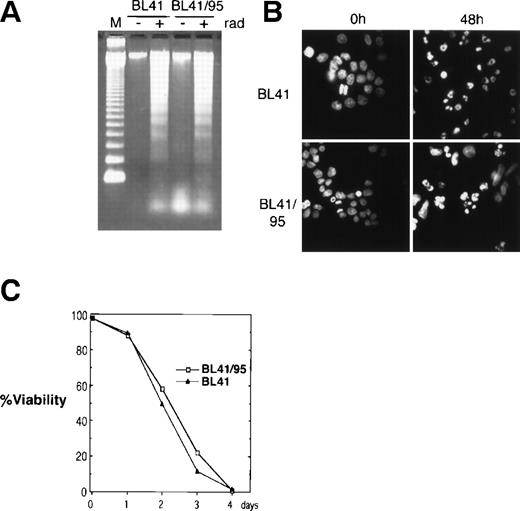
The EBV-negative Burkitt’s lymphoma cell line BL41 and its EBV-positive derivative BL41/95 was used for cocultivation with human fetal fibroblasts (HF). The BL41/95 is an in vitro EBV-infected BL41 cell line that carries integrated copies of EBV that express the EBNA1 and 2 (as detected by immunofluorescence) but does not produce virus20 21 (Barbro Ehlin-Eriksson, personal communication, December 1998). Apoptosis was reproducibly induced by irradiating cells at 150 Gy. Radiation induced DNA ladder formation as analyzed by DNA gel electrophoresis. Staining of nuclei with the DNA fluorochrome Hoechst 33258 showed nuclear condensation of DNA and fragmentation of nuclei into apoptotic bodies (Fig 1A and B). Irradiation resulted in 100% cell death after 4 days, as analyzed by trypan blue exclusion (Fig 1C).
Induction of apoptosis in BL41 and BL41/B95 cells. (A) Analysis of DNA fragmentation 48 hours after irradiation with 150 Gy by DNA gel electrophoresis. M, 123-bp DNA ladder (GIBCO). (B) Apoptotic morphology was examined with Hoechst 33258 DNA staining. (C) Viability of irradiated cells was analyzed by trypan blue exclusion.
Induction of apoptosis in BL41 and BL41/B95 cells. (A) Analysis of DNA fragmentation 48 hours after irradiation with 150 Gy by DNA gel electrophoresis. M, 123-bp DNA ladder (GIBCO). (B) Apoptotic morphology was examined with Hoechst 33258 DNA staining. (C) Viability of irradiated cells was analyzed by trypan blue exclusion.
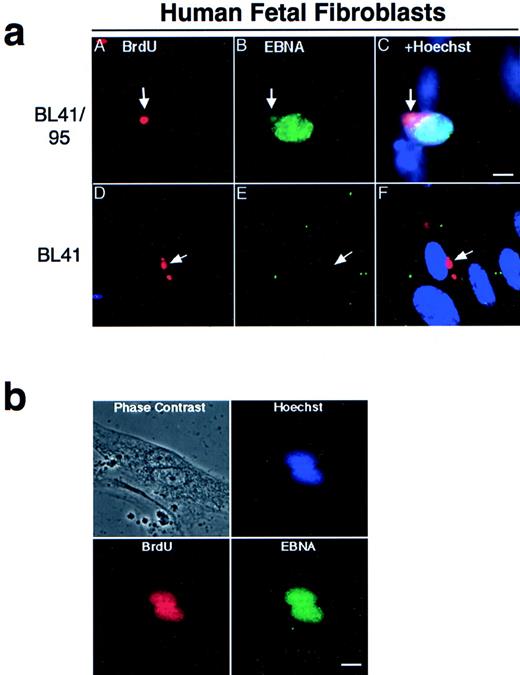
Apoptotic bodies cultivated with human fetal fibroblasts became phagocytosed and internalized within 1 hour, as previously reported.9 To follow the fate of internalized DNA, the BL-41 and BL41/95 were labeled with 2, 3 Bromo-deoxy-uridine (BrdU) for 48 hours before irradiation. The irradiated cells and HF cells were cocultivated for 1 week, as previously described.22 The presence of BL41 and BL41/95 DNA in HF cells was detected by immunostaining with antibodies against BrdU. In both cell lines, apoptotic bodies were still detectable in a perinuclear location in the cytoplasm of the HF cells after 1 week of cocultivation (Fig 2a). Double staining with KF human serum against the EBNA complex 1-6 showed that nuclei of HF cells containing a BL41/95 apoptotic body stained positive for EBNA (Fig 2a). The presence of EBNA1 in cocultures of HF cells was shown by staining with human serum preadsorbed with an EBNA1-negative, EBNA2-6–positive cell line E95-A-BL28. Control experiments in which fibroblasts were cultivated with the original EBV-negative BL41 cell line were always negative (Fig 2a). EBV did not infect HF cells, as shown by the lack of positive EBNA staining after incubation with B95-8 virus23that could infect and transform B cells in parallel cultures (Table 1).
(a) Presence of apoptotic bodies in HF cells after 1 week of cocultivation. The EBV-positive BL41/95 (A through C) and -negative BL41 (D through F) were labeled for 48 hours with BrdU before irradiation and cultivation with HF cells. DNA from the lymphomas was detected with antibodies against BrdU (A and D). Cells were double-stained with human serum against EBV nuclear antigens (EBNA; B and E). (C) and (F) show Hoechst 33258, anti-EBNA, and anti-BrdU stainings in the same picture. Size bar = 5 μm. (b) Fibroblasts pulsed with BrdU before being cultivated with irradiated BL41/95 cells. Nuclei of fibroblastic origin were detected with antibodies against BrdU. Cells were double-stained with human serum against EBNAs. The picture shows overlapping signals of EBNA, BrdU, and Hoechst 33258 staining in fibroblast nuclei. Size bar = 8 μm.
(a) Presence of apoptotic bodies in HF cells after 1 week of cocultivation. The EBV-positive BL41/95 (A through C) and -negative BL41 (D through F) were labeled for 48 hours with BrdU before irradiation and cultivation with HF cells. DNA from the lymphomas was detected with antibodies against BrdU (A and D). Cells were double-stained with human serum against EBV nuclear antigens (EBNA; B and E). (C) and (F) show Hoechst 33258, anti-EBNA, and anti-BrdU stainings in the same picture. Size bar = 5 μm. (b) Fibroblasts pulsed with BrdU before being cultivated with irradiated BL41/95 cells. Nuclei of fibroblastic origin were detected with antibodies against BrdU. Cells were double-stained with human serum against EBNAs. The picture shows overlapping signals of EBNA, BrdU, and Hoechst 33258 staining in fibroblast nuclei. Size bar = 8 μm.
Induction of EBNA1 and EBER Expression in HF Cells After Coculture With Apoptotic Bodies From Lymphoid Cell Lines
| Donor Cell . | EBV Status . | HF Phenotype After Cocultivation . | ||
|---|---|---|---|---|
| EBNA1 . | EBNA2 . | EBER1 + 2 . | ||
| BL41/95 | Integrated | 2.5% ± 0.4% | — | 1.2% ± 0.3% |
| EHR-A-BL41 | Integrated | 1.9% ± 0.4% | — | 1.4% ± 0.4% |
| IB-4 | Integrated | 3.0% ± 0.2% | — | 3.4% ± 0.8% |
| Namalwa | Integrated | 4.3% ± 0.4% | — | 2.9% ± 0.3% |
| Raji | Integrated/ episomal | 4.0% ± 0.3% | — | ND |
| Jijoye M13 | Episomal | <1/500,000 | — | <1/500,000 |
| P3H3 | Episomal | <1/500,000 | — | <1/500,000 |
| Rael | Episomal | <1/500,000 | — | <1/500,000 |
| CBMI Ral Sto | Episomal | <1/500,000 | — | <1/500,000 |
| BL41 | Negative | — | — | — |
| Ramos | Negative | — | — | — |
| BL28 | Negative | — | — | — |
| B95.8 virus | Virus | — | — | — |
| No donor | — | — | — | — |
| Donor Cell . | EBV Status . | HF Phenotype After Cocultivation . | ||
|---|---|---|---|---|
| EBNA1 . | EBNA2 . | EBER1 + 2 . | ||
| BL41/95 | Integrated | 2.5% ± 0.4% | — | 1.2% ± 0.3% |
| EHR-A-BL41 | Integrated | 1.9% ± 0.4% | — | 1.4% ± 0.4% |
| IB-4 | Integrated | 3.0% ± 0.2% | — | 3.4% ± 0.8% |
| Namalwa | Integrated | 4.3% ± 0.4% | — | 2.9% ± 0.3% |
| Raji | Integrated/ episomal | 4.0% ± 0.3% | — | ND |
| Jijoye M13 | Episomal | <1/500,000 | — | <1/500,000 |
| P3H3 | Episomal | <1/500,000 | — | <1/500,000 |
| Rael | Episomal | <1/500,000 | — | <1/500,000 |
| CBMI Ral Sto | Episomal | <1/500,000 | — | <1/500,000 |
| BL41 | Negative | — | — | — |
| Ramos | Negative | — | — | — |
| BL28 | Negative | — | — | — |
| B95.8 virus | Virus | — | — | — |
| No donor | — | — | — | — |
Expression of EBV markers, EBNA1, EBNA2, and EBER1 + 2, in HF cells after cocultivation for 3 weeks with irradiated EBV-positive or -negative lymphoid cell lines. EBV-carrying cell lines could be divided into those with integrated EBV genomes and those with episomal genomes. EBNA1 and 2 expression was analyzed by immunofluorescence staining and EBER1 + 2 expression was detected by in situ hybridization analysis. The percentage of positive cells was estimated by counting a minimum of 500 cells in triplicate samples. Experiments were repeated at least three times for each cell line.
Abbreviation: ND, not detected.
One explanation for our findings is that the EBNA1-positive nuclei were derived from entire lymphoid nuclei taken up by the fibroblast cells. To exclude this possibility, fibroblasts were pulsed with BrdU before cocultivation with irradiated (unlabeled) BL41/95 cells. Double-staining of cells with BrdU and EBNA1 antibodies showed EBNA1 staining in nuclei that also were positive for BrdU. Sporadic cells could be detected containing more than one EBNA1-staining nuclei. These nuclei were always of fibroblastic origin, as shown by positive BrdU staining (Fig 2b).
Integrated EBV-DNA but not episomal induces expression of EBV-encoded genes in fibroblasts.
We next investigated whether cocultivation of apoptotic bodies derived from other EBV-carrying cell lines could induce EBNA expression to HF cells. Apoptosis was elicited by irradiation in 9 EBV-carrying cell lines and 3 EBV-negative cell lines. The resulting apoptotic bodies were cultivated with HF cells for 3 weeks. Subsequently, the cells were tested for the presence of the EBV-encoded RNAs, EBER1 and 2, by in situ hybridization analysis and for EBNA1 expression by immunofluorescence staining (Table 1 and Fig 3G and H). The donor cell lines could be divided into two groups: apoptotic bodies from 5 of the cell lines (Table 1, upper part) induced EBNA1 and EBER expression in HF, whereas the other 4 could not. The lines that could act as EBV donors induced EBNA1 and EBER expression in 1% to 5% HF cells. The other cell lines induced little or no EBNA or EBER expression in the recipient cells (<5 of 500,000 cells in 5 experiments). The cell lines that could induce expression of EBV markers in HF cells carried integrated EBV genomes,24-27 whereas the ineffective cell lines contained episomal EBV-DNA (Table 1).28
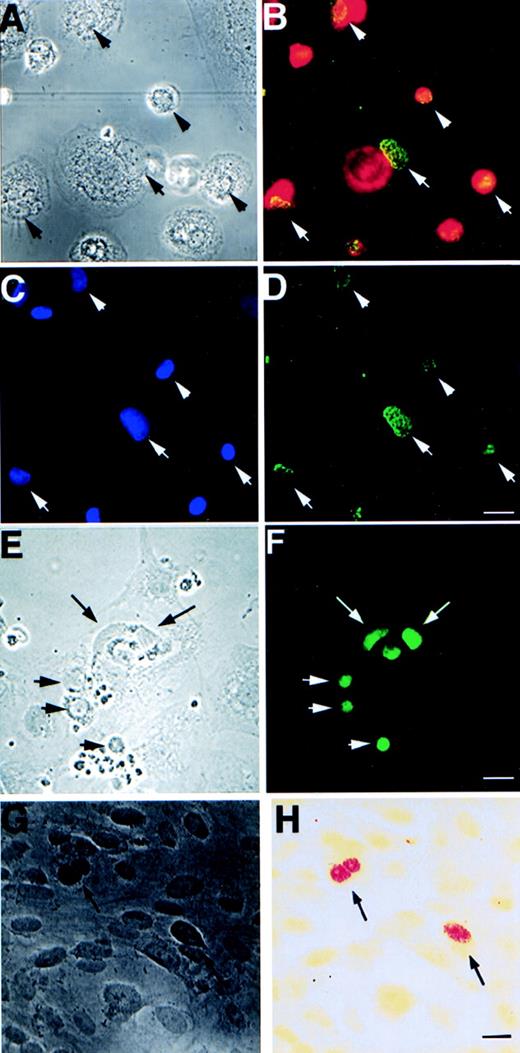
Expression of EBV-specific genes, EBER1+2 and EBNA1-6, in different cell types after 3 weeks of cultivation with apoptotic bodies from EBV-carrying Namalwa line. (A) Phase contrast and (B) immunofluorescence staining showing coexpression of macrophage-specific marker CD68 (red) and EBNA (green) in the same cells. (C) shows the location of macrophage nuclei by Hoechst 33258 DNA staining that overlap with EBNA as shown in (D). Arrows mark the location of the nuclei that are located in the periphery of the cytoplasm of the macrophages. (E) Phase contrast and (F) fluorescence showing EBNA expression in bovine aortic endothelial cells. (G) Phase contrast and (H) bright field view of in situ hybridization analysis of EBER1 and 2 expression in HF cells. Arrows depict positive nuclei. Size bar = 20 μm.
Expression of EBV-specific genes, EBER1+2 and EBNA1-6, in different cell types after 3 weeks of cultivation with apoptotic bodies from EBV-carrying Namalwa line. (A) Phase contrast and (B) immunofluorescence staining showing coexpression of macrophage-specific marker CD68 (red) and EBNA (green) in the same cells. (C) shows the location of macrophage nuclei by Hoechst 33258 DNA staining that overlap with EBNA as shown in (D). Arrows mark the location of the nuclei that are located in the periphery of the cytoplasm of the macrophages. (E) Phase contrast and (F) fluorescence showing EBNA expression in bovine aortic endothelial cells. (G) Phase contrast and (H) bright field view of in situ hybridization analysis of EBER1 and 2 expression in HF cells. Arrows depict positive nuclei. Size bar = 20 μm.
The induction of EBNA1 in HF cells was not restricted to radiation-induced apoptosis, because a similar effect was observed with cells treated with etoposide (Table 2). Interestingly, cocultivation of HF cells with Namalwa cells exposed to hypo-osmotic shock did not affect EBNA1 expression in HF cells. This finding indicates that transfer of expression of EBV-encoded genes is mediated by EBV-carrying apoptotic but not necrotic cells.
Expression of EBNA1-6 in Human Fetal Fibroblasts After 3 Weeks of Coculture With Apoptotic/Necrotic Namalwa Cells
| Induction of Cell Death . | EBNA1-6 Expression in HF Cells . |
|---|---|
| Radiation | 4.1% |
| Etoposide | 2.5% |
| Osmotic shock | ND |
| Induction of Cell Death . | EBNA1-6 Expression in HF Cells . |
|---|---|
| Radiation | 4.1% |
| Etoposide | 2.5% |
| Osmotic shock | ND |
Expression of EBNA1-6 in human fetal fibroblasts after 3 weeks of coculture with Namalwa cells treated with irradiation, etoposide, or hypo-osmotic shock.
Abbreviation: ND, not detectable.
Host-dependent expression of EBNA2.
We next investigated whether the expression pattern of EBV-encoded genes depended on the phenotype of the donor or the host. Earlier findings have shown that EBNA2 is only expressed in immunoblastic lines.29 EBNA2 was expressed in all donor lines studied, with the exception of Rael, P3H3, and EHR-A-BL41.29-31 We investigated whether the expression pattern of EBNA2 was downregulated in EBNA1- and EBER-expressing HF cells. No detectable EBNA2 staining could be observed with either the PE2 anti-EBNA2 monoclonal antibody or adsorbed EBNA-2 specific human serum (Table 1). This finding is consistent with the lack of EBNA2 expression in EBV-carrying non-B cells, including somatic cell hybrids between B and non-B cells with a dominating fibroblastic or epithelial phenotype.32
High efficiency transfer of expression EBV to macrophages and endothelial cells.
We have also tested whether integrated EBV can be transferred with similar efficiency to other cell types. Apoptotic bodies derived from Namalwa cells were cultured with either bovine aortic endothelial (BAE) cells, human monocytes, or vascular smooth muscle cells for 3 weeks. Both monocytes and BAE cells showed a high percentage of EBNA1-6–positive cells (20% to 50% positive EBNA1-6 staining after 3 weeks of cocultivation; Fig 3A through F and Table 3). Smooth muscle cells exhibit a significantly lower frequency of uptake and expression of EBNA1-6 (<0.01%). The high efficiency with which monocytes and endothelial cells are induced to express EBNA1 may relate to their normally high phagocytotic activity.
Expression of EBNA1-6 After Culture With Apoptotic Bodies From the Namalwa Cell Line
| Recipient Cell Type . | EBNA1-6 . | EBNA2 . |
|---|---|---|
| Human monocytes | 51% | — |
| BAE cells | 20% | — |
| Human fetal fibroblasts | 4% | — |
| Human smooth muscle cells | <0.01% | — |
| Recipient Cell Type . | EBNA1-6 . | EBNA2 . |
|---|---|---|
| Human monocytes | 51% | — |
| BAE cells | 20% | — |
| Human fetal fibroblasts | 4% | — |
| Human smooth muscle cells | <0.01% | — |
Expression of EBNA1-6 in human fetal lung fibroblasts, BAE cells, human monocytes, or human vascular smooth muscle cells cocultivated for 3 weeks with irradiated Namalwa cells. Cells were analyzed by immunofluorescent staining using human serum against EBNA1-6 and EBNA2. The table shows values from a representative experiment. The experiments were performed three times (except for the cultivation with macrophages, which was performed twice).
Presence of DNA from apoptotic cells in the nuclei of recipient cells.
The maintenance of high EBNA1 and EBER expression over more than 3 weeks, the integration dependent expression of these genes, and the recipient cell type-related expression pattern (EBNA1+, EBNA2−) suggested that integrated EBV sequences were transferred to the phagocytotic host during cocultivation. We therefore examined whether DNA from apoptotic lymphoid cell lines was transferred to the nuclei of the recipient cells. FISH analysis was used to follow the fate of EBV as well as genomic DNA after phagocytosis by bovine endothelial cells. Coculture of cells from two different species permitted us to distinguish between the DNA of the donor cells from that of the recipient cells. Total human genomic DNA was labeled with tetra-methyl-rhodamine-dUTP and used as a probe to identify DNA of human origin. Hybridization of human DNA to Namalwa interphase nuclei resulted in a uniform hybridization pattern with no detectable cross-reactivity to bovine nuclei (Fig 4A). The presence of EBV-DNA was assayed by FISH analysis using a biotin-labeled BamHI W fragment as a probe (a 3-kb fragment that is repeated 6 to 10 times in the EBV genome).24,33 TheBamHI W probe gave two distinct signals in Namalwa nuclei, as previously reported,24 33 whereas all BAE nuclei were negative (Fig 4A).
Samples were hybridized with the BamHI W fragment of EBV, a sequence approximately 3-kb long that is repeated 6 to 10 times in the EBV genome. After hybridization, the biotinylatedBamHI W probe was detected using the fluorescein-conjugated avidin-biotin amplification system (green signals). Human total genomic DNA was directly labeled with tetra-methyl-rhodamine-dUTP (red signal). The presence of yellow signals indicated overlap of the human and the EBV probes. Nuclei were counterstained with the DNA fluorochrome, DAPI (blue). Digital images were captured in gray scale and subsequently pseudo-colored using computer imaging software. (A) Namalwa (left) and BAE nuclei (right) hybridized simultaneously with the EBV BamHI W probe (green) and human genomic DNA probe (red). The Namalwa nucleus shows two distinct signals with the EBV probe and uniform hybridization with the human probe, whereas the bovine nucleus was negative for both probes. (B through E) These figures show four examples from the two-color FISH analysis of the presence of human genomic DNA (red) and EBV-DNA (green) in BAE cells cultured with irradiated Namalwa cells for 1 week. Yellow signals indicate overlap of the signals from the EBVBamHI W and the human probes. (F) Simultaneous analysis of presence of EBV-DNA and human DNA in a BAE nucleus after cultivation with the EBV-negative cell line BL41 shows the presence of human DNA but not of EBV DNA in the BAE nucleus. (G) BAE nucleus cultured with irradiated Namalwa cells showing uptake of human DNA (red). A positive signal was analyzed by digital confocal microscopy. Images were sampled in the z-axis and subsequently processed with 3-D rendering software (Openlab) to generate three-dimensional pictures. The nuclei and incoming DNA could be viewed at different angles (0°, 44°, 56°, and 72°) and depths that showed that the positive signal was localized within the nuclear cage.
Samples were hybridized with the BamHI W fragment of EBV, a sequence approximately 3-kb long that is repeated 6 to 10 times in the EBV genome. After hybridization, the biotinylatedBamHI W probe was detected using the fluorescein-conjugated avidin-biotin amplification system (green signals). Human total genomic DNA was directly labeled with tetra-methyl-rhodamine-dUTP (red signal). The presence of yellow signals indicated overlap of the human and the EBV probes. Nuclei were counterstained with the DNA fluorochrome, DAPI (blue). Digital images were captured in gray scale and subsequently pseudo-colored using computer imaging software. (A) Namalwa (left) and BAE nuclei (right) hybridized simultaneously with the EBV BamHI W probe (green) and human genomic DNA probe (red). The Namalwa nucleus shows two distinct signals with the EBV probe and uniform hybridization with the human probe, whereas the bovine nucleus was negative for both probes. (B through E) These figures show four examples from the two-color FISH analysis of the presence of human genomic DNA (red) and EBV-DNA (green) in BAE cells cultured with irradiated Namalwa cells for 1 week. Yellow signals indicate overlap of the signals from the EBVBamHI W and the human probes. (F) Simultaneous analysis of presence of EBV-DNA and human DNA in a BAE nucleus after cultivation with the EBV-negative cell line BL41 shows the presence of human DNA but not of EBV DNA in the BAE nucleus. (G) BAE nucleus cultured with irradiated Namalwa cells showing uptake of human DNA (red). A positive signal was analyzed by digital confocal microscopy. Images were sampled in the z-axis and subsequently processed with 3-D rendering software (Openlab) to generate three-dimensional pictures. The nuclei and incoming DNA could be viewed at different angles (0°, 44°, 56°, and 72°) and depths that showed that the positive signal was localized within the nuclear cage.
The transfer of EBV and human DNA was then analyzed in bovine endothelial cells cultured with apoptotic Namalwa cells. Using the human DNA- and EBV-specific probes simultanously, it was shown that approximately 17% of the bovine endothelial nuclei contained human DNA and 15% of the bovine nuclei analyzed a positive signal with theBamHI W (Fig 4 and Table 4). Similar frequencies were also observed in nuclei from BAE cells cultured with apoptotic IB4 cells (Table 4). In the bovine nuclei showing positive hybridization with both probes, signals overlapped in the same nuclear compartment (Fig 4B through D). Transfer of human DNA was also detectable in 3% of the nuclei after coculture of BAE cells and the EBV-negative BL41 line (Fig 4F). As expected, no positive signal could be detected in these cells with the BamHI W probe. To exclude the possibility that the positive signal was originating from cytoplasmic apoptotic DNA, positive signals were also analyzed by digital confocal microscopy. Images were sampled in the z-axis and subsequently processed with 3-D rendering software to generate three-dimensional pictures (Fig 4G). The nuclei and incoming DNA could be viewed at different angles and depths that showed that the positive signal was residing within the nuclear cage.
Frequency of DNA Transfer From Apoptotic Bodies to Bovine Endothelial Cells
| Probes . | BL414-150 + BAE . | Namalwa4-150 + BAE . | IB-44-150 + BAE . |
|---|---|---|---|
| Human DNA | 3% | 17% | 12% |
| EBV BamHI W | 0% | 15% | 11% |
| Probes . | BL414-150 + BAE . | Namalwa4-150 + BAE . | IB-44-150 + BAE . |
|---|---|---|---|
| Human DNA | 3% | 17% | 12% |
| EBV BamHI W | 0% | 15% | 11% |
Frequency of transfer of human and EBV-DNA in BAE cells after 3 weeks of culture with apoptotic bodies derived from BL41 (EBV-negative), Namalwa cells, or IB-4 cells (both with integrated copies of EBV). Two-color FISH analysis was used to estimate the percentage of nuclei that were positive for the human-specific or EBV-specific probes.
Irradiated cells.
DISCUSSION
We have shown that cultivation of apoptotic bodies derived from EBV-carrying cell lines with either fibroblasts, monocytes, or endothelial cells resulted in the uptake of DNA (as shown in the BAE cells) and expression of EBV-specific markers in the recipient cells. Expression of EBNA1 as well as EBER1+2 could be detected up to 5 weeks after the start of cocultivation experiments. The EBNA expression pattern was host-dependent, because EBNA1 but not EBNA2 was detected in fibroblast cells. This finding is consistent with previous findings showing that EBNA2 is only expressed in cells of lymphoblastoid origin.
BrdU-labeled apoptotic bodies could still be detected in a perinuclear location of the fibroblast recipient cells after 1 week of culture. Fibroblast cells containing an apoptotic body from the EBV-carrying cell line BL41/95 also expressed EBNA1, whereas fibroblasts cultivated with EBV-negative cell lines did not. Labeling recipient cells with BrdU before cocultivation showed that recipient cells only contained nuclei of fibroblast origin. This argues against the possibility that whole nuclei of the donor cell are phagocytosed to generate a functional hybrid cell with the recipient cell. FISH analysis showed that genomic as well as EBV-DNA from the apoptotic bodies was transferred into the nuclear compartment of the phagocytosing cell. The transferred DNA was stable over time, because similar frequencies could be detected with human and EBV-specific probes after 1, 2, and 3 weeks of cocultivation (data not shown).
Cultivation of lymphoid cell lines with either integrated or episomal copies of EBV showed that integrated EBV-DNA is transferred to the recipient cell, whereas episomal DNA is not. It is not clear why integration of EBV-DNA seems to be a requirement for EBV-DNA transfer. One explanation is that the episomal forms may be more prone to degradation by nucleases activated during apoptosis than integrated viral copies. FISH analysis showed that integrated EBV-DNA in Namalwa is transferred at a relative high frequency to bovine endothelial cells. The Namalwa cell line contains two integrated copies (172 kb) of EBV on the human chromosome 1 and constitutes a minute fraction of the total genome. It is therefore striking that the frequency of transfer of EBV-DNA after cocultivation with apoptotic bodies induced in the Namalwa line is at a level comparable to that of total Namalwa DNA. It suggests that the transfer of different fragments from the apoptotic chromatin and/or integrated EBV-DNA may be preferentially transferred after phagocytosis.
EBV infects B lymphocytes after binding to the complement receptor 2 (CR2), which is only present on B cells and immature T cells. However, EBV-DNA and/or its expression has been detected in several additional cell types in vivo, such as fibroblasts from rheumatoid arthritis patients, T-cell lymphomas, and a variety of carcinomas and sarcomas.34-38 Our findings raise the question of whether apoptotic bodies derived from EBV-carrying B lymphocytes may serve as the source of viral transfer to cells that lack receptors for the EBV virus in vivo. This could play an important role in the persistence of the EBV virus in the human host. In addition, the extended expression of viral genes in antigen presenting cells such as endothelial cells and macrophages may play an important role in eliciting an immune response to viral antigens. Finally, we speculate that similar mechanisms of horizontal DNA transfer may be of importance in conditions characterized by high levels of apoptosis, eg, in tumors treated with irradiation or chemotherapy.
Supported by grants from the Swedish Cancer Society and the Jeannska Foundation.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Lars Holmgren, PhD, Microbiology and Tumor Biology Center, PO Box 280, Karolinska Institute, S-171 77, Stockholm, Sweden; e-mail: Lars.Holmgren@mtc.ki.se.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal