Abstract
The prognostic value of immunophenotype in adult acute lymphoblastic leukemia (ALL) has varied based on the methods used, surface markers studied, and therapy administered. From April 1991 to September 1996, samples of leukemic marrow or blood from 259 eligible and evaluable adult ALL patients entering dose-intensive Cancer and Leukemia Group B (CALGB) front-line treatment protocols were prospectively studied for immunophenotypic classification by multiparameter flow cytometry (MFC) in a central laboratory. A B-lineage (B-LIN) phenotype was expressed in 79% of cases, with one third coexpressing myeloid antigens. A T-lineage (T-LIN) phenotype was expressed in 17% of cases, with one quarter coexpressing myeloid antigens. Since the advent of more intensive CALGB therapy which incorporated cyclophosphamide and the early use of L-asparaginase into the backbone of daunorubicin, vincristine and prednisone, together with central nervous system prophylaxis for adult ALL, no significant differences in response rates, remission duration, or survival have been seen in those patients coexpressing myeloid antigens. The T-LIN phenotype was associated with younger age (P = .01), a higher male to female ratio (P = .01), higher white blood cell count (P = .001) and hemoglobin (P < .001) levels, presence of a mediastinal mass (P < .001), and longer survival (P = .01) and disease-free survival (DFS) (P = .01) when compared to patients with a B-LIN phenotype. The 3-year probability of survival and DFS (95% confidence interval [CI]) of T-LIN adult ALL was 0.62 (0.46 to 0.76) and 0.62 (0.44 to 0.77), respectively. Comparatively, the 3-year probability of survival and DFS (95% CI) of B-LIN adult ALL was 0.42 (0.35 to 0.50) and 0.39 (0.31 to 0.47), respectively. The number of T markers expressed in T-LIN ALL cases was shown to have prognostic significance. In particular, patients expressing six or more markers compared with patients expressing three or fewer markers had longer DFS (P = .003) and survival (P = .004). The presence of the Philadelphia chromosome was significantly associated with B-LIN ALL cases which coexpressed CD19+, CD34+, and CD10+ (49%;P = .003), whereas the majority of t(4;11) cases were CD19+, CD34+ but CD10−. The knowledge gained from this study of MFC of a large number of patients will permit a reduction in the number of antigens to be evaluated in future studies. Overall, this should lead to cost savings without loss of valuable information. A rational approach for future studies would be to use four-color flow cytometry (instead of the current three-color) to help further streamline the study of immunophenotype of adult ALL by MFC.
ACUTE LYMPHOBLASTIC leukemia (ALL) is biologically and clinically a heterogeneous group of diseases characterized by malignant proliferation and accumulation of immature lymphoid cells within the bone marrow, blood, and lymphoid organs. Historically, prognostic data were obtained from routine physical examination, serum biochemical profiles, bone marrow morphology, and peripheral blood counts. More recently, information obtained from karyotype, molecular genetics, and surface immunophenotype have contributed to a better understanding of this complex disease and its treatment.1-3 Immunophenotyping by flow cytometry aids in distinguishing ALL from acute myeloid leukemia (AML) in approximately 10% to 15% of cases in which morphology and cytochemistry are inadequate to make a definitive diagnosis.
Immunophenotyping of ALL is accomplished by use of panels of specific monoclonal antibodies (MoAbs) that recognize distinct epitopes of cellular membrane antigens. Normal lymphocytes are derived from pluripotent stem cells that become committed to the lymphoid developmental pathway. As normal B-cell and T-cell ontogeny occurs, these committed cells acquire and lose various surface molecules that represent typical changes in cytokine-induced metabolic, and adhesive function which occur at various stages of differentiation.4-6
The neoplastic process that leads to leukemogenesis is characterized by cellular and molecular dysregulation. The net result is that the leukemic cell is phenotypically different from any normal cell, but the degree of difference is extremely variable. Although the functions of many of these surface molecules still remain unknown, fluorochrome-conjugated antibodies against them, when used in combination, allow resolution of a unique repertoire with variable expression that can differentiate between normal and leukemic cells.
Protocol 8364: “Immunologic diagnostic studies in adult ALL” was activated by the Cancer and Leukemia Group B (CALGB) on June 27, 1983 to allow specific immunophenotyping questions to be answered independent of a given therapy protocol. Initial studies of ALL patients prospectively studied and uniformly treated on CALGB treatment protocols (8011, 8411, 8513, and 8811) were among the first to establish the incidence of specific lineages in adult ALL patients. Approximately 50% to 60% had B-lineage disease, 15% had T-lineage disease, and the remaining 25% had unusual immunophenotypes including expression of myeloid lineage-associated antigens.7-9 These and other early studies analyzed the expression of a single antigen at a time by direct immunofluorescence microscopy or uniparameter flow cytometry.
In the Spring of 1991, ALL immunophenotyping studies began to be performed using multiparameter flow cytometry (MFC), thus allowing definitive assignment of surface antigens to leukemic cells or normal cells. In uniparameter flow cytometry, cells are stained with a single antibody and the mononuclear cell population is analyzed. In MFC, a single tube of cells is simultaneously stained with three or more different antibodies to define a distinct cluster pattern of up to eight different populations. Each of the following characteristics allow identification of leukemia cells: expression of antigens that are not typically coexpressed on normal cells, lack of expression of antigens that are typically coexpressed on normal cells, or abnormal antigen density. The data used for clinical correlations and prognosis by MFC are considered more accurate than data obtained by uniparameter methods because they more closely represent the true phenotype of the leukemic cells and allows them to be objectively resolved from normal cells. Thus, the immunophenotypic pattern defined by multiparameter techniques may more precisely reflect the biology of ALL and provide prognostic information.
MATERIALS AND METHODS
Patients
Patients in this report were eligible to be enrolled on CALGB 8364 within 1 month before registration on a front-line CALGB treatment protocol (8811, 9111, 9311, 9511) for untreated ALL. In general, beginning with CALGB 8811, more intensive chemotherapy programs for adult ALL were instituted that incorporated cyclophosphamide and early, more intensive L-asparaginase administration into the backbone of daunorubicin, vincristine, and prednisone, together with central nervous system (CNS) prophylaxis.10-13 Before initiation of therapy and again at time of relapse, leukemic cells were collected in heparin from bone marrow and/or blood and shipped overnight at ambient temperature to the central CALGB laboratory. Unstained bone marrow aspirate and blood smears were concurrently submitted for central pathologic review of cytologic and cytochemical studies to confirm a diagnosis of ALL. All cases were myeloperoxidase negative.
Multiparameter Flow Cytometry Studies
Since April 1, 1991, CALGB immunophenotyping studies in ALL have been performed in the laboratory of Carleton Stewart, PhD, at Roswell Park Cancer Institute.
Procedure.
Submitted specimens were filtered through a 75-μm mesh and then washed twice in a phosphate-buffered saline (PBS)-heparin solution. Specimens were incubated with 200 μg/mL murine IgG per milliliter of specimen to block Fc receptors. Trios of diagnostic MoAbs shown in Table 1 were individually labeled with the fluorochromes fluorescein (F), phycoerythrin (PE), and tandem complexes (TC) of phycoerythrin Texas-red-biotin or phycoerythrin-cyanine 5 (Cy5)-biotin or directly conjugated with TC or peridinin chlorophyll a Protein (Per CP). If a biotinyated antibody was used (before availability of direct conjugates), the TC-avidin was added to the cells for an additional 15 minutes of incubation. A detailed explanation of staining, data acquisition, and analysis has been described elsewhere.14 15 Light-activated ethidium-monoxide–treated aliquots were used to detect dead cells. Isotype and autofluorescence controls were performed. Leukemia gates were used for analysis. Data were obtained using a dedicated Becton Dickinson FACScan flow cytometer (Becton Dickinson, San Jose, CA). Events are recorded on disk and files analyzed using the Winlist multiparameter analysis software package (Verity Software House, Topsham, ME). Results of analysis with appropriate clinical comments were transferred electronically to a report template and the original bidimensional data plots for each antibody panel were reviewed and interpreted by the principal investigators (M.S.C., S.R.F., and C.C.S.). Hard copies of final reports were signed manually and then electronically transferred via an Ethernet link between the Laboratory of Flow Cytometry at Roswell Park Cancer Institute and the CALGB Data Management Center in Durham, NC.
Immunophenotyping Panels for ALL
| Panel . | Purpose . | Label-CD Cluster . |
|---|---|---|
| 1 | Defines leukocyte gate | FL-CD45 |
| PE-CD14 | ||
| TC-HLADr | ||
| 2 | Defines major T-cell subsets and their malignant counterparts | FL-CD3 PE-CD4 |
| TC-CD8 | ||
| 3 | Defines uncommitted progenitor cells (34+, 38−, DR−) from committed progenitor cells (34+, 38+, DR−) | FL-CD38 PE-CD34 TC-HLADR |
| 4 | Detects CD10 on B cells, or T cells; detects CLL (19+, 5+) | FL-CD5 PE-CD19 |
| TC-CD10 | ||
| 5 | Detects immature T- or B-cell malignancies | FL-CD5 |
| PE-CD34 | ||
| TC-CD19 | ||
| 6 | Helps distinguish B-ALL from B-lymphoproliferative disorders (11c+) | FL-CD20 PE-CD11c |
| TC-CD22 | ||
| 7 | Detects CD19+ B-myeloid cells | FL-CD33 |
| PE-CD13 | ||
| TC-CD19 | ||
| 8 | Verifies or excludes CD19 expression on T-ALL | FL-CD7 PE-CD19 TC-CD2 |
| 9 | Detects surface Ig expression on B cells | FL-IgD |
| PE-IgM | ||
| TC-CD20 | ||
| 10 | Detects light-chain clonal excess for B-cell malignancies | FL-κ PE-λ |
| TC-CD20 | ||
| 11 | Isotype control for nonspecific binding | FL-IgG1 |
| PE-IgG2a | ||
| TC-IgG2b |
| Panel . | Purpose . | Label-CD Cluster . |
|---|---|---|
| 1 | Defines leukocyte gate | FL-CD45 |
| PE-CD14 | ||
| TC-HLADr | ||
| 2 | Defines major T-cell subsets and their malignant counterparts | FL-CD3 PE-CD4 |
| TC-CD8 | ||
| 3 | Defines uncommitted progenitor cells (34+, 38−, DR−) from committed progenitor cells (34+, 38+, DR−) | FL-CD38 PE-CD34 TC-HLADR |
| 4 | Detects CD10 on B cells, or T cells; detects CLL (19+, 5+) | FL-CD5 PE-CD19 |
| TC-CD10 | ||
| 5 | Detects immature T- or B-cell malignancies | FL-CD5 |
| PE-CD34 | ||
| TC-CD19 | ||
| 6 | Helps distinguish B-ALL from B-lymphoproliferative disorders (11c+) | FL-CD20 PE-CD11c |
| TC-CD22 | ||
| 7 | Detects CD19+ B-myeloid cells | FL-CD33 |
| PE-CD13 | ||
| TC-CD19 | ||
| 8 | Verifies or excludes CD19 expression on T-ALL | FL-CD7 PE-CD19 TC-CD2 |
| 9 | Detects surface Ig expression on B cells | FL-IgD |
| PE-IgM | ||
| TC-CD20 | ||
| 10 | Detects light-chain clonal excess for B-cell malignancies | FL-κ PE-λ |
| TC-CD20 | ||
| 11 | Isotype control for nonspecific binding | FL-IgG1 |
| PE-IgG2a | ||
| TC-IgG2b |
Antibody panels.
Ten panels containing three conjugated antibodies each were used for ALL immunophenotyping studies, and their composition is shown in Table1 along with a brief rationale for their combination. Clusters of both normal and abnormal cells were explicitly resolved, and the pattern of these cell clusters served to provide another dimension for visualizing aberrant antigen expression.
Definitions.
The criterion for surface marker positivity was coexpression of an antigen by at least 10% of the leukemia blast population. Ten percent positivity was selected as a cutoff to eliminate the possibility that coexpression was caused by a nonspecific binding process or to dead cells. B-lineage (B-LIN) antigen expression was defined as CD19 or CD20 positivity; T-lineage (T-LIN) antigen expression as either (1) CD2 or CD7 positivity with CD1 or CD3 or CD4 or CD5 or CD8 positivity, or (2) CD5 positivity without CD19 or CD20 positivity; myeloid (My) antigen expression as CD13 and/or CD33 positivity coexpressed with either CD19 or CD2 and/or CD7; and stem cell phenotype as CD34 expression in the absence of positivity with any other lymphoid or myeloid lineage markers. Cases expressing combinations of antigens were classified as BMy, TMy, BTMy, BT, or unclassified ALL. For analysis purposes, the BMy cases were considered to be B-LIN and the TMy cases were considered to be T-LIN. Cases of FAB-L3 (Burkitt type ALL) were not included in our analyses, nor were cases that were strictly myeloid (ie, AML-MO).
Cytogenetics.
Many of these ALL patients also underwent genetic studies. Cytogenetic analysis of adult ALL specimens was performed as part of CALGB study 8461 (a prospective study of chromosomes in acute leukemia) as previously described.16 The companion cytogenetics study was not mandatory for patient enrollment on CALGB ALL clinical therapy trials or CALGB 8364. Sixty-one percent of the samples submitted for patients registered to both studies were evaluable by central review of karyotypes and available for this report. In addition, molecular detection of the Philadelphia (Ph) chromosome using reverse-transcriptase polymerase chain reaction (PCR) to detect the BCR-ABL fusion gene was a main objective of CALGB study 8762.17 In our statistical analyses, Ph+patients are those who were classified by either cytogenetic or molecular methods, unless otherwise specified. A “better-risk B-LIN” subgroup was defined as patients with B/BMy immunophenotype who were studied by cytogenetic and/or molecular methods and known not to be Ph+, BCR/ABL+, or having the t(4;11) cytogenetic abnormality.
Statistical analysis.
The relationship between immunophenotypic subgroups and the endpoints of disease-free survival (DFS) and overall survival were the main statistical analyses undertaken for this study. Some of the analyses were confirmatory (eg, the comparison of B-LIN and T-LIN), while other analyses were exploratory in an attempt to define new prognostic groups based on marker expression.
The duration of DFS was defined as the time from achieving a complete remission (CR) to relapse (bone marrow, blood, CNS, or testicular), death, or date of last follow-up. Survival was defined as the time from study entry to the date of last follow-up. Patients still at risk were censored at the date of last follow-up for the analysis of these endpoints. Probabilities of surviving and remaining in CR were estimated by the Kaplan-Meier method.18 Ninety-five percent confidence intervals (CI) for these probabilities were obtained by the method of Simon and Lee.19 Differences in survival or DFS between patient subgroups were tested using the logrank statistic, adjusted for multiple comparisons where appropriate.20 21Median follow-up time was estimated from the survival times of patients still at risk. Differences in proportions among patient subgroups were analyzed using Fisher’s exact test. All reported P values are nominal two-sided values. The analyses were based on all data available as of January 1998.
RESULTS
Patient Accrual
Between April 1, 1991 and September 30, 1996, 351 adult ALL patients were registered to CALGB 8364 and underwent MFC immunophenotyping studies. Of these 351 patients, 22 with ALL-L3 were treated on CALGB 9251, a protocol for Burkitt-type leukemia and diffuse, small noncleaved cell lymphoma, and excluded from this analysis. There were 4 additional L3 cases that were registered and treated on one of the four front-line ALL trials, but recognized as Burkitt-type leukemia after central review and, therefore, also excluded. In addition, patients were excluded from these analyses if they were found ineligible for the planned CALGB treatment study (n = 18) or if their specimens were found to be not evaluable (n = 31), with normal marker expression (n = 4), or not received at the central laboratory or evaluated at time of analysis (n = 13). Thus, a total of 259 evaluable samples were analyzed by MFC. These patients were treated on the following protocols: 8811 (n = 12), 9111 (n = 148), 9311 (n = 74), and 9511 (n = 25); the results of these studies have been reported in detail or in part.10-13 During the time period stated above, 96% of the patients accrued to these four studies had samples submitted for MFC immunophenotyping. These studies were comparable with respect to CR rates (82% to 85%) and estimated 3-year DFS (40% to 46%).
ALL Lineage Frequency
Lineage assignment by MFC in patients with adult ALL entering consecutive, similarly intensive CALGB front-line treatment protocols (8811, 9111, 9311, 9511) was determined. Features of a B-lineage phenotype were expressed in 79% of cases, with one third of this group coexpressing myeloid antigens. Seventeen percent of cases had a T-lineage phenotype, with one quarter of the group coexpressing myeloid antigens. BT, BTMy, stem cell, and unclassifiable phenotype comprised 1% or less of the remaining groups, respectively (Table2), and were dropped from further evaluation.
ALL Lineage Frequency (CALGB 8364)
| Lineage Assignment . | No. of Cases . |
|---|---|
| B antigens only | 138 (53%) |
| B + myeloid antigens | 68 (26%) |
| B + T antigens | 2 (1%) |
| B + T + myeloid antigens | 3 (1%) |
| T antigens only | 33 (13%) |
| T + myeloid antigens | 11 (4%) |
| Stem cell | 3 (1%) |
| Unclassified | 1 (<1%) |
| Total | 259 (100%) |
| Lineage Assignment . | No. of Cases . |
|---|---|
| B antigens only | 138 (53%) |
| B + myeloid antigens | 68 (26%) |
| B + T antigens | 2 (1%) |
| B + T + myeloid antigens | 3 (1%) |
| T antigens only | 33 (13%) |
| T + myeloid antigens | 11 (4%) |
| Stem cell | 3 (1%) |
| Unclassified | 1 (<1%) |
| Total | 259 (100%) |
Frequency of Individual Antigen Expression on B-LIN and T-LIN ALL
In addition to lineage analysis, the frequency of individual marker expression for the B-LIN and T-LIN immunophenotypic subgroups was evaluated (Table 3).
Frequency (%) of Individual Marker Expression on B-LIN and T-LIN ALL
| Marker . | B-LIN (n = 206) . | T-LIN (n = 44) . |
|---|---|---|
| CD1+ | 0% | 63% |
| CD2+ | 6% | 70% |
| CD3+ | 0% | 41% |
| CD4+ | <1% | 61% |
| CD5+ | 1% | 84% |
| CD7+ | <1% | 93% |
| CD8+ | 0% | 55% |
| CD10+ | 72% | 16% |
| CD11c+ | 1% | 2% |
| CD13+ | 32% | 25% |
| CD19+ | 98% | 0% |
| CD20+ | 33% | 0% |
| CD22+ | 87% | 6% |
| CD33+ | 3% | 2% |
| CD34+ | 81% | 27% |
| CD38+ | 84% | 78% |
| HLA DR | 96% | 9% |
| κ+ | 1% | 0% |
| λ+ | 1.5% | 0% |
| IgD+ or IgM+ | 0%3-150 | 0%3-150 |
| Marker . | B-LIN (n = 206) . | T-LIN (n = 44) . |
|---|---|---|
| CD1+ | 0% | 63% |
| CD2+ | 6% | 70% |
| CD3+ | 0% | 41% |
| CD4+ | <1% | 61% |
| CD5+ | 1% | 84% |
| CD7+ | <1% | 93% |
| CD8+ | 0% | 55% |
| CD10+ | 72% | 16% |
| CD11c+ | 1% | 2% |
| CD13+ | 32% | 25% |
| CD19+ | 98% | 0% |
| CD20+ | 33% | 0% |
| CD22+ | 87% | 6% |
| CD33+ | 3% | 2% |
| CD34+ | 81% | 27% |
| CD38+ | 84% | 78% |
| HLA DR | 96% | 9% |
| κ+ | 1% | 0% |
| λ+ | 1.5% | 0% |
| IgD+ or IgM+ | 0%3-150 | 0%3-150 |
Four patients were B-LIN based on CD19 dim expression; 1 patient was not evaluable for CD19 but expressed CD20 and was thereby classified as B-LIN.
Surface IgD or IgM expression was found on 0 of 103 B-LIN and 0 of 18 T-LIN adult ALL cases evaluated.
In the B-LIN (B/BMy) group, antigen expression in decreasing order of frequency was as follows: CD19 (98%), HLA DR (96%), CD22 (87%), CD38 (84%), CD34 (81%), CD10 (72%), CD20 (33%), CD13 (32%), and occasionally CD2 (6%) or CD33 (3%), and rarely or never lambda (1.5%), kappa (1%), CD5 (1%), CD11c (1%), CD4 (<1%), CD7 (<1%), CD1 (0%), CD3 (0%), CD8 (0%), IgD (0%), or IgM (0%).
In the T-LIN (T/TMy) group, antigen expression in decreasing order of frequency was as follows: CD7 (93%), CD5 (84%), CD38 (78%), CD2 (70%), CD1 (63%), CD4 (61%), CD8 (55%), CD3 (41%), CD34 (27%), CD13 (25%), CD10 (16%), HLA DR (9%), and occasionally CD22 (6%), CD11c (2%), or CD33 (2%), and never CD19 or CD20.
Clinical Correlates Associated With Immunophenotype
Clinical and biological features by immunophenotype.
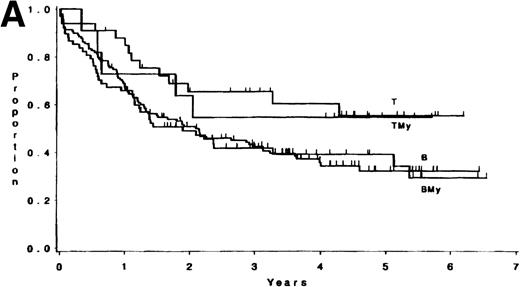
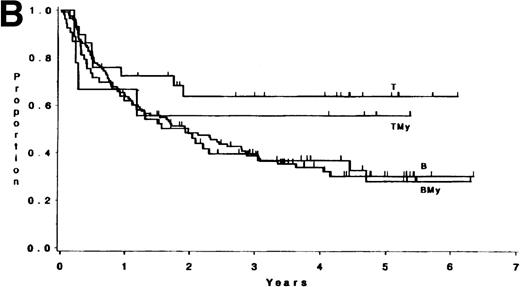
The median follow-up time for these patients has been 3.8 years (range, 1 month to 6.5 years); only 3 patients have had less than 1 year of follow-up. There were no differences in survival or DFS between patients with ALL expressing B antigens only and those expressing B plus myeloid antigens (P = .84, and P = .82, respectively; Fig 1). Similarly, there were no differences observed between patients with ALL expressing T antigens only and those expressing T plus myeloid antigens (p = .81, and p = .53, respectively; Fig 1). Hence, B and BMy patients were combined, as were T and TMy patients, for comparison purposes. Three groups were studied: B/BMy (n = 206), T/TMy (n = 44), and a “Better risk B” subgroup (n = 53), which consisted of only the B/BMy patients who had adequate cytogenetics and excluded patients with cytogenetic or molecular evidence of the Ph chromosome (Ph+) or t(4;11)(q21;q23). Statistical comparisons of these features between T-LIN cases and each B group are given in Table4.
(A) Survival for 250 adults with ALL according to expression of only B (n = 138) or T (n = 33) antigens or B + myeloid (n = 68) or T + myeloid (n = 11) antigens. There were no differences between the B and BMy patients (P = .84) and the T and TMy patients (P = .81). (B) DFS for 212 adults with ALL according to expression of only B (n = 121) or T (n = 29) antigens or B + myeloid (n = 53) or T + myeloid (n = 9) antigens. There were no differences between the B and BMy patients (P = .82), and the T and TMy patients (P = .53).
(A) Survival for 250 adults with ALL according to expression of only B (n = 138) or T (n = 33) antigens or B + myeloid (n = 68) or T + myeloid (n = 11) antigens. There were no differences between the B and BMy patients (P = .84) and the T and TMy patients (P = .81). (B) DFS for 212 adults with ALL according to expression of only B (n = 121) or T (n = 29) antigens or B + myeloid (n = 53) or T + myeloid (n = 9) antigens. There were no differences between the B and BMy patients (P = .82), and the T and TMy patients (P = .53).
Clinical and Biological Features by Immunophenotype
| Feature . | B-LIN (n = 206) . | P . | Better Risk B (n = 53) . | P . | T-LIN (n = 44) . |
|---|---|---|---|---|---|
| Median age (range) | 34 (17-78) | .01 | 28 (17-78) | .50 | 28 (16-81) |
| Sex: | |||||
| Males | 105 (51%) | .01 | 34 (64%) | .39 | 32 (73%) |
| Females | 101 | 19 | 12 | ||
| Median WBC count (range) | 15.5 (0.3-307) | <.001 | 13.1 (0.7-286) | <.001 | 48.2 (0.8-326) |
| Median platelets (range) | 47 (4-361) | .17 | 68 (4-259) | .93 | 61 (9-258) |
| Median hemoglobin (range) | 9.6 (3.6-18.5) | <.001 | 9.8 (3.6-16.3) | .01 | 11.1 (6.7-16.5) |
| Mediastinal mass: | |||||
| Yes | 1 (1%) | <.001 | 0 (0%) | <.001 | 17 (40%) |
| No | 194 | 52 | 25 | ||
| Median yr DFS (range) | 1.9 (1.3-2.6) | .01 | 1.9 (1.1-4.5) | .06 | >2.0 (>1.2->4.3) |
| Median yr survival (range) | 1.9 (1.3-3.0) | .01 | 2.5 (1.3-5.1) | .08 | >3 (>1.8->4.3) |
| 3-yr probability of DFS (95% CI) | .39 (.31-.47) | .42 (.28-.56) | .62 (.44-.77) | ||
| 5-yr probability of DFS (95% CI) | .29 (.18-.43) | .32 (.16-.53) | .62 (.36-.83) | ||
| 3-yr probability of survival (95% CI) | .42 (.35-.50) | .47 (.33-.60) | .62 (.46-.76) | ||
| 5-yr probability of survival (95% CI) | .34 (.23-.47) | .36 (.21-.55) | .55 (.31-.77) |
| Feature . | B-LIN (n = 206) . | P . | Better Risk B (n = 53) . | P . | T-LIN (n = 44) . |
|---|---|---|---|---|---|
| Median age (range) | 34 (17-78) | .01 | 28 (17-78) | .50 | 28 (16-81) |
| Sex: | |||||
| Males | 105 (51%) | .01 | 34 (64%) | .39 | 32 (73%) |
| Females | 101 | 19 | 12 | ||
| Median WBC count (range) | 15.5 (0.3-307) | <.001 | 13.1 (0.7-286) | <.001 | 48.2 (0.8-326) |
| Median platelets (range) | 47 (4-361) | .17 | 68 (4-259) | .93 | 61 (9-258) |
| Median hemoglobin (range) | 9.6 (3.6-18.5) | <.001 | 9.8 (3.6-16.3) | .01 | 11.1 (6.7-16.5) |
| Mediastinal mass: | |||||
| Yes | 1 (1%) | <.001 | 0 (0%) | <.001 | 17 (40%) |
| No | 194 | 52 | 25 | ||
| Median yr DFS (range) | 1.9 (1.3-2.6) | .01 | 1.9 (1.1-4.5) | .06 | >2.0 (>1.2->4.3) |
| Median yr survival (range) | 1.9 (1.3-3.0) | .01 | 2.5 (1.3-5.1) | .08 | >3 (>1.8->4.3) |
| 3-yr probability of DFS (95% CI) | .39 (.31-.47) | .42 (.28-.56) | .62 (.44-.77) | ||
| 5-yr probability of DFS (95% CI) | .29 (.18-.43) | .32 (.16-.53) | .62 (.36-.83) | ||
| 3-yr probability of survival (95% CI) | .42 (.35-.50) | .47 (.33-.60) | .62 (.46-.76) | ||
| 5-yr probability of survival (95% CI) | .34 (.23-.47) | .36 (.21-.55) | .55 (.31-.77) |
P values are from comparisons with T-LIN group and are unadjusted for multiple comparisons. “Better risk B” subgroup consists of only those B/BMy patients evaluated by cytogenetics and/or molecular methods and known not to have the Philadelphia chromosome, BCR/ABL fusion gene, or t(4;11).
Compared with B-LIN patients T-LIN patients were younger (P = .01), had a higher male to female ratio (P = .01), more often had a mediastinal mass (P < .001), had higher white blood cell (WBC) counts (P = .001) and hemoglobin (P < .001) levels, and longer survival (P = .01) and DFS (P = .01). With a median follow-up of 3.8 years, the 3-year probability of survival and DFS of T-LIN adult ALL is .62 and .62 compared with .42 and 0.39 for the B-LIN adult ALL group, respectively. The potential impact that cytogenetics plays with respect to prognosis in ALL was evaluated when the same clinical and biological features were reanalyzed using a “Better risk B” subgroup compared with the T-LIN group (Table 4). In this analysis, T-LIN patients were still more likely to show a mediastinal mass (P < .001) and a higher WBC count (P = .001) and hemoglobin (P = .01), although DFS (P = .06) and survival (P = .08) were of marginal statistical significance.
T-lineage ALL.
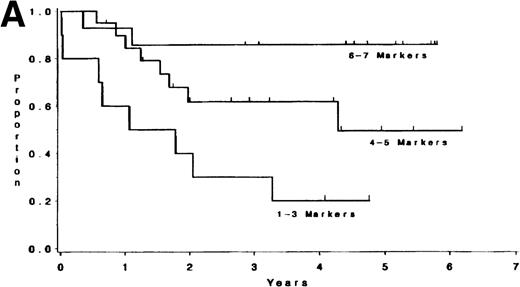
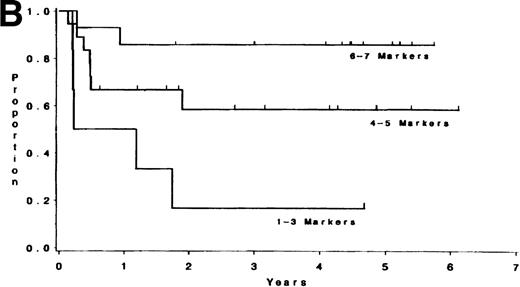
Adult T-lineage ALL currently treated with more intensive chemotherapy regimens now show more favorable results than adult B-lineage ALL.10 22 When we analyzed individual T-cell markers in our T-lineage group, those patients with CD1, CD2, CD4, and CD5 expression were all associated with a significant improvement in survival (P = .006, P = .04, P = .05, andP = .05, respectively) compared with those patients not expressing these antigens (data not presented). The 44 individual T-LIN patients expressed a varying number of T markers. Ten patients expressed 1-3 markers, 20 patients expressed 4-5 markers, and 14 patients expressed 6-7 T-cell markers. Of the 10 patients with 1-3 markers, 6 were classified as pre-T ALL defined by expression of CD7 and lack of expression of CD1, CD2, CD4, and CD8. The 1-3 marker group had a shorter survival compared to those patients with at least 6 markers positive (P = .004). Furthermore, the same relationship between these two groups was also found for DFS (P = .003) (see Table 5 and Fig2). The group expressing 4-5 T-cell markers had an intermediate outcome. The incidence of individual T-cell markers was reviewed among the three groups. CD5 and CD7 were coexpressed in the majority of T-cell ALL patients tested. CD1, CD2, CD3, CD4, and CD8 were seldom expressed in the 1-3 marker group (25%, 10%, 0%, 20%, and 0% respectively) compared with the 4-5 marker group (62%, 85%, 40%, 60%, and 50%, respectively), and the 6-7 marker group (91%, 93%, 71%, 93%, and 100%, respectively).
CR Rate, DFS, and Survival Versus Number of T-Cell Markers Expressed
| Group (no. of markers) . | No. of Patients . | CR (%) . | Probability of DFS at 3 yr (95% CI) . | P5-150 . | Probability of Survival at 3 yr (95% CI) . | P5-150 . |
|---|---|---|---|---|---|---|
| 1-3 | 10 | 6 (60%) | 0.17 (0.05-0.45) | — | 0.30 (0.12-0.57) | — |
| 4-5 | 20 | 18 (90%) | 0.58 (0.29-0.81) | .13 | 0.62 (0.36-0.82) | .13 |
| 6-7 | 14 | 14 (100%) | 0.86 (0.59-0.96) | .003 | 0.86 (0.60-0.96) | .004 |
| Group (no. of markers) . | No. of Patients . | CR (%) . | Probability of DFS at 3 yr (95% CI) . | P5-150 . | Probability of Survival at 3 yr (95% CI) . | P5-150 . |
|---|---|---|---|---|---|---|
| 1-3 | 10 | 6 (60%) | 0.17 (0.05-0.45) | — | 0.30 (0.12-0.57) | — |
| 4-5 | 20 | 18 (90%) | 0.58 (0.29-0.81) | .13 | 0.62 (0.36-0.82) | .13 |
| 6-7 | 14 | 14 (100%) | 0.86 (0.59-0.96) | .003 | 0.86 (0.60-0.96) | .004 |
Adjusted P value by logrank comparing 1-3 marker group with 4-5 or 6-7 marker group.
(A) Survival for T-LIN patients by the number of T-cell markers expressed. Patients expressing 6-7 markers (n = 14) had longer survival than those patients expressing 3 or fewer markers (n = 10; adjusted P = .004). Patients expressing 4-5 markers (n = 20) had intermediate survival that was not significantly different from the 6-7 marker group (adjustedP = .24) or the 1-3 marker group (adjustedP = .13). (B) DFS for T-LIN patients by the number of T-cell markers expressed. Patients expressing 6-7 markers (n = 14) had longer DFS than those patients expressing 3 or fewer markers (n = 6; adjusted P = .003). Patients expressing 4-5 markers (n = 18) have intermediate DFS that was not significantly different from the 6-7 marker group (adjusted P = .31) or the 1-3 marker group (adjusted P = .13).
(A) Survival for T-LIN patients by the number of T-cell markers expressed. Patients expressing 6-7 markers (n = 14) had longer survival than those patients expressing 3 or fewer markers (n = 10; adjusted P = .004). Patients expressing 4-5 markers (n = 20) had intermediate survival that was not significantly different from the 6-7 marker group (adjustedP = .24) or the 1-3 marker group (adjustedP = .13). (B) DFS for T-LIN patients by the number of T-cell markers expressed. Patients expressing 6-7 markers (n = 14) had longer DFS than those patients expressing 3 or fewer markers (n = 6; adjusted P = .003). Patients expressing 4-5 markers (n = 18) have intermediate DFS that was not significantly different from the 6-7 marker group (adjusted P = .31) or the 1-3 marker group (adjusted P = .13).
CR rate in adult T-LIN ALL also appears to be influenced by the number of T-cell markers expressed. The 1-3 marker group showed CR in 6 of 10 (60%) patients, compared with 18 of 20 (90%) in the 4-5 marker group and 14 of 14 (100%) in the 6-7 marker group (P = .02).
B-lineage ALL.
A B-lineage immunophenotype was found in 79% of the 259 evaluable ALL patients. In addition to analysis by lineage or individual markers, MFC allows analysis by groups of antigens expressed on individual cell populations. This technique was used in an attempt to subclassify 206 patients with B-lineage (CD19+) ALL. CD34 and CD10 were evaluated because these antigens have been previously reported as having prognostic importance in ALL.23-25 In our current study, analysis of individual antigen markers demonstrated that only CD34-positivity in the B-LIN group was associated with shorter survival (P = .06) and DFS (P = .02) when compared with CD34− patients. In Table 6, four B-lineage ALL populations (CD10+34+, CD10+34−, CD10−34+, CD10−34−) are defined, and selected clinical and laboratory features are demonstrated. For age, time to CR, DFS, and survival, no statistically significant differences were seen among the four subgroups. However, the CD10−34+ group was observed to have significantly higher WBC counts than the CD10+34+ and CD10+34−groups (both at P < .05), adjusting for multiple comparisons. With respect to the presence of the Ph chromosome, the CD10+34+ group had a significantly higher fraction of positive cases (49%) when compared with the three other groups (P = .003).
Selected Clinical and Laboratory Features in Subgroups of CD19+ ALL
| Feature . | CD10+34+ . | CD10+34− . | CD10−34+ . | CD10−34− . | P Value . |
|---|---|---|---|---|---|
| No. of patients | 124 | 24 | 43 | 13 | |
| Median age | 34 | 32 | 35 | 48 | .28 |
| (range) | 17-78 | 17-64 | 17-78 | 19-75 | |
| Median WBC count | 12.9 | 8.5 | 26.9 | 11.8 | .04 |
| (range) | 0.3-307 | 0.7-110 | 0.8-251 | 1.7-72 | |
| Response: | |||||
| CR (%) | 105 (85%) | 23 (96%) | 34 (79%) | 11 (85%) | .32 |
| No CR (%) | 19 (15%) | 1 (4%) | 9 (21%) | 2 (15%) | |
| Time to CR: | |||||
| ≤30 d (%) | 75 (71%) | 15 (65%) | 22 (65%) | 9 (82%) | .64 |
| >30 d (%) | 30 (29%) | 8 (35%) | 12 (35%) | 2 (18%) | |
| Median DFS (yr) | 1.9 | >2.3 | 1.1 | >.7 | .24 |
| Median survival (yr) | 2.1 | 3.7 | 1.1 | 1.1 | .20 |
| 3-yr probability of DFS (95% CI) | .34 (.25-.45) | .55 (.33-.75) | .37 (.22-.55) | .55 (.26-.80) | |
| 5-yr probability of DFS (95% CI) | .24 (.11-.45) | .55 (.26-.81) | .27 (.13-.47) | .55 (.21-.85) | |
| 3-yr probability of survival (95% CI) | .40 (.31-.50) | .67 (.45-.83) | .34 (.21-.49) | .43 (.20-.70) | |
| 5-yr probability of survival (95% CI) | .31 (.16-.52) | .48 (.24-.72) | .34 (.18-.54) | .43 (.16-.75) | |
| Ph chromosome: | |||||
| Negative | 35 | 11 | 24 | 7 | .003 |
| Positive6-150 | 34 (49%) | 2 (15%) | 7 (23%) | 0 (0%) |
| Feature . | CD10+34+ . | CD10+34− . | CD10−34+ . | CD10−34− . | P Value . |
|---|---|---|---|---|---|
| No. of patients | 124 | 24 | 43 | 13 | |
| Median age | 34 | 32 | 35 | 48 | .28 |
| (range) | 17-78 | 17-64 | 17-78 | 19-75 | |
| Median WBC count | 12.9 | 8.5 | 26.9 | 11.8 | .04 |
| (range) | 0.3-307 | 0.7-110 | 0.8-251 | 1.7-72 | |
| Response: | |||||
| CR (%) | 105 (85%) | 23 (96%) | 34 (79%) | 11 (85%) | .32 |
| No CR (%) | 19 (15%) | 1 (4%) | 9 (21%) | 2 (15%) | |
| Time to CR: | |||||
| ≤30 d (%) | 75 (71%) | 15 (65%) | 22 (65%) | 9 (82%) | .64 |
| >30 d (%) | 30 (29%) | 8 (35%) | 12 (35%) | 2 (18%) | |
| Median DFS (yr) | 1.9 | >2.3 | 1.1 | >.7 | .24 |
| Median survival (yr) | 2.1 | 3.7 | 1.1 | 1.1 | .20 |
| 3-yr probability of DFS (95% CI) | .34 (.25-.45) | .55 (.33-.75) | .37 (.22-.55) | .55 (.26-.80) | |
| 5-yr probability of DFS (95% CI) | .24 (.11-.45) | .55 (.26-.81) | .27 (.13-.47) | .55 (.21-.85) | |
| 3-yr probability of survival (95% CI) | .40 (.31-.50) | .67 (.45-.83) | .34 (.21-.49) | .43 (.20-.70) | |
| 5-yr probability of survival (95% CI) | .31 (.16-.52) | .48 (.24-.72) | .34 (.18-.54) | .43 (.16-.75) | |
| Ph chromosome: | |||||
| Negative | 35 | 11 | 24 | 7 | .003 |
| Positive6-150 | 34 (49%) | 2 (15%) | 7 (23%) | 0 (0%) |
The P values are based on 3 degrees of freedom tests.
Ph chromosome positive refers to patients that are BCR/ABL+ by molecular assay and/or have a t(9;22) cytogenetic karyotype.
Correlation of immunophenotype with cytogenetics.
Cytogenetic abnormalities were analyzed in 111 of the 250 patients with B-LIN or T-LIN immunophenotypes. The most common cytogenetic abnormalities seen (ie, 5 cases or more) and their corresponding lineage immunophenotype are described in Table7. The majority (36 of 38, 95%) of the cases with karyotypes of either t(4;11) or t(9;22) were associated with a B-LIN immunophenotype. These patients had shorter survival (P = .005) and DFS (P = .02) than B-LIN patients with other cytogenetic abnormalities or a normal karyotype. Of interest, two t(9;22) cases (not shown in Table 7) were found to have a mixed (B + T) lineage immunophenotype. The majority of patients had more than one karyotypic abnormality found. When studied only in those cases where they were expressed as the sole chromosomal abnormality, relatively specific surface antigen combinations were associated with the t(9;22) and t(4;11) karyotypes. Ph+ cases usually demonstrate CD19+ (11 of 12), CD10+ (9 of 12), CD34+ (12 of 12); these results are consistent with the previous finding that the B-LIN subgroup coexpressing CD10+/34+ was associated with a significantly higher than expected number of Ph+ cases (49%) compared with the three other subgroups. The majority of t(4;11) cases showed CD19+ (13 of 13) and CD34+ (9 of 13), but no CD10+ (0 of 13). The lack of CD10 expression in adult ALL patients with the t(4;11) karyotype has been independently noted by Ludwig et al from German multicenter trials.26
Clonal Cytogenetic Abnormalities and ALL Immunophenotype
| Abnormality . | B . | BMy . | T . | TMy . | B-LIN . | T-LIN . | Total . |
|---|---|---|---|---|---|---|---|
| No. of cases | 69 | 24 | 14 | 4 | 93 | 18 | 111 |
| t(9;22)7-150 | 13 | 10 | 0 | 0 | 23 | 0 | 23 |
| Any 11q23 | 14 | 1 | 0 | 0 | 15 | 0 | 15 |
| t(4;11) | 12 | 1 | 0 | 0 | 13 | 0 | 13 |
| +21 | 9 | 4 | 0 | 1 | 13 | 1 | 14 |
| +8 | 7 | 3 | 0 | 0 | 10 | 0 | 10 |
| 14q32 | 6 | 2 | 0 | 0 | 8 | 0 | 8 |
| −7 | 3 | 3 | 0 | 0 | 6 | 0 | 6 |
| del 9p | 4 | 0 | 1 | 0 | 4 | 1 | 5 |
| Other abnormality | 15 | 4 | 3 | 2 | 19 | 5 | 24 |
| Normal | 15 | 6 | 10 | 1 | 21 | 11 | 32 |
| Abnormality . | B . | BMy . | T . | TMy . | B-LIN . | T-LIN . | Total . |
|---|---|---|---|---|---|---|---|
| No. of cases | 69 | 24 | 14 | 4 | 93 | 18 | 111 |
| t(9;22)7-150 | 13 | 10 | 0 | 0 | 23 | 0 | 23 |
| Any 11q23 | 14 | 1 | 0 | 0 | 15 | 0 | 15 |
| t(4;11) | 12 | 1 | 0 | 0 | 13 | 0 | 13 |
| +21 | 9 | 4 | 0 | 1 | 13 | 1 | 14 |
| +8 | 7 | 3 | 0 | 0 | 10 | 0 | 10 |
| 14q32 | 6 | 2 | 0 | 0 | 8 | 0 | 8 |
| −7 | 3 | 3 | 0 | 0 | 6 | 0 | 6 |
| del 9p | 4 | 0 | 1 | 0 | 4 | 1 | 5 |
| Other abnormality | 15 | 4 | 3 | 2 | 19 | 5 | 24 |
| Normal | 15 | 6 | 10 | 1 | 21 | 11 | 32 |
B-LIN combines B and BMy cases, whereas T-LIN combines T and TMy cases. Many patients were found to have more than one karyotypic abnormality and are therefore included in more than one category. Patients with t(8;14) were not included in this study.
t(9;22) confirmed by cytogenetic analysis only. There were 4 B-LIN cases lacking cytogenetic evidence of t(9;22) that were BCR-ABL+. Additionally, 1 T-LIN case and 16 B-LIN cases were BCR-ABL+ but not evaluated cytogenetically.
Immunophenotype of ALL at relapse.
A total of 37 evaluable pairs of pretreatment and relapsed specimens have been analyzed by MFC. The immunophenotypes are shown in Table8. In 25 of 37 patients (68%), the same pretreatment immunophenotypic lineage markers were seen at time of relapse. In the remaining one third, the change at time of relapse consisted primarily of a minor shift with loss or gain of a myeloid marker. As the number of relapsed cases studied increases, future subset analysis may help in determining if any clinical or lab correlations are associated with specific immunological marker shifts. Because approximately two thirds of patients do not change their pretreatment immunophenotypic lineage, the possibility of evaluating minimal residual disease by MFC in serial samples during and following therapy remains a promising area of research.
Sequential Immunophenotyping Studies Before Treatment and at Relapse of ALL in 37 Patients
| Pretreatment . | Relapse . | No. of Cases (n = 37) . | Average No. of Markers . | |
|---|---|---|---|---|
| Gained . | Lost . | |||
| B | B | 15 | 2 | 1 |
| B + My | B + My | 8 | 2 | 1 |
| T | T | 2 | 0 | 2 |
| B | B + My | 6 | 2 | 0 |
| B + My | B | 2 | 0 | 1 |
| T | CD34, CD45 dim | 1 | 0 | 2 |
| T | Normal | 1 | 0 | 7 |
| B + T | B | 1 | 2 | 3 |
| B + T + My | T + My | 1 | 1 | 1 |
| Pretreatment . | Relapse . | No. of Cases (n = 37) . | Average No. of Markers . | |
|---|---|---|---|---|
| Gained . | Lost . | |||
| B | B | 15 | 2 | 1 |
| B + My | B + My | 8 | 2 | 1 |
| T | T | 2 | 0 | 2 |
| B | B + My | 6 | 2 | 0 |
| B + My | B | 2 | 0 | 1 |
| T | CD34, CD45 dim | 1 | 0 | 2 |
| T | Normal | 1 | 0 | 7 |
| B + T | B | 1 | 2 | 3 |
| B + T + My | T + My | 1 | 1 | 1 |
DISCUSSION
Adult ALL is a heterogeneous disease with leukemic blasts showing various immunophenotypes and variable responsiveness to systemic therapies. Karyotype and molecular genetics, in addition to surface immunophenotype, have refined the diagnosis and prognosis of adult ALL beyond the information available from physical examination, peripheral blood counts, serum biochemical profiles, and bone marrow morphology. Immunophenotyping is essential for confirmation of the diagnosis of ALL. Immunophenotyping permits recognition of misdiagnosed cases of AML (M0), which typically may occur in 10% to 15% of cases of ALL being evaluated by morphology and standard immunohistochemical stains alone (C. Stewart, personal communication, 1998). Immunophenotyping also provides a biologic definition of the disease that may be useful in assigning prognosis and choosing appropriate therapy. In recent years the use of uniform criteria (including age, WBC count at presentation, DNA index, early response to therapy, cytogenetics, CNS status, and immunophenotype) to assign risk-based therapy for patients with pediatric ALL has been advocated.27
In the current study, a total of 259 evaluable adult ALL patients treated on similar intensive CALGB front-line treatment protocols had immunophenotype classifications prospectively assigned by MFC performed in a central lab. The presence of myeloid antigen (CD13 or CD33) expression was found in earlier CALGB treatment studies (8011, 8411, 8513) to predict a lower CR rate and shorter survival.8Studies by other investigators treating adult ALL patients with “standard” therapy also described a negative prognostic impact associated with myeloid antigen coexpression.28-30 Since the advent of more intensive chemotherapy programs for adult ALL patients on CALGB protocols (beginning with CALGB 8811 in which cyclophosphamide and more intensive early L-asparaginase was incorporated into the backbone of daunorubicin, vincristine and prednisone together with CNS prophylaxis), along with the exclusion of cases of AML (M0) in these latter CALGB trials, no significant differences in response rates, remission duration, or survival have been seen in patients coexpressing CD13 and/or CD33. Thus, a once-poor prognostic indicator appears to have been eliminated by better therapy. However, the potential contribution that immunophenotyping by MFC has had in excluding poor-prognostic AML (M0) cases from the current analyses of adult ALL cannot be excluded. In general, the clinical course of these patients appears to be determined by their primary lymphoid lineage (ie, B or T). For this reason, B/BMy (B-LIN) and T/TMy (T-LIN) groups are paired together, respectively, for the current analysis.
Furthermore, the use of a more intensive CALGB remission induction program has led to improved overall DFS of T-LIN compared with B-LIN subgroups. Current data also show that an increased number of T-cell antigens expressed by T-LIN ALL (ie, 6 or more compared to 3 or less T-cell markers) correlates with improved DFS and survival in the T-LIN group. The improved survival seen between these groups may be explained in part to a higher percentage of expression of the “good risk” individual T-cell markers of CD1, CD2, and CD4 in the 6-7 marker group compared with the 1-3 marker group.
A valuable outcome from the current study is the possibility for future reduction in the number of antibodies required to evaluate adult ALL. For example, because expression of myeloid antigens has no significance in outcome, their continued use may be less important, except for controlling for myeloid cell contamination. In fact, our data suggest that the deletion of CD33 from future proposed antibody panels is warranted. Of 79 patients coexpressing myeloid antigens (Table 2), 8 (10%) expressed CD33 and 76 (96%) expressed CD13, while 5 patients expressed both. Thus, only 3 of the 8 cases of CD33+ adult ALL did not also coexpress CD13 (ie, they were CD33+, CD13−). Furthermore, we have deleted antibodies against those antigens with low expression and thus not potentially useful targets for future specific MoAb immunotherapy for ALL; these include antibodies against CD14, CD11c, CD33, IgD, IgM, kappa, and lambda. Morphology is the most important factor used in making a diagnosis of ALL-L3. However, surface Ig staining by immunohistochemistry is found to be positive in the majority of ALL-L3 cases and, therefore, should continue to be included as a routine assay in the initial evaluation of adult ALL.
Future studies can use four-color (instead of three-color) MFC to decrease the redundant use of the same antibody in some panels by excluding antibodies with low prognostic value and concurrently switching to four-color MFC; one could reduce the number of antibodies used from 30 to 16, and the number of tubes to process from 10 to 4, as summarized in Table 9. This could not only significantly reduce costs, but also streamline future MFC immunophenotyping of adult ALL.
Useful Combination of Antibodies for Immunophenotyping ALL
| Purpose . | Label-CD Cluster . |
|---|---|
| By plotting side scatter (SSC)v CD45, malignant T and B cells can often be resolved from normal cells by their degree of CD45 expression. B cells are often HLADR+, but not T cells. CD20 expression can aid in determining the maturity of B-cell malignancy while CD5 expression in the absence of CD20 expression is associated with T-cells. | FITC-CD20 PE-CD5 PP-HLADR APC-CD45 |
| Designed to evaluate myeloid CD13+ coexpression on B-lineage (CD22) v T-lineage CD2+/CD7+ malignancy. T-lineage malignancy often exhibits over or underexpression of CD2 and/or CD7. | FITC-CD7 PE-CD22 PP-CD2 APC-CD13 |
| Designed to evaluate CD19+ subgroups of B-lineage ALL with respect to CD10, CD38, and CD34 expression. | FITC-CD38 PE-CD19 PP-CD10 APC-CD34 |
| This combination is designed to evaluate subgroups of T-lineage ALL with respect to CD1/CD4/CD8 and CD3 expression. Overexpression or underexpression of one or more of these markers is often found in T-lineage malignancy. | FITC-CD1 PE-CD4 PP-CD8 APC-CD3 |
| Purpose . | Label-CD Cluster . |
|---|---|
| By plotting side scatter (SSC)v CD45, malignant T and B cells can often be resolved from normal cells by their degree of CD45 expression. B cells are often HLADR+, but not T cells. CD20 expression can aid in determining the maturity of B-cell malignancy while CD5 expression in the absence of CD20 expression is associated with T-cells. | FITC-CD20 PE-CD5 PP-HLADR APC-CD45 |
| Designed to evaluate myeloid CD13+ coexpression on B-lineage (CD22) v T-lineage CD2+/CD7+ malignancy. T-lineage malignancy often exhibits over or underexpression of CD2 and/or CD7. | FITC-CD7 PE-CD22 PP-CD2 APC-CD13 |
| Designed to evaluate CD19+ subgroups of B-lineage ALL with respect to CD10, CD38, and CD34 expression. | FITC-CD38 PE-CD19 PP-CD10 APC-CD34 |
| This combination is designed to evaluate subgroups of T-lineage ALL with respect to CD1/CD4/CD8 and CD3 expression. Overexpression or underexpression of one or more of these markers is often found in T-lineage malignancy. | FITC-CD1 PE-CD4 PP-CD8 APC-CD3 |
Specific chromosomal abnormalities such as t(9;22) and t(4;11), which are known to carry a poor prognosis, are found in only a subset of adult ALL patients. The B-LIN subgroup of CD10+/CD34+ had a statistically higher than expected number of Ph+ cases than the three other CD19+ B-LIN subgroups. The CD19+/CD34+/CD10+ immunophenotype is especially helpful in those patients in whom karyotyping is not possible since it identified a group of patients where molecular studies (PCR or fluorescence in situ hybridization) for BCR-ABL must be done.
Two recent CALGB treatment protocols for ALL have used prognostic factors validated by ongoing correlative studies to target therapy for poor-prognostic groups. Study 9251 was devised to improve the outcome in patients with Burkitt-type, surface Ig+ ALL (FAB-L3).31 This study requires immunophenotype as an entry criterion and represents one attempt to tailor therapy based on immunophenotype. In some centers that treat childhood ALL, there are separate study protocols for B-precursor, B-cell, and T-cell ALL, and additional provisions for certain high-risk genotypes within the B-precursor immunophenotype.27,32 A recent CALGB attempt at lineage-specific therapy of adult ALL (CALGB study 9311: “A Phase II Trial of Lineage-Specific Consolidation Therapy for Adult ALL: Anti-B4–Blocked Ricin for B-Lineage ALL and High-Dose Cytarabine for Non–B-lineage ALL”) sought to explore the impact of these two different intensification regimens in patients stratified by immunophenotypic lineage assignments.12 The anti-B4–blocked ricin immunotoxin targets the CD19 surface antigen found on the vast majority of B-LIN adult ALL. Unfortunately, anti-B4–blocked ricin immunotoxin did not show a significant impact in eradicating minimal residual disease as measured by qualitative PCR assays for BCR-ABL and semiquantitative PCR assays for Ig heavy-chain gene rearrangements. However, other MoAbs with possibly enhanced antitumor activity that are currently being evaluated in clinical trials may be incorporated into consolidative therapy in future adult ALL regimens. Identification of common immunologic phenotypes will permit the choice of optimal combinations of MoAb preparations that may find application in systemic passive immunotherapy. Similarly, knowledge of the immunophenotypes in adult ALL may guide the selection of the most appropriate antibodies to purge bone marrow specimens of residual leukemic cells as part of autologous bone marrow transplantation protocols.
In conclusion, our study of MFC in a large number of adult ALL patients treated on dose-intensive CALGB treatment protocols has resulted in several important observations. These include, first, myeloid antigen (CD13 and/or CD33) coexpression is not associated with a worse prognosis in adult ALL. Second, compared with the B-LIN group, intensively treated T-LIN patients have longer overall DFS and survival. Third, an increased number of T-cell antigens expressed in T-LIN adult ALL (ie, 6 or more compared to 3 or less T-cell markers) correlates with improved CR rate, DFS, and survival. Fourth, CD34+/CD10+ coexpression in the B-LIN (CD19+) group shows a higher incidence of Ph-positivity than other CD34/CD10 subgroups. In contrast, adult B-LIN ALL patients with a t(4;11) karyotype have a CD34+/CD10−phenotype. Lastly, knowledge gained from this study will permit reduction in the number of antigens required for future immunophenotyping. This, along with the institution of four-color MFC, can translate into cost savings without loss of valuable information. Ultimately, the knowledge gained from this study, added to other known prognostic variables, may someday lead to individualized risk-based therapy of adult ALL.
ACKNOWLEDGMENT
The following institutions participated in the study: University of Alabama, Birmingham, AL: Robert Diasio, MD, supported by CA47545; Bowman Gray School of Medicine, Winston-Salem, NC: M. Robert Cooper, MD, supported by CA03927; University of North Carolina, Chapel Hill, NC: Thomas Shea, MD, supported by CA47559; University of Chicago Medical Center, Chicago, IL: Nicholas J. Vogelzang, MD, supported by CA41287; Dana-Farber Cancer Institute, Boston, MA: George P. Canellos, MD, supported by CA32291; Dartmouth-Hitchcock Medical Center, Hanover, NH: L. Herbert Maurer, MD, supported by CA04326; Duke University Medical Center, Durham, NC: Jeffrey Crawford, MD, supported by CA47577; University of Iowa Hospitals, Iowa City, IA: Gerald Clamon, MD, supported by CA47642; Long Island Jewish Medical Center, New Hyde Park, NY: Marc Citron, MD, supported by CA11028; University of Maryland Cancer Center, Baltimore, MD: Ernest Borden, MD, supported by CA31983; University of Massachusetts Medical Center, Worcester, MA: F. Marc Stewart, MD, supported by CA37135; Massachusetts General Hospital, Boston, MA: Michael Grossbard, MD, supported by CA12449; McGill Cancer Center, Montreal, Quebec, Canada: Brian Leyland-Jones, MD, supported by CA31809; Medical University of South Carolina, Charleston, SC: Mark Green, MD, supported by CA03927; University of Minnesota, Minneapolis, MN: Bruce Peterson, MD, supported by CA16450; University of Missouri, Ellis Fischel Cancer Center, Columbia, MO: Michael C. Perry, MD, supported by CA12046; Mount Sinai Hospital, New York, NY: James Holland, MD, supported by CA04457; New York Hospital, Cornell Medical Center, New York, NY: Ted Szatrowski, MD, supported by CA07968; North Shore University Hospital, Manhasset, NY: Daniel R. Budman, MD, supported by CA35279; Rhode Island Hospital, Providence, RI: Louis A. Leone, MD, supported by CA08025; Roswell Park Memorial Institute, Buffalo, NY: Ellis Levine, MD; supported by CA02599; SUNY Health Science Center at Syracuse, Syracuse, NY: Stephen Graziano, MD, supported by CA21060; University of Tennessee, Memphis, TN: Alvin Mauer, MD, supported by CA47555; University of California at San Diego, San Diego, CA: Stephen Seagren, MD, supported by CA11789; University of California at San Francisco, San Francisco, CA: Alan Venook, MD, supported by CA60138; Washington University, Barnes Hospital, St Louis, MO: Daniel Ihde, MD, supported by CA47546; Walter Reed Army Medical Center, Washington, DC: Nancy Dawson, MD, supported by CA26806.
The research for CALGB 8364 was supported, in part, by grants from the National Cancer Institute (CA31946) to the Cancer and Leukemia Group B (Richard L. Schilsky, Chairman). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Myron S. Czuczman, MD, Roswell Park Cancer Institute, Elm and Carlton St, Buffalo, NY 14263.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal