Abstract
Vascular endothelial cell growth factor (VEGF) is a major regulator of angiogenesis. We report here that treatment of endothelial cells with VEGF leads to upregulation of tissue factor mRNA and protein expression on the cell surface. Reporter gene studies show that transcriptional activation of the tissue factor gene by VEGF is mediated by a GC-rich promoter element containing overlapping binding sites for Sp1 and EGR-1. As shown by immunofluorescence and electrophoretic mobility shift assays, upon VEGF treatment EGR-1 rapidly accumulates in the nucleus and binds to its respective recognition site in the tissue factor promoter. Sp1 occupies this element in unstimulated cells and seems to be partially displaced by increasing amounts of EGR-1. Transfection of endothelial cells with an EGR-1 expression plasmid mimics the upregulation of tissue factor transcription observed after VEGF treatment. In contrast, NFκB, the major transcription factor involved in tissue factor upregulation by inflammatory stimuli, is not activated by VEGF. These data show that VEGF induces a response in endothelial cells largely distinct from inflammatory stimuli, and suggest that EGR-1 is a major mediator of the activation of the tissue factor and possibly other VEGF-responsive genes.
TISSUE FACTOR (TF) is a member of the cytokine-receptor superfamily. It functions as the high-affinity receptor/cofactor for plasma factor VII/VIIa1,2 and is the primary cellular initiator of blood coagulation. Consistent with a protective role in the hemostatic response, TF is expressed constitutively in several extravascular cell types surrounding blood vessels and at boundaries of organs, but is normally absent in vascular endothelial cells. However, it is rapidly induced in response to inflammatory stimuli including lipopolysaccharide (LPS), tumor necrosis factor-α (TNF-α), and interleukin-1 (IL-1).3 4
On the other hand, the constitutive expression of TF was shown to be a characteristic feature of various neoplastic cells. Data obtained from experimental tumor models and clinical studies suggest that TF may facilitate tumor growth in vivo by enhancing angiogenesis. Indeed, tumor cells transfected to overexpress TF established larger and more vascularized tumors.5 Moreover, TF has been reported to be detected on vascular endothelial cells and tumor cells within human breast cancer tissue, but is absent on vessels of benign fibrocystic breast disease.6 Additional evidence indicates that the same tumor cells that express TF also produce vascular endothelial cell growth factor (VEGF), and that TF may regulate VEGF production by those cells.7 Taken together, these data are consistent with a highly complex interaction between tumor cells and endothelial cells in the tumor microenviroment, and suggest that tumor cells activate nearby endothelial cells and regulate blood vessel growth in vivo. It is not clear whether vessel growth and expression of TF by endothelial cells is mediated by the same angiogenic protein, eg, VEGF, or by some other cytokine produced by the tumor. However, VEGF is a likely candidate because more vascularized, TF-positive tumors secrete increasing amounts of VEGF,5,7 and VEGF from murine fibrosarcoma induces expression of TF in endothelial cells.8
To date, five angiogenic and endothelial cell–specific growth factors of the VEGF family have been isolated: VEGF-A, VEGF-B, VEGF-C, VEGF-D, and placental growth factor, PlGF.9,10 Three high-affinity tyrosine kinases receptors for VEGF-A, VEGF-C, and PlGF have been identified on endothelial cells: VEGF receptor-1, encoded by the flt-1 gene; VEGF receptor-2, encoded by the flk-1/KDR gene; and VEGF receptor-3, encoded by the flt-4 gene. Some intracellular signaling pathways triggered by these receptors have been described recently.11-13 However, the transcription factors involved in the angiogenic response to VEGF have not yet been defined.
TF gene expression has been characterized for different cell types and appears to be controlled by transcription factors that are constitutively active as well as factors induced by external signals. In case of inflammatory stimuli, such as LPS, TNF-α or IL-1β, induction of the TF promoter in human monocytic cells and endothelial cells is mediated by a single NFκB site binding c-Rel/p65 heterodimers in combination with two AP-1 sites.3,4DNA-binding motifs for NFκB were found in the promoters of more than 50 genes that are known to be activated upon inflammation.14 NFκB is kept in a premade inactive form in the cytoplasm and rapidly translocates to the nucleus after stimulation. In contrast, serum and phorbol ester induction of the TF gene in epithelial cells is controled by a GC-rich promoter region containing overlapping Sp1/EGR-1 sites.15 Recently, the same region was shown to be responsible for increased expression of TF in hypoxic mononuclear phagocytes and epithelial cells.16Sp1 is constitutively expressed and mediates a basal promoter activity. EGR-1, also known as Zif268, NGFI-A, Krox 24, or TIS8,17-19is expressed with the kinetics of an immediate-early transcription factor. The egr-1 gene can be induced by diverse signals that initiate growth and differentiation without requirements for de novo protein synthesis.20,21 Putative nucleotide recognition elements for EGR-1, which usually overlap with Sp1 binding sites, appear in the promoters of a number of pathologically important genes, including transforming growth factor-β1 (TGF-β1),22TF,3 urokinase-type plasminogen activator,23PDGF-A and PDGF-B,24 as well as in the EGR-1 promoter itself.25
We have now investigated the mechanism(s) responsible for TF upregulation by VEGF. A VEGF-responsive region in the TF promoter has been characterized. This region is distinct from the promoter elements involved in TNF-α– or LPS–induced TF gene expression. The transcription factor EGR-1 has been identified as a major component of a VEGF-inducible complex binding to this region. Furthermore, EGR-1 overexpression has been found to mimic TF gene activation by VEGF. The data presented define for the first time a transcription factor induced by VEGF that may be generally important for genes upregulated by this cytokine.
MATERIALS AND METHODS
Cell culture.
Human umbilical vein endothelial cells (HUVEC) and human skin microvascular endothelial cells (HSMEC) were prepared as previously described.26 27 HUVEC and HSMEC were cultured at 37°C and 5% CO2 in medium 199 containing 20% iron-supplemented bovine calf serum (SCS) (HyClone, Logan, UT), 1 U/mL heparin, 50 μg/mL ECGS, 2 mmol/L glutamine, 100 U/mL penicillin, and 0.1 mg/mL streptomycin. Cells were used for experiments up to passage number 5. Recombinant human VEGF was obtained from PromoCell (Heidelberg, Germany) or was a gift of Dr Matthias Clauss (Max-Planck-Institut für Physiologische und Klinische Forschung, Bad Nauheim, Germany). Induction of cells was performed by addition of VEGF165 or VEGF121 at 1.25 nmol/L and TNF-α (Genzyme Inc, Cambridge, MA) at 100 U/mL to confluent cells for 4 hours (clotting assay) or 6 hours (reporter gene assay).
Clotting assay.
Cells were seeded in 6-well plates at 80% to 90% confluency and grown overnight. Cells were scraped from the plates and analyzed for TF activity as described.4,8 Briefly, after induction for 4 hours with VEGF or TNF-α, cells were washed twice and then scraped in 1mL clotting buffer (12 mmol/L sodium acetate, 7 mmol/L diethylbarbitate, and 130 mmol/L sodium chloride; pH 7.4). One hundred microliters of resuspended cells were mixed with 100 μL of citrated plasma (Sigma, St Louis, MO), and clotting times were measured after recalcification with 100 μL of 20 mmol/L CaCl2 solution at 37°C. TF equivalents were determined by using a standard curve obtained from rabbit brain thromboplastin (Sigma). For studies with neutralizing antibodies (American Diagnostica Inc, Greenwich, CT; no. 4508) cells were incubated for 30 minutes before starting the clotting assay. Mithramycin A, a DNA-binding drug known to preferentially bind to GC-rich DNA sequences,28-30 was used to estimate the functional importance of a GC-rich region within the TF promoter. For this purpose, cells were preincubated with medium containing mithramycin (Sigma) for 30 minutes before addition of VEGF or TNF-α.
Northern blot analysis.
Total cellular RNA was isolated from confluent cell cultures by scraping cells in TRIzol reagent (GIBCO, Grand Island, NY), extracting DNA and proteins with chloroform, and precipitating the RNA by isopropanol. Twenty micrograms of total RNA was fractioned on a 1.3% agarose-6.4% formaldehyde gel, transferred to a nylon membrane (Amersham Life Science Inc, Little Chalfont, UK), and covalently linked by UV irradiation.
Membranes were prehybridized in 5X NET (20X NET: 3 mol/L sodium chloride, 300 mmol/L TRIS/chloride, 20 mmol/L EDTA, pH 7.5), 5X Denhardt’s solution, 0.5% sodium dodecyl sulfate (SDS), and 100 μg/mL salmon sperm DNA at 56°C for 4 hours. Then α-32P-radiolabeled probe equal to 106 cpm/mL was added and hybridization was allowed to proceed overnight. A 641-bp fragment of the human TF cDNA was used as a specific probe. The probe was labeled by random priming using the Prime-it II Random Primer Kit from Stratagene Cloning Systems (La Jolla, CA). The membrane was washed at 56°C twice with 2X NET, 0.2% SDS for 30 minutes, once with 1X NET, 0.2% SDS, and again with 2X NET, 0.2% SDS. Bound radioactivity was visualized by exposure to XAR-5 films at −70°C using intensifying screens (Eastman Kodak Co, Rochester, NY).
Electrophoretic mobility shift assays.
Nuclear extracts from confluent endothelial cell cultures were prepared as previously described,4 31 except that a modified buffer C was used during nuclear extraction [20 mmol/L HEPES-KOH, pH 7.9, 420 mmol/L NaCl, 400 mmol/L (NH4)2SO4, 1.5 mmol/L MgCl2, 0.2 mmol/L EDTA, 0.5 mmol/L dithiothreitol (DTT), 25% glycerol]. Protease inhibitors (0.5 mmol/L phenylmethylsulfonyl fluoride, 25 μmol/L Na-p-tosyl-L-lysine chloromethyl ketone, 50 μmol/L N-tosyl-l-phenylalanine chloromethyl ketone) were added at all steps during extract preparation. For the electrophoretic mobility shift assay, 3 μg of nuclear proteins was incubated with radioactively labeled oligonucleotide equal to 105 cpm in binding buffer (20 mmol/L HEPES-KOH, pH 7.9, 1 mmol/L EDTA, 5 mmol/L MgCl2, 50 mmol/L KCl, 1 mmol/L DTT, 10% glycerol) and 2 μg poly (dI/dC) (Boehringer Mannheim, Mannheim, Germany), giving a total volume of 20 μL, for 30 minutes at room temperature. The double-stranded synthetic oligonucleotides were radioactively labeled by filling in the overhangs with Klenow enzyme in the presence of (α-32P)dATP and subsequently purified over a 10% polyacrylamide gel. The sequences of the probes used were as follows: hTF AP-1, 5′-aattGCGGTTGAATCACTGGGGTGAGTCATCCCTTGCAGG-3′; hTF NFκB, 5′-aattCCCGGAGTTTCCTACCGG-3′; hIg NFκB, 5′-aattCAGAGGGGGATTTCCCAGAG-3′.
The oligonucleotides spanning the GC-rich region of the human TF promoter are listed in Figure 5. Where indicated, 2 μg of polyclonal rabbit antibodies, all obtained from Santa Cruz Biotechnology (Santa Cruz, CA) [Egr-1: sc-189x, AP-2: sc-184x, AP-1 (c-Fos): sc-52x], were added to the binding reactions for 30 minutes at 4°C before the addition of the radioactive probe. Protein-DNA complexes were resolved by native 5.5% polyacrylamide gel electrophoresis in 0.5X TBE.
Transfection of HUVEC.
A fragment of the porcine TF promoter (−330 to +118) designated as wild-type promoter was cloned into a luciferase expression vector, pUBT-luc.4,32 The NFκB, AP-1, and SP1/Egr-1 deletion fragments were obtained from polymerase chain reaction (PCR) fragments of the −330/+118 wild-type TF promoter, resulting in the substitution of the sequences from position −161 to −137, from −209 to −156 or from −134 to −43 with anXbaI linker sequence, respectively.4 The minimal promoter construct was created from an XbaI/HindIII restriction enzyme fragment of the ΔNFκB construct containing the sequences from −160 to +118 of the TF promoter, which was then ligated into a luciferase expression vector pUBT-luc. All constructs were sequenced to show the fidelity of PCR and subcloning procedures. One of the −330 to +118-bp constructs was found to contain a spontaneous point mutation (G → A) at the position −51 before the consensus EGR-1 site −51(G/A)GCGGGGGCG −42 and was included in the experiments. The expression vectors pCB.EGR-1 (containing the full-length murine EGR-1 cDNA coding sequence) and the pCB.EGR-1Δ331-374 (lacking the first and part of the second EGR-1 zinc finger domains) were generously provided by Dr Vikas P. Sukhatme (Harvard Medical School, Boston, MA).33
Twenty-four hours before transfection, HUVEC were seeded in 6-well tissue-culture plates to reach 80% to 90% confluency the next morning. Transient transfections were performed by using the Lipofectamine reagent (GIBCO) according to the protocol. Cells were incubated with transfection mixture containing 3 μg DNA (including a cytomegalovirus [CMV]-β-gal construct as internal control) and 8 μL Lipofectamine in a total volume of 1 mL medium 199 per well for 2 hours and 10 minutes. Induction by VEGF was performed the next day for 6 hours. Luciferase and β-gal assays were performed with cellular lysates of transfected cells as previously described.34 35 All experimental values were determined from duplicate wells, and several identical experiments were performed with plasmids from different DNA preparations to assess the influence of DNA quality. Relative promoter activities were calculated relative to the wild-type promoter activity (100%), and results from three independent experiments were presented as means ± SD.
Cell enzyme-linked immunosorbent assay (ELISA).
Cells were seeded into a 96-well plate to reach confluency the next day. Cells were treated with TNF-α (100 U/mL) or VEGF (1.25 nmol/L) for various periods of time before fixation with 0.1% glutaraldehyde in phosphate-buffered saline (PBS). The antibodies specific for VCAM-1, ICAM-1, and E-selectin (R&D Systems, Minneapolis, MN; αVCAM-1: BBA-5, αICAM-1: BBA-3, αELAM-1: BBA-1) were diluted in PBS, 0.1% Tween 20, 1% skimmed milk, and incubated with the fixed cells for 1 hour at 37°C. Goat anti-mouse IgG conjugated to peroxydase (Amersham) was used at a dilution of 1:2,000 and incubated for 1 hour at 37°C. For development, o-phenylene diamine (Sigma) was added and incubated for 10 to 20 minutes in the dark at room temperature. The reaction was stopped by 3 mol/L H2SO4 and the optical density (OD) was measured at 492 nm.
Immunofluorescence.
Immunofluorescence assay was performed mainly as previously described.36 Briefly, cells were grown in LabTek tissue-culture chamber slides (Nunc, Inc, Naperville, IL) for at least 24 hours before fixation. After appropriate stimulation, cells were washed twice with PBS, fixed for 10 minutes at room temperature with 3.7% formaldehyde, 2% sucrose in PBS, and permeabilized for 5 minutes with 0.5% Triton X-100 (Serva, Heidelberg, Germany) in PBS. Primary antibodies (polyclonal rabbit IgG from Santa Cruz Biotechnology; Egr-1: sc-189x and sc-110x; SP1: sc-59x; p65: sc-109x) were diluted in PBS, 1% bovine serum albumin (BSA) and incubated with the cells for 1 hour at room temperature. To confirm the specificity of anti–EGR-1 antibodies, the immunizing peptide (sc-189) was preincubated with antibodies for 15 minutes before addition to the cells. Cells were washed in PBS and incubated with rhodamine- or fluorescein isothiocyanate (FITC)-labeled goat anti-rabbit IgG (Accurate Scientific, Westbury, NY) for 1 hour at room temperature. To visualize cell nuclei, a fluorescent groove-binding probe for DNA (4’,6-Diamidino-2-Phenylindole, DAPI; Sigma) was added to the secondary antibody solution at 100 ng/mL. Slides were washed in PBS and mounted with mounting fluid (Difco, Detroit, MI). The immunofluorescence results were analyzed with a Bio-Rad MRC 600 confocal laser scanning microscope (Bio-Rad, Hercules, CA).
Statistical analysis.
The results obtained were analyzed by one-way analysis of variance and the Student’s paired t-test.
RESULTS
VEGF upregulates TF activity and TF mRNA in endothelial cells.
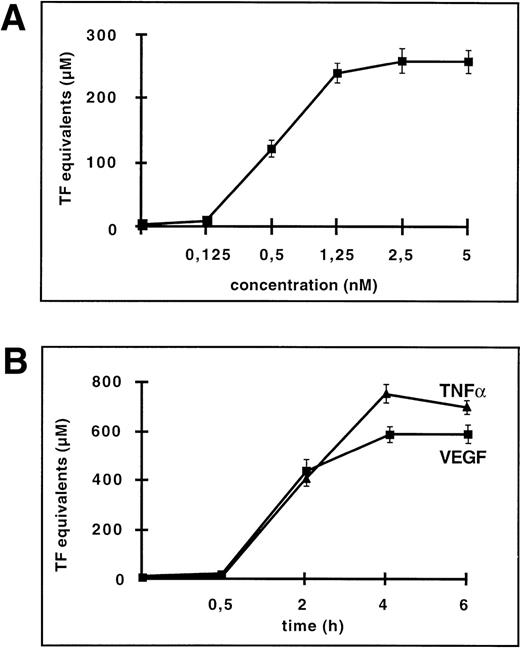
VEGF has been shown to modulate procoagulant activity on the surface of human monocytes and endothelial cells.8 However, the mechanisms of VEGF action have not been defined. Therefore, we tested the effect of VEGF on TF expression in comparison to a known inducer of TF, TNF-α. TF activity, as monitored by a one-stage clotting assay, was very low in unstimulated HUVEC, and VEGF165 caused a dose-dependent increase in TF activity reaching plateau values at 1.25 nmol/L (Fig 1A). Accordingly, all of the following experiments were performed in the presence of this concentration of VEGF165. Depending on the experiment, TF activity was upregulated from 10- to 80-fold, which is comparable to the induction observed with TNF-α (100 U/mL). HUVEC induced by VEGF165 or TNF-α reached maximal TF activity at 4 hours (Fig 1B). To confirm that the procoagulant activity detected in the clotting assay is indeed mediated by TF, anti-TF monoclonal antibodies were preincubated with the cells before the clotting reaction. These antibodies completely abolished VEGF-induced procoagulant activity (data not shown).
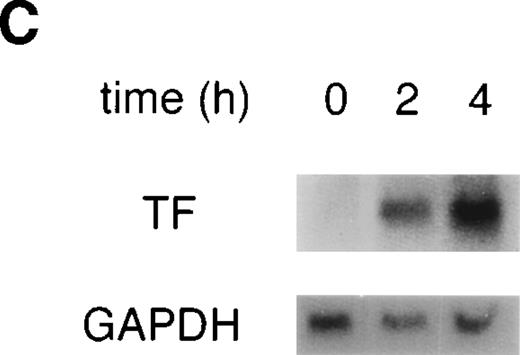
VEGF upregulates TF activity and mRNA in HUVEC. (A) VEGF induces TF activity in a dose-dependent manner. TF activity equivalents were determined in a one-stage clotting assay. Incubation of cells with increasing concentrations of VEGF in the range of 0.125 to 5 nmol/L for 4 hours. (B) Incubation with 1.25 nmol/L VEGF for 0.5 , 1, 2, 4, and 6 hours. For comparison, kinetics of TF induction by TNF- (100 U/mL) is shown. Each value is the mean ± SD of triplicate in one experiment representative of five performed with similar results. (C) VEGF upregulates TF mRNA in HUVEC: Northern blot analysis of total RNA (20 μg) extracted from unstimulated cells and cells treated with VEGF for 2 and 4 hours. This experiment is representative of two experiments done with similar results.
VEGF upregulates TF activity and mRNA in HUVEC. (A) VEGF induces TF activity in a dose-dependent manner. TF activity equivalents were determined in a one-stage clotting assay. Incubation of cells with increasing concentrations of VEGF in the range of 0.125 to 5 nmol/L for 4 hours. (B) Incubation with 1.25 nmol/L VEGF for 0.5 , 1, 2, 4, and 6 hours. For comparison, kinetics of TF induction by TNF- (100 U/mL) is shown. Each value is the mean ± SD of triplicate in one experiment representative of five performed with similar results. (C) VEGF upregulates TF mRNA in HUVEC: Northern blot analysis of total RNA (20 μg) extracted from unstimulated cells and cells treated with VEGF for 2 and 4 hours. This experiment is representative of two experiments done with similar results.
It is known that the human VEGF165 isoform binds to both VEGF receptor-1 (Flt-1) and VEGF receptor-2 (Flk-1/KDR), whereas VEGF121 interacts selectively only with Flk-1/KDR.10,37 Because HUVEC express both types of VEGF receptors,13 38 we investigated whether both VEGF isoforms would induce TF. Treatment of endothelial cells with VEGF121 or VEGF165 at 1.25 nmol/L resulted in comparable TF upregulation (data not shown). Thus, VEGF binding to the Flk-1/KDR receptor seems to be sufficient to mediate TF activation. In all of the following experiments VEGF165 was used.
To evaluate whether the upregulation observed on the protein level is based on induction on the mRNA level, total RNA was isolated from HUVEC exposed to 1.25 nmol/L VEGF for 2 and 4 hours, and TF mRNA levels were determined by Northern blotting (Fig 1C). TF mRNA was not detectable in unstimulated HUVEC and was strongly upregulated by VEGF treatment at both time-points.
Furthermore, to show that the effect of VEGF on endothelial TF expression is not restricted to umbilical-vein–derived endothelial cells, we tested HSMEC after treatment with increasing concentrations of VEGF. A similar dose-response curve and kinetics of TF induction were detected by clotting assay (data not shown). Thus, our data clearly demonstrate that TF is generally induced on vascular endothelial cells by the angiogenic factor VEGF to an extent comparable to the inflammatory stimulus TNF-α.
VEGF does not upregulate adhesion molecule expression on the surface of endothelial cells.
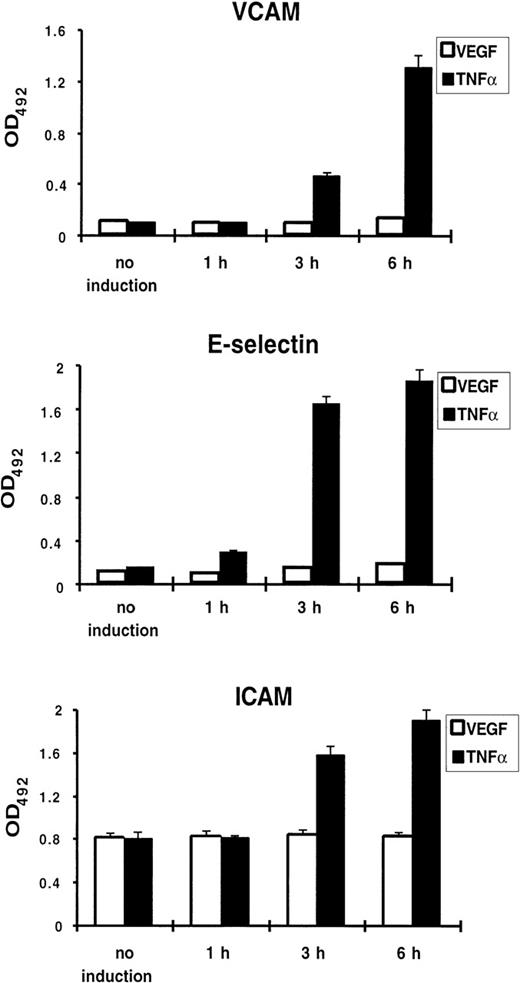
Because TNF-α treatment of endothelial cells results in upregulation of a number of genes involved in the inflammatory response, including the endothelial-leukocyte adhesion molecules VCAM-1, E-selectin, and ICAM-1, we tested whether VEGF induces expression of these proteins. Using a cell ELISA, we failed to detect any increase of VCAM-1, E-selectin, or ICAM-1 expression on the surface of VEGF-treated HUVEC, while TNF-α used in parallel caused a strong upregulation (Fig 2).
VEGF does not upregulate adhesion molecule expression on the surface of endothelial cells. Cell ELISA was performed with anti–VCAM-1, anti–E-selectin, and anti–ICAM-1 antibodies. HUVEC were treated by TNF- (100 U/mL) or VEGF (1.25 nmol/L) for 1, 3, and 6 hours. Each value is the mean ± SD of triplicate in one experiment representative of three performed with similar results.
VEGF does not upregulate adhesion molecule expression on the surface of endothelial cells. Cell ELISA was performed with anti–VCAM-1, anti–E-selectin, and anti–ICAM-1 antibodies. HUVEC were treated by TNF- (100 U/mL) or VEGF (1.25 nmol/L) for 1, 3, and 6 hours. Each value is the mean ± SD of triplicate in one experiment representative of three performed with similar results.
NFκB is not involved in VEGF-induced TF upregulation.
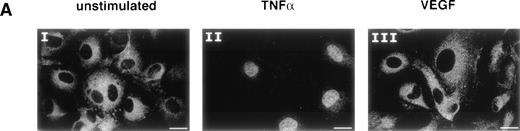
Mackman et al39 and our group4 have previously analyzed the regulation of the TF gene by inflammatory mediators. These studies showed that the NFκB-like site within the TF promoter is required for transcriptional upregulation during the inflammatory response. Therefore, we investigated whether NFκB would play an important role in the induction of the TF gene by VEGF. The observation that VEGF does not induce adhesion molecule expression already suggested that NFκB may not be involved, because NFκB activation is known to be a major trigger of adhesion molecule induction. By using immunofluorescence staining with anti-p65 antibodies, we first analyzed whether VEGF induces NFκB translocation from the cytoplasm into the nucleus. Unstimulated HUVEC showed a clear cytoplasmic localization of the p65 subunit of NFκB (Fig 3A, I). One-hour treatment with TNF-α (100 U/mL) resulted in translocation of p65 into the nucleus (Fig 3A, II), while VEGF treatment (1.25 nmol/L) from 10 minutes to 4 hours had no detectable effect (Fig 3A, III).
NFκB is not induced by VEGF. (A) Immunofluorescence staining of p65 (NFκB subunit) in unstimulated (I), TNF-–treated (II), or VEGF-treated (III) HUVEC. TNF- (100 U/mL) and VEGF (1.25 nmol/L) were added for 1 hour before cell fixation. Scale bar, 25 μmol/L. (B) Electophoretic mobility shift assay with oligonucleotides containing the NFκB recognition sites from the TF and Ig promoters. Nuclear extracts were prepared from unstimulated HUVEC and cells induced for 1 hour with VEGF (1.25 nmol/L) or TNF- (100 U/mL). These data are representative of three experiments performed with similar results.
NFκB is not induced by VEGF. (A) Immunofluorescence staining of p65 (NFκB subunit) in unstimulated (I), TNF-–treated (II), or VEGF-treated (III) HUVEC. TNF- (100 U/mL) and VEGF (1.25 nmol/L) were added for 1 hour before cell fixation. Scale bar, 25 μmol/L. (B) Electophoretic mobility shift assay with oligonucleotides containing the NFκB recognition sites from the TF and Ig promoters. Nuclear extracts were prepared from unstimulated HUVEC and cells induced for 1 hour with VEGF (1.25 nmol/L) or TNF- (100 U/mL). These data are representative of three experiments performed with similar results.
Taking into account the possibility that VEGF could induce activation of NFκB subunits other than p65, we performed in addition electrophoretic mobility shift assays with the TF NFκB as well as the consensus NFκB sites that have been shown to bind c-Rel/p65 and p50/p65 complexes, respectively. Nuclear extracts from VEGF-treated cells failed to form protein-DNA complexes with the respective binding sites. In contrast, NFκB binding activity was significantly activated by TNF-α (Fig 3B). These results show that VEGF treatment of endothelial cells does not lead to NFκB activation.
Functional importance of a GC-rich region for VEGF-induced expression of TF: Mithramycin strongly inhibits TF activity.
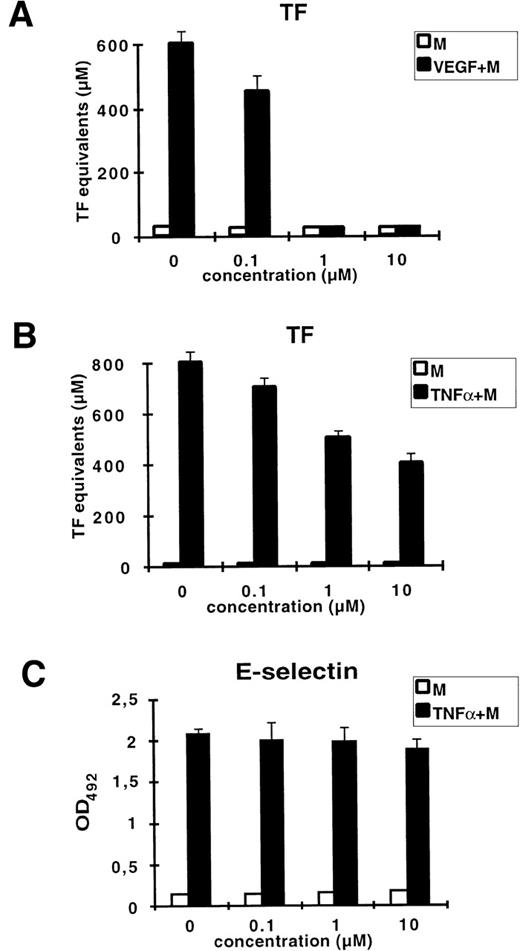
Mithramycin is a DNA-binding drug known to preferentially bind to GC-rich DNA sequences. It has been shown to specifically inhibit the transcription of genes highly dependent on GC-rich regulatory elements within their promoters such as c-myc, H-ras, dihydrofolate reductase, and VEGF.28-30 We examined whether mithramycin could inhibit VEGF– or TNF-α–induced upregulation of TF (Fig 4). Treatment of HUVEC with mithramycin at 1 μmol/L resulted in complete inhibition of VEGF-induced TF activity. At the same time, induction by TNF-α was blocked by only 50% at the highest concentration of mithramycin used (10 mmol/L). At concentrations of mithramycin ranging from 10 nmol/L to 10 μmol/L, cell morphology and viability were not altered (data not shown). To show that the observed inhibition of TF upregulation is not due to a more general effect of this drug on DNA synthesis, we tested whether mithramycin would block the induction of E-selectin by TNF-α. The E-selectin promoter does not contain GC-rich regions functionally important for TNF-α regulation. We could not detect any decrease of TNF-α–induced E-selectin expression in presence of mithramycin. Thus, these data suggest that the VEGF-responsive element of the TF promoter is located within a GC-rich region that is separate from the NFκB recognition site.
Mithramycin strongly inhibits VEGF-induced TF activity. HUVEC were preincubated for 30 minutes with increasing concentrations of mithramycin (M) before stimulation with VEGF (A) or TNF- (B) for 4 hours. TF activity equivalents were measured in a one-stage clotting assay. (C) Mithramycin has no effect on TNF-–induced E-selectin expression measured by ELISA. X-axis: concentration of mithramycin. Each value is the mean ± SD of triplicate in one experiment representative of two performed with similar results.
Mithramycin strongly inhibits VEGF-induced TF activity. HUVEC were preincubated for 30 minutes with increasing concentrations of mithramycin (M) before stimulation with VEGF (A) or TNF- (B) for 4 hours. TF activity equivalents were measured in a one-stage clotting assay. (C) Mithramycin has no effect on TNF-–induced E-selectin expression measured by ELISA. X-axis: concentration of mithramycin. Each value is the mean ± SD of triplicate in one experiment representative of two performed with similar results.
A VEGF-inducible DNA binding complex containing EGR-1 interacts with the GC-rich region at −85 to −70 of the human TF promoter.
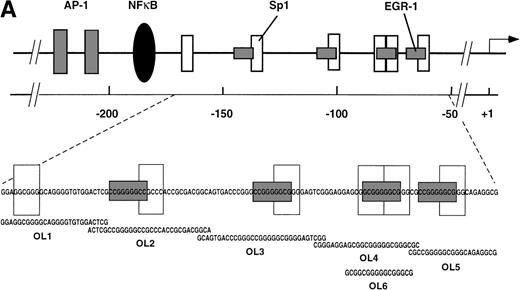
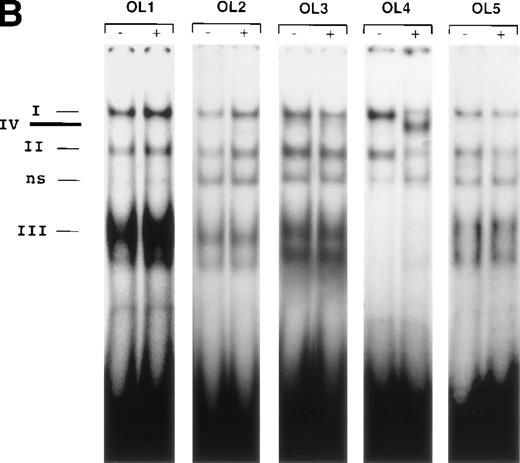
The results presented thus far show that NFκB is not involved in VEGF-mediated TF upregulation. Moreover, the inhibition of VEGF effects by mithramycin points to the importance of a GC-rich promoter region in TF upregulation by VEGF. To identify possible VEGF-induced proteins interacting with this region we used several overlapping oligonucleotides, covering the entire GC-rich region of the human TF promoter (−171 to −52) (Fig5A). Nuclear extracts from unstimulated cells and cells treated with VEGF for 1 hour were incubated with each of the six radiolabeled oligonucleotides (Figs 5B and 6). OL1, OL3, and OL5 exhibited a similar pattern of protein binding of three distinct complexes, that were specific, constitutive, and not altered by treatment with VEGF. Complexes I and II were attributed to Sp1 binding and were supershifted by anti-Sp1 antibodies4 (and data not shown). Complex III was not characterized in detail because it was detected in both unstimulated and VEGF-stimulated extracts.
(A) Transcription factor binding elements within the region of −230 to −50 bp of the human TF promoter. The start site of transcription is indicated by an arrow. Numbering is from the human TF sequence as given in Mackman et al.59 Oligonucleotides spanning the EGR-1/Sp1 sites (designated OL1 to OL6) were used in electrophoretic mobility shift assays. (B) Binding of nuclear proteins to the GC-rich region. Oligonucleotides OL1 to OL6 were radiolabeled and incubated with nuclear extracts from uninduced cells (−) and cells stimulated with VEGF (+) for 1 hour. Protein-DNA complexes I, II, III, and IV are indicated; ns, nonspecific binding. OL4 and OL6 showed identical protein/DNA complexes. Shown is one experiment that is representative of four experiments with similar results.
(A) Transcription factor binding elements within the region of −230 to −50 bp of the human TF promoter. The start site of transcription is indicated by an arrow. Numbering is from the human TF sequence as given in Mackman et al.59 Oligonucleotides spanning the EGR-1/Sp1 sites (designated OL1 to OL6) were used in electrophoretic mobility shift assays. (B) Binding of nuclear proteins to the GC-rich region. Oligonucleotides OL1 to OL6 were radiolabeled and incubated with nuclear extracts from uninduced cells (−) and cells stimulated with VEGF (+) for 1 hour. Protein-DNA complexes I, II, III, and IV are indicated; ns, nonspecific binding. OL4 and OL6 showed identical protein/DNA complexes. Shown is one experiment that is representative of four experiments with similar results.
OL4 and OL6, a shorter version of OL4, showed an identical pattern of protein binding: in addition to complexes I and II described above, a single inducible complex (complex IV) was detected with VEGF-stimulated cells. Sp1 binding (complexes I and II) seemed to decrease upon induction of complex IV. OL6 includes Sp1 binding sites as well as an overlapping putative EGR-1 recognition site GCGGGGGCG21overlapping the Sp1 sites. A similar inducible complex, but of lower intensity, was observed with OL2, which also contains in addition to an Sp1 recognition element an EGR-1 site that differs from the consensus sequence at positions 1 and 9.
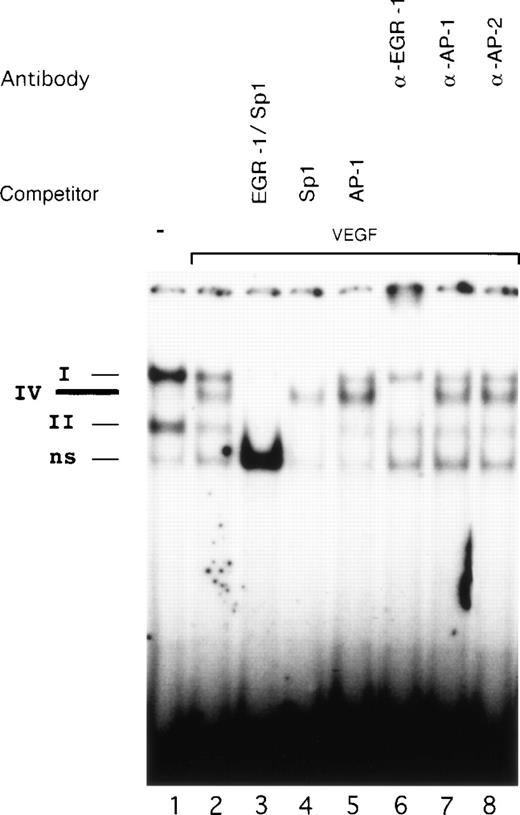
Competition studies were performed using various unlabeled oligonucleotides to probe the binding specificity of complexes I, II, and IV with OL6 (Fig 6). All three complexes were competed by excess of cold OL6 (lane 3). Competition with OL1 (Sp1 consensus) blocked formation of complexes I and II, leaving complex IV intact (lane 4). An oligonucleotide covering AP-1 sites from the TF promoter did not influence the formation of any of the three complexes (lane 5).
Identification of the VEGF-induced factor binding to OL6. Radioactively labeled OL6 was incubated with nuclear extracts from unstimulated cells (lane 1) and cells treated with VEGF (lanes 2 to 8) in the absence or presence of unlabeled oligonucleotides and specific antibodies. The Sp1 (complexes I and II), VEGF-induced (complex IV), and nonspecific (ns) protein/DNA complexes are indicated. VEGF-stimulated extracts were analyzed by competition with a 100-fold molar excess of unlabeled oligonucleotides OL6 (lane 3), OL1 (lane 4), or AP-1 (see Materials and Methods, lane 5). Supershift experiments were performed using 3 μL of anti–EGR-1 (lane 6), anti–AP-1 (lane 7), or anti–AP-2 (lane 8) antibodies. Shown is one experiment that is representative of three experiments with similar results.
Identification of the VEGF-induced factor binding to OL6. Radioactively labeled OL6 was incubated with nuclear extracts from unstimulated cells (lane 1) and cells treated with VEGF (lanes 2 to 8) in the absence or presence of unlabeled oligonucleotides and specific antibodies. The Sp1 (complexes I and II), VEGF-induced (complex IV), and nonspecific (ns) protein/DNA complexes are indicated. VEGF-stimulated extracts were analyzed by competition with a 100-fold molar excess of unlabeled oligonucleotides OL6 (lane 3), OL1 (lane 4), or AP-1 (see Materials and Methods, lane 5). Supershift experiments were performed using 3 μL of anti–EGR-1 (lane 6), anti–AP-1 (lane 7), or anti–AP-2 (lane 8) antibodies. Shown is one experiment that is representative of three experiments with similar results.
The constitutive expression of Sp1 in endothelial cells suggests that this transcription factor occupies the binding sites in the absence of a VEGF-inducible factor. Our electrophoretic mobility shift data confirm Sp1 binding in unstimulated cells. Upon exposure to VEGF, however, the intensity of Sp1 binding decreased substantially and seemed to be replaced by a VEGF-inducible protein interacting with the same GC-rich element of the TF promoter.
To identify the proteins present in the VEGF-inducible complex IV, we used antibodies directed against EGR-1, AP-1, and AP-2 in supershift experiments (Fig 6, lanes 6 to 8). AP-2 is a zinc-finger transcription factor that also interacts with a GC-rich nucleotide sequence very similar to the EGR-1 site and has been shown to play a significant role in the upregulation of the VEGF gene by TGF-α.40Preincubation with anti–EGR-1 antibodies completely abolished formation of complex IV (lane 6), whereas antibodies to AP-1 and AP-2 (lanes 7 and 8) failed to produce supershifts. Therefore, we conclude that VEGF-inducible complex IV represents binding of EGR-1. We assume that the identity of the OL6 sequence to the EGR-1 consensus binding site and possible additional influences of surrounding nucleotides may explain the preferential binding of EGR-1 to OL6.
Analysis of the VEGF-responsive region within the TF promoter by reporter gene studies.
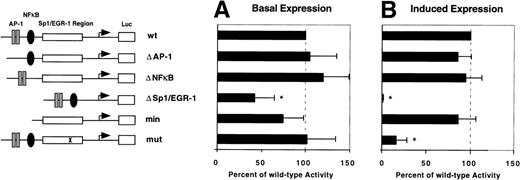
To confirm the role of EGR-1 elements in the transcriptional activation of the TF gene by VEGF, a series of deletion constructs based on the −330/+118 basal TF promoter construct were transiently transfected into HUVEC. Six plasmids were analyzed: wt, covering the wild-type TF promoter from −330 to +118 bp; Δ AP-1, Δ NFκB, and Δ Sp1/EGR-1, which contain substitutions of the respective transcription factor binding sites; min, a minimal promoter construct, which is limited to the GC-rich promoter region including Sp1 and EGR-1 sites; mut, a mutated promoter construct containing a single point mutation (G → A) at the position 5′ to the consensus EGR-1 site. This point mutation is located within the region to which VEGF-induced EGR-1 was found to bind in the electrophoretic mobility shift assay. Values of all other reporter constructs are given in percent of wt activity. Deletion of AP-1 or NFκB sites as well as the point mutation did not affect basal promoter activity (Fig 7A). However, deletion of the entire GC-rich region reduced basal promoter activity by more than 60%. The basal expression of the minimal promoter construct was slightly reduced, although this difference was not statistically significant. These results are in good agreement with previously published data showing that Sp1 mainly regulates basal activity of the TF promoter.15
Identification of the VEGF-responsive region within the TF promoter. Reporter plasmids used in this study are shown schematically (see also Materials and Methods). Luciferase activity of reporter constructs transiently transfected in HUVEC in the absence (A) or presence (B) of VEGF was calculated relative to wild-type (wt) promoter activity designated as 100%. In each experiment values were determined from duplicate wells and normalized for transfection efficiency. Results from three independent experiments are shown as mean values ± SD. *P < .01 versus wt activity.
Identification of the VEGF-responsive region within the TF promoter. Reporter plasmids used in this study are shown schematically (see also Materials and Methods). Luciferase activity of reporter constructs transiently transfected in HUVEC in the absence (A) or presence (B) of VEGF was calculated relative to wild-type (wt) promoter activity designated as 100%. In each experiment values were determined from duplicate wells and normalized for transfection efficiency. Results from three independent experiments are shown as mean values ± SD. *P < .01 versus wt activity.
Luciferase activity of the wild-type reporter was induced twofold to threefold by VEGF. This may not reflect the full degree of TF gene upregulation in vivo, because we observe a partial activation of TF expression in primary endothelial cells by the transfection procedure leading to increased basal levels. Deletion of NFκB and AP-1 sites had no influence on the level of upregulation (Fig 7B). However, deletion of all Sp1 and EGR-1 sites abolished VEGF induction completely. In contrast, the minimal promoter construct covering all Sp1 and EGR-1 sites showed a level of upregulation comparable to the wild-type promoter. Interestingly, the point mutation immediately 5′ to the consensus EGR-1 significantly reduced promoter responsiveness to VEGF. These results show that the VEGF-responsive site is located within the GC-rich region of the TF promoter and suggest that the consensus EGR-1 site in the 3′ part of this region is functionally important for maximal induction of the TF promoter.
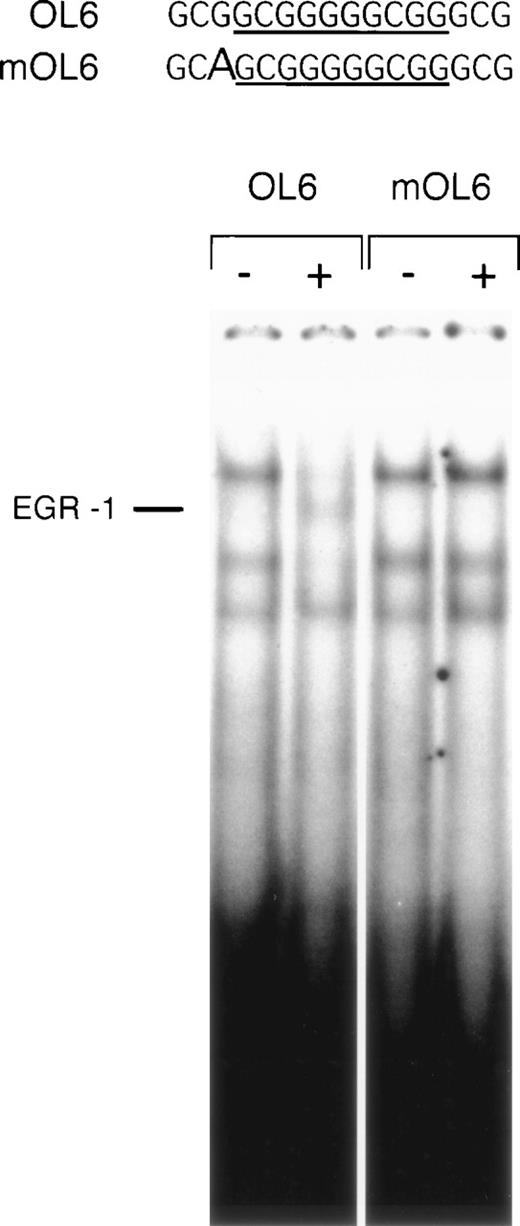
A point mutation affects DNA binding of EGR-1.
The influence of nucleotides surrounding an EGR-1 recognition element on the affinity of EGR-1 binding has not been subject to detailed analysis. However, based on the results of our reporter studies, we expected the point mutation 5′ to the EGR-1 consensus site to interfere with EGR-1 binding to DNA. We performed electrophoretic mobility shift assays with oligonucleotide mOL6, containing the same G → A substitution present in the reporter construct. It may be of importance to note that this position is conserved in the sequences of human and porcine TF promoters.4 mOL6 formed Sp1 complexes identical to the wild-type oligonucleotide OL6. However, VEGF-inducible EGR-1 binding was no longer detectable with the mutant oligonucleotide (Fig 8). Competition studies showed that this mutated oligonucleotide was able to compete the formation of the EGR-1 complex, but at significantly higher concentration than the wild-type (data not shown). These results confirm that a single nucleotide substitution immediately 5′ of the consensus EGR-1 site significantly decreases the affinity of EGR-1 binding to its recognition site and, thus, interferes with VEGF induction of TF expression.
A point mutation 5′ to the EGR-1 consensus site abrogates EGR-1 binding. Sequences of the wild-type (OL6) and mutant (mOL6) are shown at the top. The consensus EGR-1 site is underlined and the G → A substitution is indicated by a bold letter. Nuclear extracts from unstimulated (−) and VEGF-stimulated (+) cells were incubated with radiolabeled OL6 or mOL6. The position of the EGR-1 complex is indicated. Shown is one experiment that is representative of three experiments with similar results.
A point mutation 5′ to the EGR-1 consensus site abrogates EGR-1 binding. Sequences of the wild-type (OL6) and mutant (mOL6) are shown at the top. The consensus EGR-1 site is underlined and the G → A substitution is indicated by a bold letter. Nuclear extracts from unstimulated (−) and VEGF-stimulated (+) cells were incubated with radiolabeled OL6 or mOL6. The position of the EGR-1 complex is indicated. Shown is one experiment that is representative of three experiments with similar results.
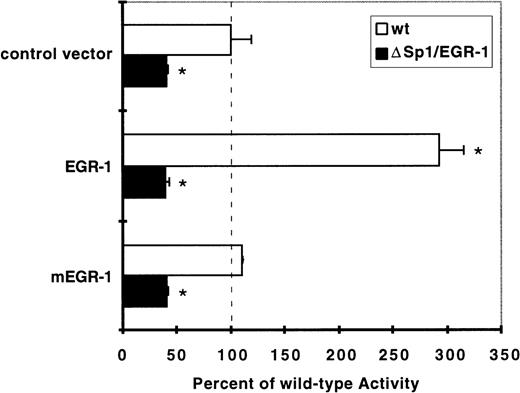
Over-expression of EGR-1 results in trans-activation of the TF promoter in HUVEC.
To further confirm the functional importance of EGR-1 in the transcriptional regulation of TF gene expression, we performed cotransfection experiments. Expression vectors encoding full-length EGR-1 or a transcriptionally inactive mutant, mEGR-1, lacking the first and part of the second zinc finger domains,33 were transiently transfected together with the wild-type TF reporter or the ΔSp1/EGR-1 construct (Fig 9). Cotransfection of full-length EGR-1 markedly increased the activity of the wild-type promoter in a manner comparable to VEGF induction. However, the mutant EGR-1 did not lead to upregulation of luciferase expression. In addition, the ΔSp1/EGR-1 construct did not display any EGR-1–mediated response. Taken together, these findings show that EGR-1 is able to mediate transcriptional activation of the TF gene via the VEGF-responsive element, and substantiate the functional role of EGR-1 in the transcriptional regulation of the TF gene by VEGF.
Overexpression of full-length EGR-1, but not a transcriptionally inactive mutant, results in activation of the TF promoter in HUVEC. EGR-1–mediated activation of the TF promoter is prevented by deletion of the Sp1/EGR-1 sites. Luciferase activity of wild-type (wt, white bars) or deleted (▵Sp1/EGR-1, black bars) reporter constructs in the presence of the EGR-1 expression plasmid or the inactive mutant (mEGR-1) was calculated relative to the wt promoter activity (mean ± SD from three independent experiments performed in duplicates; *P < .01 versus basal wt activity).
Overexpression of full-length EGR-1, but not a transcriptionally inactive mutant, results in activation of the TF promoter in HUVEC. EGR-1–mediated activation of the TF promoter is prevented by deletion of the Sp1/EGR-1 sites. Luciferase activity of wild-type (wt, white bars) or deleted (▵Sp1/EGR-1, black bars) reporter constructs in the presence of the EGR-1 expression plasmid or the inactive mutant (mEGR-1) was calculated relative to the wt promoter activity (mean ± SD from three independent experiments performed in duplicates; *P < .01 versus basal wt activity).
Subcellular localization of EGR-1 protein in endothelial cells.
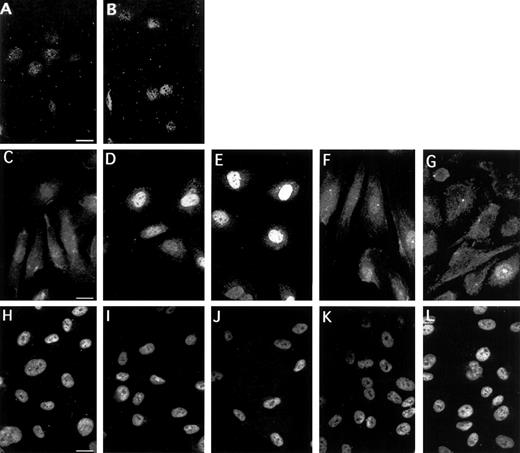
To substantiate the importance of EGR-1 for the VEGF response, we analyzed the subcellular localization and expression of EGR-1 protein in unstimulated and VEGF-induced endothelial cells (Fig 10A through G). The great majority of unstimulated cells under normal growth conditions had diffuse staining in the cytoplasm, as well as weak staining in the nucleus (Fig10C). Fifteen minutes of treatment with VEGF were sufficient to trigger a clear increase in nuclear staining (Fig 10D). Maximal nuclear staining was seen between 1 and 2 hours of stimulation with VEGF. Six hours after the addition of VEGF to the cells, the signal distribution returned to the level seen in uninduced cells. At 8 hours, weak cytoplasmic staining was detectable in most cells (Fig 10G). We tested two anti–EGR-1 antibodies (Santa Cruz Biotechnology) and obtained comparable results. After preincubation of anti–EGR-1 antibodies with the peptide to which the antibodies were raised (Fig 10A and B), only a very low level of nuclear staining was still visible and cytoplasmic staining was completely abolished, confirming the specificity of the antibodies used. Nuclear localization of induced EGR-1 protein was confirmed by costaining with DAPI (data not shown). In contrast, staining for Sp1 was constitutively present in the nucleus and is not influenced by VEGF treatment (Fig 10H through L). These data combined with other data described above show that VEGF treatment of endothelial cells results in rapid accumulation of EGR-1 transcription factor in the nucleus, thereby activating expression of the TF gene.
Subcellular localization of EGR-1 and Sp1 in HUVEC. Immunofluorescence staining of EGR-1 in unstimulated endothelial cells (C) and cells stimulated with VEGF for 15 minutes (D), 1 hour (E), 6 hours (F), and 8 hours (G). Preabsorbtion of the anti–EGR-1 antibodies with an appropriate peptide abolished both cytoplasmic and nuclear staining of unstimulated (A) and VEGF-stimulated cells (B). Sp1 staining was exclusively nuclear and identical for all time points (H through L). The picture is representative of four experiments with similar results.
Subcellular localization of EGR-1 and Sp1 in HUVEC. Immunofluorescence staining of EGR-1 in unstimulated endothelial cells (C) and cells stimulated with VEGF for 15 minutes (D), 1 hour (E), 6 hours (F), and 8 hours (G). Preabsorbtion of the anti–EGR-1 antibodies with an appropriate peptide abolished both cytoplasmic and nuclear staining of unstimulated (A) and VEGF-stimulated cells (B). Sp1 staining was exclusively nuclear and identical for all time points (H through L). The picture is representative of four experiments with similar results.
DISCUSSION
Several recent findings suggest that TF may have function(s) distinct from the activation of the coagulation cascade. This protein has a capacity to transmit intracellular signals41 and appears to participate in embryonic blood vessel development,42metastasis,43,44 and tumor-associated angiogenesis.5 45
Detection of TF on the endothelium of tumor-induced vessels raised the question whether tumor cells per se stimulate the production of TF. One possibility would be that expression of TF is mediated by the direct angiogenic factor VEGF, because many tumor cells produce VEGF and endothelial cells express its high-affinity receptors. For this reason we were interested to understand the molecular mechanisms that would lead to TF activation in endothelial cells in response to VEGF.
Our data show that TF expression induced by VEGF was as strong as the expression induced by TNF-α. However, whereas inflammatory cytokines are known to trigger the TF gene in endothelial cells mainly via activation of NFκB, the present report shows that the mechanisms leading to induction of TF expression by the angiogenic factor VEGF are different from those of inflammatory stimuli and do not involve NFκB.
Several lines of evidence indicate that the main factor involved in VEGF-mediated TF expression is the transcription factor EGR-1. Firstly, strong inhibition of TF activity by mithramycin, a drug that binds to GC-rich DNA sequences, initially suggested the functional importance of the GC-rich region within the TF promoter in response to VEGF. This region is separate from the NFκB recognition site and spans several Sp1/EGR-1 overlapping binding sites. Secondly, a VEGF-inducible DNA binding complex was detected with an oligonucleotide covering the sequence of the human TF promoter from −85 to −70. In unstimulated endothelial cells this element is occupied by Sp1. Supershift data using anti–EGR-1 antibodies clearly show that the VEGF-inducible complex contains the transcription factor EGR-1. It is intriguing that Sp1 binding to this region decreased substantially and seems to be replaced by EGR-1 upon exposure of the cells to VEGF. Thus, it appears that the induction of the TF gene by VEGF in endothelial cells involves the interplay of the zinc-finger transcription factors Sp1 and EGR-1 with the indicated promoter element. A similar competition for binding was proposed for the regulation of PDGF-A and PDGF-B expression in endothelial cells by PMA.24 Thirdly, reporter gene studies confirmed that the VEGF-responsive site is localized within the GC-rich region of TF promoter. The consensus EGR-1 site in the 3′ part of this region seems to be functionally important for maximal induction of the TF gene by VEGF because a point mutation 5′ to the consensus EGR-1 site significantly diminished upregulation by VEGF. This position which is conserved in the sequences of the human, porcine, and murine promoters seems to be important for the optimal interaction of VEGF-induced EGR-1 with its recognition site, because in electrophoretic mobility shift assays VEGF-inducible EGR-1 binding was no longer detectable with the corresponding mutant oligonucleotide. Another observation based on the reporter gene studies concerns the basal expression of the TF promoter in endothelial cells. Deletion of the entire GC-rich region reduced basal promoter activity by about 60%. As mentioned above, in unstimulated endothelial cells this element is occupied by constitutively expressed Sp1 factors. These results are in accordance with data from Cui et al15showing that Sp1 plays a role for basal activity of the TF promoter in the epithelial-like HeLa cells.
Finally, overexpression of full-length EGR-1, but not of an inactive mutant, results in trans-activation of the TF promoter comparable to VEGF induction. Addition of VEGF to cells overexpressing functional EGR-1 did not result in further activation of the TF promoter (data not shown). These findings substantiate the significant role of EGR-1 in VEGF-induced transcriptional regulation of the TF gene in endothelial cells.
A multitude of data published on EGR-1 supports its central role as a multifunctional transcription factor important for growth and differentiation. EGR-1 mRNA is induced within 15 to 30 minutes by a wide range of extracellular signals, including growth factors, and does not require de novo protein synthesis. Some data suggest that multiple, independent, possibly additive pathways for the induction of this gene must exist. When neural PC12 cells were exposed simultaneously to maximally inducing levels of PMA and EGF, EGR-1/TIS induction was synergistically elevated.46 Similarly, when endothelial cells were treated with a combination of VEGF and TNF-α, TF activity was more than additively upregulated46 (and D.M., E.H., unpublished data, 1998). However, the specific signaling pathways and their possible cross-talk leading to EGR-1 activation in endothelial cells still need to be defined. Another aspect of EGR-1 is the cell-type–specific variation of its function. For example, the activity of the PDGF-A promoter is reduced by EGR-1 in NIH3T3 cells but activated in human embryonic kidney-derived 293 cells.47Second, the dramatic enhancement in transcriptional activity seen in HeLa cells with the EGR-1 construct from which the repressor domain was deleted (Δ284-330) was not detected in 3T3 cells.33Importantly, EGR-1 is one of only a small number of bifunctional transcription factors that contain both activation and repression domains. Signal-specific posttranslational modification of EGR-1, interaction with other regulating factors, and/or presence of a transcription factor inhibitor may activate or repress different target genes in a cell-type–specific manner.
In addition to the results discussed above, we have further tested the subcellular localization and expression of EGR-1 protein in endothelial cells by immunofluorescence. Its subcellular localization has been described for serum-starved fibroblasts after serum stimulation.33,48 Using immunostaining, we found that the protein starts to accumulate in the nucleus within 15 to 30 minutes, reaches a peak level at about 1 to 2 hours, and decreases gradually thereafter within a few hours. The time course of nuclear accumulation is in agreement with its proposed function as the primary inducer of TF by VEGF. Thus far, the mechanisms that mediate rapid egr-1 gene activation by growth factors are only partially understood. Our data suggest the presence of low levels of the protein in the cytoplasm of unstimulated endothelial cells. It is not clear at the moment whether that means that low amounts of EGR-1 are continuously synthesized in endothelial cells in culture or if EGR-1 protein is retained in the cytoplasm of untreated cells. It is well established that many transcription factors can be selectively relocalized from the cytoplasm to the nucleus, and the nuclear transport of these proteins involved in replication and/or transcription is an important regulatory step in cellular growth and differentiation. Examples of such regulations are: the nuclear transport of the glucocorticoid49 and progesterone50 receptors in response to binding of their cognate hormone; the nuclear translocation of NFκB,51 STAT-1,52 and NFIL-653 dependent on the continuous stimulation by external signals; some data also argue in favor of a regulated transport in the case of c-Fos54 and E2F.55However, except for NFκB, the mechanisms and factors that participate in retention of pre-existing transcription factors within the cytoplasm are not completely understood. It remains to be seen whether a potential regulated release of EGR-1 from cytoplasmic anchoring could have any physiological significance.
Our data correlate with observations made in vivo showing that vascular endothelial cells in a tumor environment express TF and suggest that VEGF secreted by tumor cells can be indeed the cause of elevated levels of TF. This may contribute to the hypercoagulability frequently seen in tumor patients. Alternatively, in the context of vessel formation, TF most likely has additional function(s) independent of its role in triggering coagulation. In this respect, Ott et al56 showed recently that an interaction of the TF cytoplasmic domain with actin-binding protein 280 supports cell adhesion and migration that may lead to vascular remodeling. In addition, strong evidence exists that polarized expression and secretion of various proteins by endothelial cells can take place upon external stimulation.57 58 It remains to be shown whether VEGF could trigger polarized distribution of TF on the cell membrane. It is conceivable that regulated and maybe polarized expression of TF on the basal side of the angiogenic endothelial cells could be required in the process of sprouting and/or invasion of capillaries. The identified molecular pathway by which VEGF induces TF expression in endothelial cells further supports the complex biological regulation and function of TF and proposes a role of EGR-1 in the angiogenic response of endothelial cells.
ACKNOWLEDGMENT
We are grateful to Dr Vikas P. Sukhatme and Dr Matthias Clauss for providing the EGR-1 plasmids and recombinant VEGF, respectively. We also thank Dr Christine Brostjan for critically reading the manuscript, and Christa Rabeck and Christoph Kaun for technical assistance in cell preparation.
D.M. and A.W. contributed equally to this study.
Supported in part by grants to E.H. from the Austrian Science Foundation (SFB05-10) and the European Commission (BMH4-98-3277).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Erhard Hofer, PhD, Department of Vascular Biology and Thrombosis Research at VIRCC, University of Vienna, Brunnerstrasse 59, A-1235 Vienna, Austria; e-mail:erhard.hofer@univie.ac.at.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal