Abstract
We have generated rat monoclonal antibodies (MoAbs) against cell surface antigens of the mouse endothelioma cell line bEND.3. Three antibodies (V.1A7, V.5C7, and V.7C7) were selected, all of which recognize a 75-kD antigen on bEND.3 cells and bind selectively to endothelial cells in cryostat sections of mouse tissues. A cDNA for the antigen was isolated from a bEND.3 pCDM8 expression library by using transient expression in COS-7 cells and immunoselection with the three MoAbs. This cDNA coded for a novel, type I membrane protein of 248 amino acids with an extracellular domain rich in threonine and serine residues (35%). The protein is sensitive to O-sialoglycoprotein endopeptidase, indicating that it belongs to the class of sialomucin-like proteins. Therefore, we suggest the name endomucin. Treatment of isolated endomucin by sialidase and O-glycosidase reduced the apparent molecular weight to 45 kD and abolished binding of all three antibodies, indicating that carbohydrates are directly or indirectly involved in the formation of the antibody epitopes. Immunohistological analysis of all examined mouse tissues showed that endomucin is an endothelial antigen found in venous endothelium as well as in capillaries, but not on arterial endothelium. Interestingly, high endothelial venules of peripheral and mesenteric lymph nodes as well as of Peyers’s patches were negative for staining with the three MoAbs.
MUCIN-LIKE MEMBRANE glycoproteins contain many serine and threonine residues that carry large amounts of O-linked glycans, forcing the molecule into an extended structure.1-4 By virtue of these features, cell surface-associated mucins have a dual function in the regulation of cell adhesion either by providing a repulsive barrier to prevent cell-cell or cell-matrix interactions5-9 or by promoting adhesion through facilitating lectin-type interactions with opposing cells.10 Members of this family of proteins can be involved in signal transduction,11-13 and recent evidence suggests that CD43, a leukocyte sialoglycoprotein,14,15 can function as a positive signaling molecule for β1 and β2 integrin-mediated cell adhesion.16
Growing evidence indicates that endothelial cell-associated mucins function as cell adhesion molecules mediating adherence with leukocytes. Leukocyte-endothelial cell interactions occur during leukocyte trafficking from the blood into lymph nodes, peripheral tissues, or sites of inflammation and are mediated at least in part by interactions between mucin-like molecules and their receptors. A well-described class of sialomucin-binding adhesion molecules are the selectins.10,17-19 All selectins recognize sialylated and fucosylated carbohydrates related to the tetrasaccharide sialyl Lewis X, and several mucin-like glycoproteins have been shown to present such structures to selectins. Glycosylation-dependent cell adhesion molecule 1 (GlyCAM-1) and CD34 are sialomucins that represent two of the principle ligands for L-selectin in the high endothelial venules (HEV) of peripheral lymph nodes.20,21 GlyCAM-1 is a secreted protein and CD34 is a transmembrane protein of endothelial cells of HEV.22 Increasing evidence indicates that sialylation, sulfation, and fucosylation of the O-linked side chains of GlyCAM-1 and CD34 are requirements for the binding to the selectins.23,24 Mucosal addressin cell adhesion molecule 1 (MAdCAM-1) is another cell adhesion molecule that contains a mucin-like domain and is expressed on HEV in Peyer’s patches and mesenteric lymph nodes and on venules in intestinal lamina propria.25,26 In the HEV of mesenteric lymph nodes, the mucin-like domain of a subpopulation of MAdCAM-1 molecules contains sulfated carbohydrate side chains that interact with L-selectin.27
The sialomucin CD43, which is found on all leukocytes except mature B cells, was recently shown to have antiadhesive functions in T-cell homing in vivo.28 T cells from CD43-deficient mice home more frequently to secondary lymphoid organs and this effect is due to CD43 interference with L-selectin–mediated interactions in vivo and in vitro. The extended structure of CD43 (length, 45 nm)3 and its strongly charged nature are likely to be responsible for the antiadhesive function of CD43. A monoclonal antibody (MoAb) against CD43 was found to block T-cell binding to lymph node HEV as well as homing in vivo, suggesting yet another mechanism for the involvement of CD43 in adhesive events.29 In summary, many sialomucins are involved in cell contact formation either directly by supporting cell adhesion or repelling the neighboring cell or indirectly by regulating adhesion events via signaling to other adhesion molecules.
Raising MoAbs against endothelial cell surface molecules, we have identified a novel endothelial-specific antigen. We have isolated a full-length cDNA for this glycoprotein by immunoselection and describe the sequence, the tissue distribution, and the biochemical properties of this antigen. The predicted 248 amino acid polypeptide is a novel type I integral membrane protein with similarities in structure to the growing family of sialomucins.
MATERIALS AND METHODS
Cell culture.
The mouse endothelioma cell line bEND.3 (kindly provided by Dr Werner Risau, Max-Planck-Institute for Physiological and Clinical Research, Bad Nauheim, Germany), the SV-40–transformed African green monkey kidney cell line COS-7, and the mouse hybridoma cell line SP2/0 were cultured in Dulbecco’s modified Eagles medium (DMEM) supplemented with 10% fetal calf serum (FCS), 100 IU/mL penicillin, 100 μg/mL streptomycin, and 2 mmol/L L-glutamin (all GIBCO Life Technologies, Karlsruhe, Germany). Serially passaged b.End.3 cells were grown to confluence in tissue culture plates (Falcon, Heidelberg, Germany) and stimulated for 24 hours at 200 U/mL with recombinant human tumor necrosis factor α (TNF-α; obtained by Knoll AG, Ludwigshafen, Germany) to be used for immunization and in cell surface enzyme-linked immunosorbent assays (ELISAs). Inducibility of endomucin was examined by immunoblots of bEND.3 cells after stimulation for 12 hours with the cytokines interleukin-1β (IL-1β; 10 pg/mL), interferon-γ (IFN-γ; 0.4 ng/mL), and TNF-α (0.05 ng/mL) according to the manufacturer’s guidelines (R&D, Abingdon, Oxon, UK). Hybridoma cells were grown in DMEM as decribed above supplemented with HAT (Boehringer Mannheim, Mannheim, Germany) according to the manufacturer’s instructions.
MoAbs.
MoAbs against endothelial cell surface antigens were produced as described previously.30 Briefly, female Lewis rats were immunized with b.END.3 cells (passage 15 to 29) that had been stimulated with 200 U/mL TNF-α for 24 hours before injection. Cells mixed with Complete Freund’s adjuvans for the first immunization and incomplete for the other immunizations were injected six times (1 to 2 × 107 cells per injection) in 1-week intervals, with 3 weeks between the fifth and sixth immunization. Three days after the last boost, cells from peripheral lymph nodes and spleen cells were fused with SP2/0 cells essentially as described.31 Hybridoma supernatants were differentially screened for indirect immunoperoxidase staining on TNF-α stimulated and nonstimulated endothelioma cells by cell surface ELISA, as described.30 Hybridoma producing antibodies against cell surface antigens were subcloned by limiting dilution. The isotype of the three MoAbs against endomucin was determined to be IgG1 for V.1A7 and V.5C7 and IgG2a for V.7C7. MoAbs EA-3 (rat IgG1)32against mouse CD31, 10E9 (rat IgG2a)33 against mouse E-selectin, 9EG7 (rat IgG2a)34 against mouse β1-integrin chain, M1/70 (rat IgG2a; called TIB128 from the ATCC, Manassas, VA) against mouse Mac-1, MECA367 (rat IgG2a)25against mouse MAdCAM-1, and polyclonal serum against human vWF (DAKO, Hamburg, Germany) were used as controls in cell surface ELISAs and for immunostaining of cryosections. Additionally, rat anti-mouse MoAb OX40 (CD134; provided by M. Puklavec, MRC Cellular Immunology Unit, Sir William Dunn School of Pathology, University of Oxford, Oxford, UK), clone MRC OX86,35 rat anti-mouse Sca-2, clone III.3A7 (U.S. and D.V., unpublished data), rat anti-CD59, clone SM8 (S.M.M., unpublished data), and rat anti-CD31, clone 390 (Serotec, Kidlington, Oxford, UK), were used as controls in the staining of wax-embedded sections.
Immunohistochemistry.
Tissue sections (5 μm) of wax-embedded normal mouse organs were washed with phosphate-buffered saline (PBS) containing 0.1% (vol/vol) Triton X-100 and incubated in phosphate buffer containing 0.1 mol/L glucose, 0.01 mol/L NaN3, and 40 U glucose oxidase (Sigma, St Louis, MO) for 15 minutes at 37°C to block endogenous peroxidase activity. Sections were blocked with 5% normal mouse serum for 30 minutes and incubated with the appropriate rat antibodies (hybridoma supernatant). Binding was detected by incubation of sections with biotinylated rabbit anti-rat IgG (Sigma) diluted in blocking buffer, followed by avidin-biotin-peroxidase complex (ABC elite; Vector Laboratories, Burlingame, CA) and 0.5 mg/mL diaminobenzidine (Polysciences Inc, Warrington, PA) with 0.024% H2O2. Sections were counter-stained with crystal violet. The primary MoAb was replaced with MoAb MRC OX86 as a negative control.
For cryosections, organs and tissues from 10- to 14-week-old NMRI mice were covered with tissue freezing medium (Jung, Nussloch, Germany), snap-frozen in liquid nitrogen, and stored at −80°C. Sections of 5 μm were made on a cryostat (Leica, Nussloch, Germany), transferred to Poly-L-Lysin–covered slides, dried overnight at room temperature, and fixed in 100% ethanol, followed by acetone treatment. Primary antibodies as supernatants or at 10 μg/μL in medium were added to the sections after washing in PBS, followed by further washing steps and incubation with peroxidase-coupled goat anti-rat or goat anti-rabbit IgG and IgM (Dianova, Hamburg, Germany), respectively, diluted in DMEM at 1:1,000. Signals were detected by incubation with 0.04% amino-ethyl-carbazole in N,N-dimethylformamid with 0.006% H2O2. For counterstaining, Mayers hematoxylin was used.
Flow cytometry.
Two-stage labeling of cell surface molecules was performed by incubation of cells (1 × 106) with the appropriate MoAb supernatant, followed after washing by the addition of purified fluorescein isothiocyanate (FITC)-conjugated goat anti-rat Ig antibody at 10 μg/mL (Serotec). Cell surface fluorescence was assayed by FACScan analysis (Becton Dickinson, San Jose, CA). Cells were gated on the scatter profiles to exclude dead cells.
Immunoprecipitations and immunoblots.
Cells were surface biotinylated with 0.5 mg/mL Sulfo-NHS-biotin (Pierce, Rockford, IL) in PBS, lysed, and subjected to immunoprecipitation as described.36 37 Immunoprecipitated proteins were separated by electrophoresis on 8% or 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose (Schleicher & Schüll, Dassel, Germany). Filters were analyzed for biotinylated proteins with peroxidase-conjugated streptavidin (Dianova) and the ECL system (Amersham, Braunschweig, Germany). Alternatively, nonlabeled cells were subjected to immunoprecipitations and precipitated antigens were separated by electrophoresis and transferred to nitrocellulose filters. Antigens were probed with hybridoma supernatant containing rat anti-endomucin MoAbs, and first antibodies were detected with peroxidase-conjugated rabbit anti-rat IgG (Dianova). Immunoreactive proteins were detected by enhanced chemoluminescence (ECL kit; Amersham Life Science, Braunschweig, Germany).
Enzyme treatments.
For treatments of purified endomucin with glycosidases, the antigen was immunoprecipitated from 5 × 106 bEND.3 cells, divided into five equal size aliquots, and treated with the appropriate enzyme while still bound to protein A Sepharose beads. The following treatments were performed on separate aliquots. (1) Beads were boiled for 5 minutes in 10 μL 0.1% SDS and then 0.05 U of endoglycosidase F (Boehringer Mannheim) in 30 μL of enzyme buffer (100 mmol/L K2HPO4/KH2PO4, 20 mmol/L EDTA, 0.5% Triton X-100, 0.1% SDS) was added. (2) Beads were incubated with 0.2 mU O-glycosidase (Boehringer Mannheim) in 40 μL enzyme buffer. (3) Beads were treated with 0.5 mU neuraminidase (fromArthrobacter ureafaciens; Boehringer Mannheim) in 40 μL enzyme buffer. (4) The same treatment as in (3) followed by the subsequent addition of 0.2 mU of O-glycosidase for 3 hours at 37°C. All other digestions were performed at 37°C overnight. The antigen was subsequently electrophoresed, transferred to nitrocellulose membranes, and detected in immunoblots with the respective anti-endomucin antibodies. Alternatively, the antigen was immunoprecipitated from surface biotinylated cells, electrophoresed, transferred to nitrocellulose, and detected by peroxidase-conjugated streptavidin (Dianova) followed by ECL. For treatment of purified endomucin or control antigens with O-sialoglycoprotein endopeptidase (Cedarlane Laboratories, Hornby, Ontario, Canada) the antigens were immunoprecipitated from (5 × 106) surface biotinylated bEND.3 cells. The precipitated material was divided in three aliquots and the different aliquots were either mock-treated or treated with neuraminidase as described above or with 2.5 μL of O-sialoglycoprotein endopeptidase (activity, 1 μL cleaves 10 μg glycophorin A per hour at 37°C) in 30 μL 50 mmol/L Tris/HCl, pH 7.4. The biotinylated antigens were subsequently electrophoresed, transferred to nitrocellulose, and analyzed as described above. For treatment of intact cells with O-sialoglycoprotein endopeptidase, 5 × 106 bEND.3 cells were washed, resuspended in RPMI/HEPES (200 μL), and incubated with or without 10 μL of O-sialoglycoprotein endopeptidase at 37°C for 30 minutes. Subsequently, cells were washed and analyzed by flow cytometry.
Expression cloning.
cDNA clones were isolated from a bEND.3 cDNA library using the COS-cell expression cloning protocol by Aruffo and Seed.38 A bEND.3 cDNA library in the pCDM8 vector was introduced into COS-7 cells by diethyl aminoethyl (DEAE)-dextran transfection. Forty-eight hours after transfection, cells were detached with EDTA, washed, and immunoselected with the rat MoAbs V.5C7, V.7C7, and V.1A7 and goat anti-rat Ig-coated plates. Plasmid DNA was recovered from the adherent COS-7 cells, amplified in E coli (strain MC1061/p3), and then reintroduced into COS-7 cells by spheroblast fusion. Three rounds of enrichment were performed to isolate positive clones.
DNA sequencing and sequence analysis.
Nucleotide sequencing was performed on both strands using α-32S-dATP with sequenase version 2.0 (USB, Little Chalfont, Bucks, UK) and automated sequencing using the dRodamine kit (Perkin-Elmer, Warrington, Cheshire, UK). Forward and reverse primers in pCDM8 and sequence-specific 18-mer oligonucleotide primers were used. Analysis of nucleic acid and protein sequence data were performed using the Wisconsin-GCG Package (Pittsburgh Supercomputing Center, Pittsburgh, PA). Sequence comparison with the mouse and human EST databases (GenBank)39 was performed using BLASTN 2.0.4 (NIH, Bethesda, MD).40
Northern blot analysis.
A mouse multiple tissue Northern blot prepared from mRNA (Origene Technologies, Rockville, MD) was hybridized with a32P-labeled cDNA probe for endomucin (Xho I fragment containing the complete insert from pCDM8) or32P-labeled β-actin probe and washed in 0.25× SSC in the presence of 0.1% SDS at 42°C.
RESULTS
Development of MoAbs against a cell surface antigen of mouse endothelial cells.
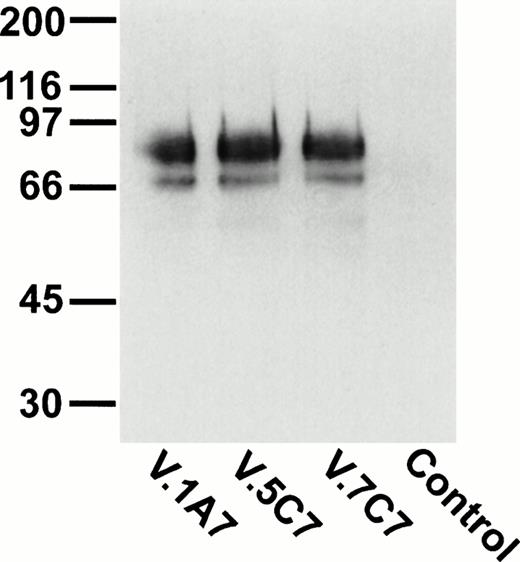
To identify novel endothelial-specific cell surface antigens, we immunized rats with intact mouse bEND.3 endothelioma cells and screened MoAbs for binding to the surface of bEND.3 cells in a cell surface ELISA, in a similar way as described before.30 Of 16 MoAbs that gave strong signals in these assays, three MoAbs, designated V.1A7, V.5C7, and V.7C7, were selected that all recognized a 75-kD cell surface antigen in immuno-precipitations with cell surface biotinylated bEND.3 cells (Fig 1A). In addition, a second weak band was detected at approximately 66 kD that is possibly a degradation product of the larger protein. Treatment of these cells with a mixture of inflammatory cytokines (IL-1β, IFN-γ, and TNF-α) for 12 hours did not alter the levels of expression of the antigens recognized by these MoAbs (data not shown).
Detection of the glycoprotein antigen for antibodies V.1A7, V.5C7, and V.7C7 by immunoprecipitation. Mouse bEND.3 cells were surface biotinylated and detergent extracts were incubated either with protein A Sepharose loaded with rabbit antirat IgG and the MoAbs V.1A7, V.5C7, or V.7C7 or a control antibody, as indicated. Specifically bound proteins were eluted with SDS-PAGE loading buffer, electrophoresed on an 8% polyacrylamide gel under reducing conditions, and transferred to nitrocellulose. Filters were incubated with peroxidase-conjugated streptavidin and analyzed by enhanced chemiluminescence.
Detection of the glycoprotein antigen for antibodies V.1A7, V.5C7, and V.7C7 by immunoprecipitation. Mouse bEND.3 cells were surface biotinylated and detergent extracts were incubated either with protein A Sepharose loaded with rabbit antirat IgG and the MoAbs V.1A7, V.5C7, or V.7C7 or a control antibody, as indicated. Specifically bound proteins were eluted with SDS-PAGE loading buffer, electrophoresed on an 8% polyacrylamide gel under reducing conditions, and transferred to nitrocellulose. Filters were incubated with peroxidase-conjugated streptavidin and analyzed by enhanced chemiluminescence.
Endothelial cells are specifically stained by all three antibodies in immunohistochemistry.
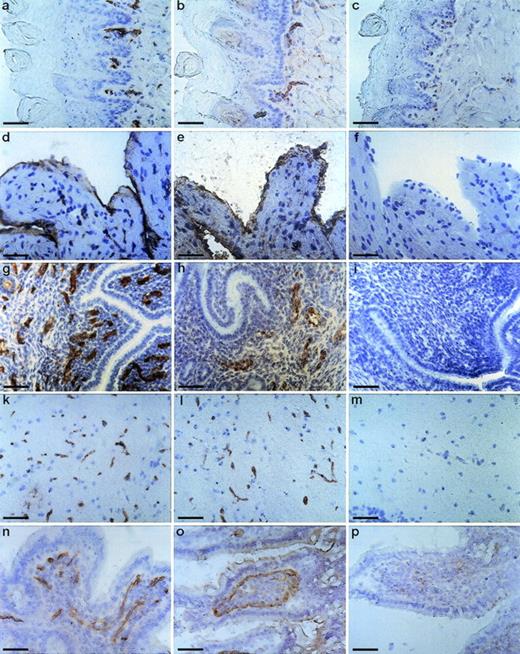
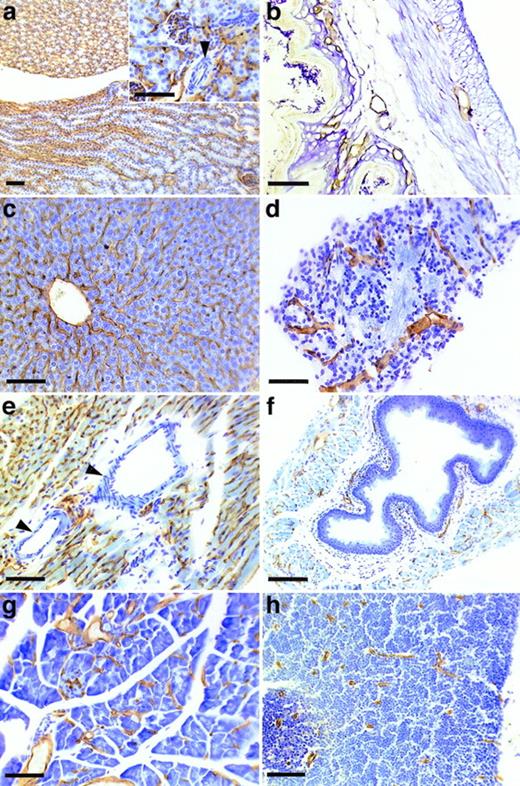
To determine the tissue distribution of the identified antigen, we stained sections of wax-embedded mouse tissues and cryostat sections of various mouse tissues with all three antibodies. An identical pattern of staining was observed with all three antibodies. Best signals were obtained with the MoAb V.5C7, and the results shown in Figs 2, 3, and4 were all obtained with this antibody. The antigen was found in all tissues examined, and in all cases its expression was specific for the vascular endothelium. Endothelial specificity was controlled by staining with antibodies against endothelial-specific antigens such as von Willebrand factor (vWF) or platelet endothelial cell adhesion molecule (PECAM-1), as is documented in Fig 2 for the uterus, brain, heart, small intestine, and tongue. The staining pattern of V.5C7 in the brain was the same as that of an antibody against the integrin chain β1, which is known to be predominantly expressed on brain capillaries (Fig 2). Intense staining was found in the glomerular capillaries and intertubular vessels in the kidney (Fig 3a). Venules and capillaries in the skin (Fig 3b), liver (Fig 3c), adrenal tissue (Fig 3d), heart (Fig3e), pancreas (Fig 3g), and thymus (Fig 3h) also stained strongly. Endothelial cells lining the aorta did not stain for the antigen (Fig3f), whereas the endocard in the heart was positive (Fig2d). Staining of arterioles was absent, as shown in the kidney and heart (Fig 3a and e).
Staining of cryostat sections of mouse tissues with the MoAb V.5C7. Cryostat sections of tongue (a, b, and c), heart (d, e, and f), uterus (g, h, and i), brain (k, l, and m), and small intestine (n, o, and p) were reacted with the MoAb V.5C7 (a, d, g, k, and n), polyclonal rabbit antibodies against von Willebrand factor (e, h, and o), the rat MoAb 9EG7 against integrin chain β1 (l), the rat MoAb EA3 against mouse PECAM-1 (b), or no first antibody (c, f, i, m, and p). First antibodies were detected by an immunoperoxidase technique. The bar represents 50 μm.
Staining of cryostat sections of mouse tissues with the MoAb V.5C7. Cryostat sections of tongue (a, b, and c), heart (d, e, and f), uterus (g, h, and i), brain (k, l, and m), and small intestine (n, o, and p) were reacted with the MoAb V.5C7 (a, d, g, k, and n), polyclonal rabbit antibodies against von Willebrand factor (e, h, and o), the rat MoAb 9EG7 against integrin chain β1 (l), the rat MoAb EA3 against mouse PECAM-1 (b), or no first antibody (c, f, i, m, and p). First antibodies were detected by an immunoperoxidase technique. The bar represents 50 μm.
Staining of sections of wax-embedded mouse tissue with the MoAb V.5C7. Sections of wax-embedded mouse kidney (a), skin (b), liver (c), adrenal tissue (d), heart (e), aorta (f), pancreas (g), and thymus (h) were incubated with MoAb V.5C7. First antibodies were detected by an immunoperoxidase technique. The arrowheads point to arterioles. The bar represents 100 μm.
Staining of sections of wax-embedded mouse tissue with the MoAb V.5C7. Sections of wax-embedded mouse kidney (a), skin (b), liver (c), adrenal tissue (d), heart (e), aorta (f), pancreas (g), and thymus (h) were incubated with MoAb V.5C7. First antibodies were detected by an immunoperoxidase technique. The arrowheads point to arterioles. The bar represents 100 μm.
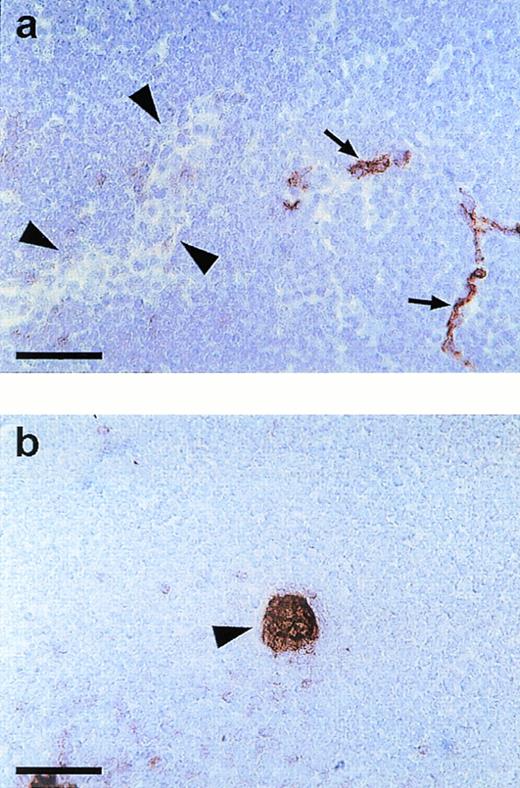
Endomucin is not expressed on HEV of lymphatic tissue. Cryostat sections of mesenteric lymph nodes were incubated with MoAb V.5C7 (a) (endomucin) or MoAb MECA 367 (b) against the vascular addressin MAdCAM-1 (MAdCAM-1). Arrowheads point to HEV and arrows point to capillaries. The bar represents 50 μm.
Endomucin is not expressed on HEV of lymphatic tissue. Cryostat sections of mesenteric lymph nodes were incubated with MoAb V.5C7 (a) (endomucin) or MoAb MECA 367 (b) against the vascular addressin MAdCAM-1 (MAdCAM-1). Arrowheads point to HEV and arrows point to capillaries. The bar represents 50 μm.
Capillaries and small venules in lymph node tissue were positive for the V.5C7 antigen, whereas HEV were clearly negative in peripheral lymph nodes and Peyer’s patches (data not shown) and in mesenteric lymph nodes (Fig 4a). Clear staining of HEV was seen with MoAb MECA79 against the peripheral node addressin (PNAd; not shown) and MoAb MECA367 against the mucosa-associated addressin MAdCAM-1, as found for Peyer’s patches (not shown) and documented on sections of mesenteric lymph nodes (Fig 4b). HEV were the only type of venular endothelium that we found to be negative for the staining with all three antibodies V.1A7, V.5C7, and V. 7C7.
The cDNA for the endothelial antigen codes for a novel sialomucin-like protein.
Using the three MoAbs V.1A7, V.5C7, and V.7C7 as panning reagents, we isolated three cDNA clones from a bEND.3 pCDM8 expression library. These clones were isolated after several rounds of panning and plasmid rescue using the COS-cell expression cloning protocol described by Aruffo and Seed.38 After the final round of enrichment using these three MoAbs, plasmid inserts from single colonies were analyzed using restriction digests.
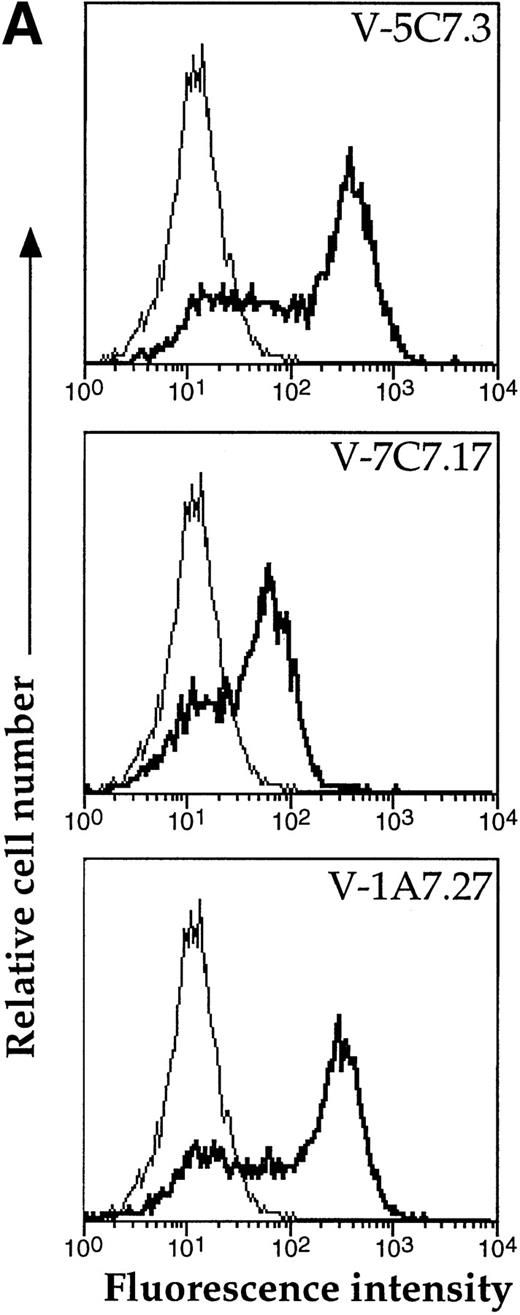
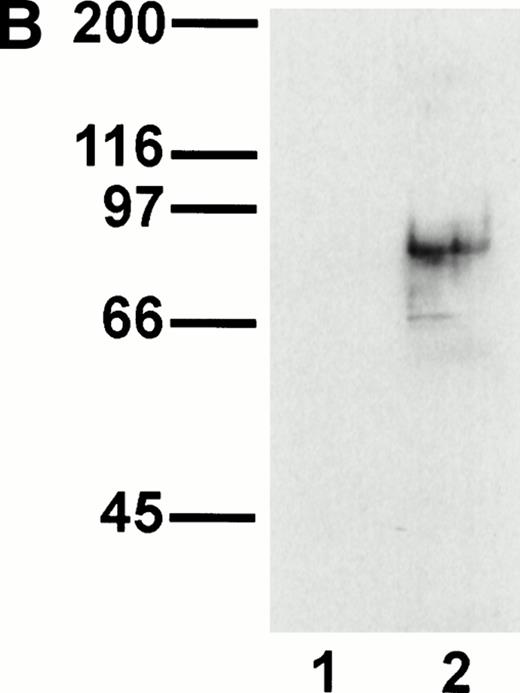
A representative clone from each group (DW-5C7.3, DW-7C7.17, and DW-1A7.27) was transfected into COS-7 cells and found positive for protein expression as determined by flow cytometry (Fig 5A). Mock-transfected COS-7 cells were unreactive for all three MoAbs (data not shown). In immunoprecipitations of surface-biotinylated COS-7 cells transfected with any of the three isolated cDNAs, each of the three MoAbs (V.1A7, V.5C7, and V.7C7) recognized a glycoprotein of similar size as that they recognize in bEND.3 cells. This is shown in Fig 5B for MoAb V.5C7 in an immunoprecipitation on COS-7 cells transfected with the cDNA clone DW-7C7.17.
Transfection of COS-7 cells with the novel cDNA confers reactivity to MoAbs V.1A7, V.5C7, and V.7C7. (A) COS-7 cells were transfected with the plasmid clones DW-5C7.3, DW-7C7.17, and DW-1A7.27 by the DEAE-dextran method. After 48 hours, the cells were detached and incubated with saturating levels of the respective MoAbs (thick line) or the control MoAb MRC OX86 (thin line), followed by a second incubation with fluorescein-conjugated goat antirat Ig antibody. Antibody binding was measured by flow cytometry. (B) Mock-transfected COS-7 cells (lane 1) or COS-7 cells transfected with the cDNA clone DW-7C7.17 (lane 2) were surface biotinylated and subjected to immunoprecipitation with MoAb V.5C7. Specifically bound proteins were analyzed and detected as described for Fig 1A. Molecular mass markers (in kilodaltons) are indicated on the left.
Transfection of COS-7 cells with the novel cDNA confers reactivity to MoAbs V.1A7, V.5C7, and V.7C7. (A) COS-7 cells were transfected with the plasmid clones DW-5C7.3, DW-7C7.17, and DW-1A7.27 by the DEAE-dextran method. After 48 hours, the cells were detached and incubated with saturating levels of the respective MoAbs (thick line) or the control MoAb MRC OX86 (thin line), followed by a second incubation with fluorescein-conjugated goat antirat Ig antibody. Antibody binding was measured by flow cytometry. (B) Mock-transfected COS-7 cells (lane 1) or COS-7 cells transfected with the cDNA clone DW-7C7.17 (lane 2) were surface biotinylated and subjected to immunoprecipitation with MoAb V.5C7. Specifically bound proteins were analyzed and detected as described for Fig 1A. Molecular mass markers (in kilodaltons) are indicated on the left.
The three positive plasmid clones contained cDNA inserts of approximately 1.2 kb and had identical restriction maps, suggesting that the same cDNA had been enriched by all three MoAbs, and each plasmid clone, when transfected into COS-7 cells, was found to express protein recognized by each of the MoAbs (data not shown). One clone (DW-7C7.17) was sequenced and the complete nucleotide sequence of 1,228 bp is shown in Fig 6 (−76 to +1152 bp). A mouse EST clone (557127) overlapping the 5′ region of the cDNA clone DW-7C7.17 was sequenced and found to be identical across this region of the cDNA insert (−76 to +424 bp) and extended the 5′ end of the cDNA clone by a further 21 bp (Fig 6; −97 to −77 bp). The DW-7C7.17 cDNA clone did not contain the complete 3′ untranslated end. However, the sequence of the mouse EST I.M.A.G.E. clone (749333) that overlapped with the 3′ region of the clone DW-7C7.17 showed complete homology in the region +962 to +1159 bp and showed the polyadenylation signal (AATAAA, +1225 bp) followed by the poly(A) tail. Highly homologous EST clones were also found in the human EST database. Searching the GenEMBL database showed an absence of homology to other known sequences, thus indicating that a novel cDNA had been cloned. The first ATG of the complete cDNA sequence (position +1 bp), which has a strong translation initiation site in the context of a Kozak41 consensus sequence (GGCACCATGC), precedes an open reading frame of 744 nucleotides encoding a protein of 248 amino acids (26,332 Daltons; Fig 6) The coding sequence is started by a putative 20 amino acid signal sequence containing a hydrophobic core, with the predicted cleavage site assigned to Ser-20/Asp21 according to von Heijne.42 Hydrophilicity (Kyte/Doolittle)43analysis suggests a type I integral membrane protein with an extracellular domain of 157 amino acids, a 23 amino acid transmembrane domain, and a cytoplasmic tail of 48 amino acids (residues 201-248). The putative extracellular domain contains two consensus sites (Asn-X-Ser/Thr) for N-glycosylation (Asn-101 and Asn-119) and is rich in serine and threonine residues (35%), suggesting that it is abundantly O-glycosylated. Using the O-GLYCBASE database,44the deduced amino acid sequence was predicted to have 25 O-linked glycosylation sites. There are three potential protein kinase C phosphorylation sites in the cytoplasmic tail (Ser-220, Ser-224, and Thr-229). By virtue of the clusters of serine and threonine residues in the extracellular domain of endomucin, computer-generated alignments of this sequence show homologies to mucins. Based on the predicted structure, it is likely that the processed protein is heavily glycosylated with O-linked glycans, which is in agreement with the fact that the apparent molecular weight of the antigen as judged by SDS-PAGE analysis is much larger than the molecular mass deduced from its amino acid sequence. Members of the sialomucin family of adhesion molecules, including CD34 and CD43, have similar features.3,45 46 We assign the name endomucin to this novel molecule due to its prominent localization on endothelial cells as well as the structural similarities with mucin-like glycoproteins.
Nucleotide and deduced amino acid sequence of endomucin. Nucleotides and amino acids are numbered on the left. The putative N-terminal signal sequence, the transmebrane region, and the polyadenylation signal are underlined. The putative N-linked glycosylation sites are in italics and double underlined. The putative protein kinase C phosphorylation sites in the cytoplasmic tail are in italics and marked by dashed lines. The sequence appears in the GenBank/EMBL sequence databases under the GenBank accession no.AF060883.
Nucleotide and deduced amino acid sequence of endomucin. Nucleotides and amino acids are numbered on the left. The putative N-terminal signal sequence, the transmebrane region, and the polyadenylation signal are underlined. The putative N-linked glycosylation sites are in italics and double underlined. The putative protein kinase C phosphorylation sites in the cytoplasmic tail are in italics and marked by dashed lines. The sequence appears in the GenBank/EMBL sequence databases under the GenBank accession no.AF060883.
Northern blot analysis.
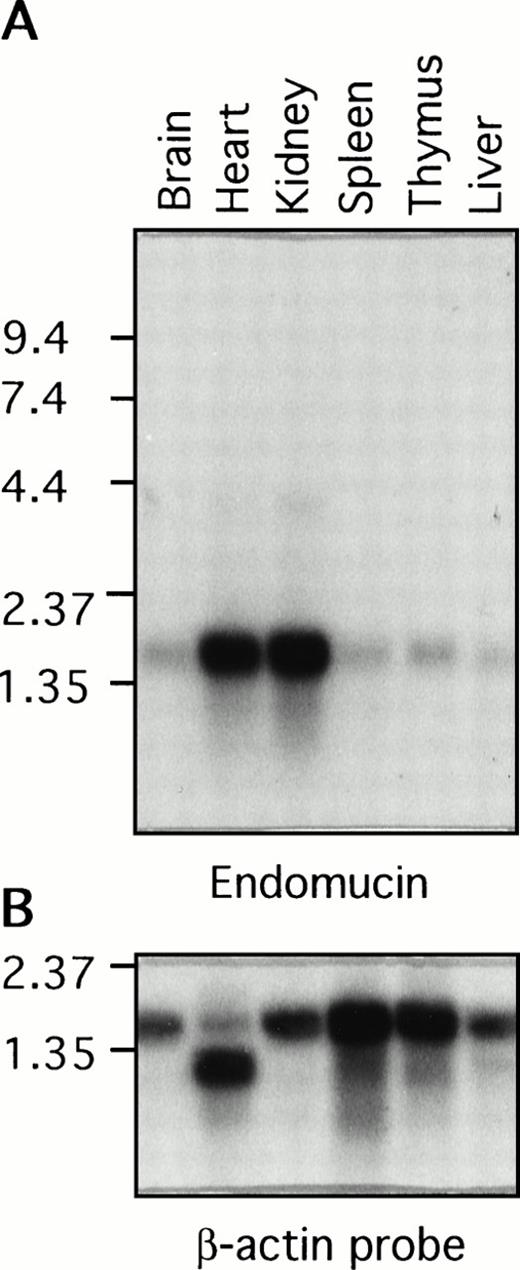
Poly (A)+ RNA isolated from various mouse tissues was examined by Northern blot analysis for the presence of endomucin transcripts (Fig 7A). A prominent endomucin transcript of approximately 1.7 kb was observed in all of the tissues examined, including the brain, heart, kidney, spleen, thymus, and liver. The transcript appeared to be highly abundant in the heart and kidney. This difference in the level of expression was not due to uneven loading of poly(A)+ RNA, because similar levels of the β-actin transcript were detected in all lanes (Fig 7B). However, it may be attributed in part to the high density of blood vessels in the heart and kidney. Overexposure of the blot showed weak hybridization with an approximately 4-kb mRNA in the heart and kidney, which may represent an incompletely spliced precursor. This broad distribution of the endomucin transcript was anticipated, because immunohistology showed constitutive expression of endomucin in all of the tissues examined (Figs 2, 3, and 4). The difference in the size between the 1.5-kb cDNA and the observed 1.7-kb mRNA transcript could be accounted for by additional 5′ untranslated sequence.
Distribution of the endomucin RNA expression. Northern blot analysis of endomucin. Northern blot containing RNA from various mouse tissues (as indicated) with 2 μg Poly(A)+ RNA per lane was hybridized with the 32P-labeled endomucin cDNA probe (A) or a probe for β-actin (B) as a control. Positions of size markers (in kilobases) are shown on the left.
Distribution of the endomucin RNA expression. Northern blot analysis of endomucin. Northern blot containing RNA from various mouse tissues (as indicated) with 2 μg Poly(A)+ RNA per lane was hybridized with the 32P-labeled endomucin cDNA probe (A) or a probe for β-actin (B) as a control. Positions of size markers (in kilobases) are shown on the left.
Sensitivity of endomucin to glycosidases and O-sialoglycoprotein endopeptidase.
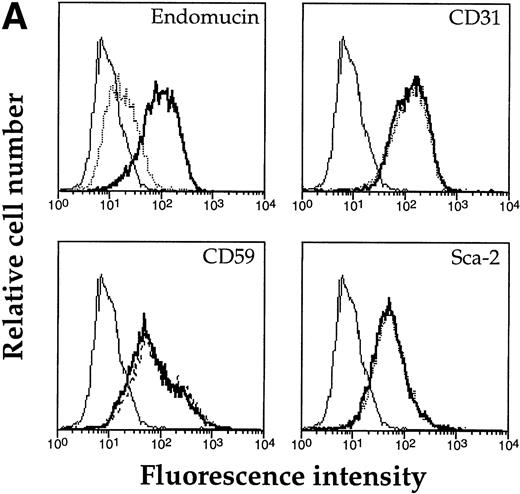
Based on the high content of serine and threonine residues of endomucin, we assumed that it would be highly O-glycosylated. O-sialoglycoprotein endopeptidase specifically cleaves highly O-glycosylated mucin-like molecules.47 48 To examine whether endomucin is sensitive to this endopeptidase, bEND.3 cells were treated with the enzyme and, subsequently, endomucin expression was analyzed by flow cytometry. As can be seen in Fig 8A, treatment was effective in producing a strong reduction of binding of the antiendomucin MoAb V.5C7. Similar results were observed for the endomucin-specific MoAbs V.7C7 and V.1A7 (data not shown). Untreated cells did not show any change. To control for the lack of contaminating proteases in the enzyme preparation, the cell surface expression of a number of glycoproteins (CD31, Sca-2, and CD59) was also analyzed and was found not to be altered on treatment with O-sialoglycoprotein endopeptidase. To exclude that the removal of the endomucin epitope was due to a contaminating neuraminidase in the O-sialoglycoprotein endopeptidase preparation, we treated immunoprecipitated endomucin with the endopeptidase. As shown in Fig 8B, endomucin was completely degraded by O-sialoglcoprotein endopeptidase, whereas treatment with neuraminidase did not alter the apparent molecular weight of endomucin. The control antigen, PECAM-1, was not sensitive to the endopeptidase. These data suggest that endomucin is modified by densely packed O-linked carbohydrate side chains rich in sialic acid.
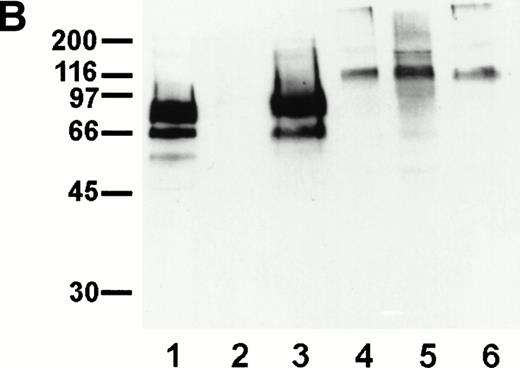
Sensitivity of endomucin to O-sialoglycoprotein endopeptidase. (A) bEND.3 cells were treated with O-sialoglycoprotein endopeptidase from Pasteurella haemolytica for 30 minutes (dotted line) or left untreated (thick line) and analyzed directly by flow cytometry as described for Fig 5A. The profiles show the binding of rat MoAb V.5C7 (endomucin), MoAb 390 (CD31), MoAb SM8 (CD59), and MoAb III.3A7 (Sca-2). Negative control staining was analyzed with MoAb MRC OX86 (thin line). (B) bEND.3 cells were surface biotinylated and subjected to immunoprecipitation with the antiendomucin antibody MoAb V.5C7 (lanes 1 through 3) or the anti–PECAM-1 antibody EA-3 (lanes 4 through 6). Immunoprecipitated antigens were either mock-treated (lanes 1 and 4), treated with O-sialoglycoprotein endopeptidase (lanes 2 and 5), or treated with neuraminidase (lanes 3 and 6). After the enzyme treatment, proteins were electrophoresed on a 10% polyacrylamide gel, transferred to nitrocellulose, and detected with peroxidase-conjugated streptavidin followed by enhanced chemoluminescence. Molecular mass markers (in kilodaltons) are indicated on the left.
Sensitivity of endomucin to O-sialoglycoprotein endopeptidase. (A) bEND.3 cells were treated with O-sialoglycoprotein endopeptidase from Pasteurella haemolytica for 30 minutes (dotted line) or left untreated (thick line) and analyzed directly by flow cytometry as described for Fig 5A. The profiles show the binding of rat MoAb V.5C7 (endomucin), MoAb 390 (CD31), MoAb SM8 (CD59), and MoAb III.3A7 (Sca-2). Negative control staining was analyzed with MoAb MRC OX86 (thin line). (B) bEND.3 cells were surface biotinylated and subjected to immunoprecipitation with the antiendomucin antibody MoAb V.5C7 (lanes 1 through 3) or the anti–PECAM-1 antibody EA-3 (lanes 4 through 6). Immunoprecipitated antigens were either mock-treated (lanes 1 and 4), treated with O-sialoglycoprotein endopeptidase (lanes 2 and 5), or treated with neuraminidase (lanes 3 and 6). After the enzyme treatment, proteins were electrophoresed on a 10% polyacrylamide gel, transferred to nitrocellulose, and detected with peroxidase-conjugated streptavidin followed by enhanced chemoluminescence. Molecular mass markers (in kilodaltons) are indicated on the left.
To examine this further, we isolated endomucin from cell surface biotinylated bEND.3 cells by immunoprecipitation. While still bound to the affinity matrix, aliquots of the antigen were separately treated with one of the enzymes (endoglycosidase F, O-glycosidase, or sialidase) or with sialidase and subsequently with O-glycosidase. The antigen was then electrophoresed, transferred to a filter, and detected by peroxidase-conjugated streptavidin and enhanced chemoluminescence. As shown in Fig 9A endoglycosidase F reduced the apparent molecular weight of endomucin to 60 kD. As expected, no effect was seen if the molecule was treated with O-glycosidase alone, because this enzyme is only active after the removal of sialic acid. After pretreatment with sialidase, O-glycosidase digestion could reduce the apparent molecular weight of endomucin to 45 kD. Removal of sialic acid by sialidase did not affect the apparent molecular weight of endomucin. We conclude that the antigen carries N-linked as well as O-linked carbohydrate side chains.
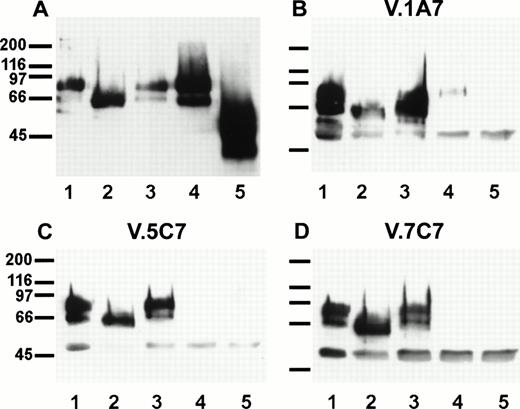
Sensitivity of the antibody epitopes on endomucin to treatment with glycosidases. bEND.3 cells either surface biotinylated (A) or unlabeled (B, C, and D) were subjected to immunoprecipitations with the antiendomucin MoAb V.5C7 (A and C), V.1A7 (B), or V.7C7 (D). Immunoprecipitated endomucin was either mock-treated (lane 1) or treated with endoglycosidase F (lane 2), O-glycosidase (lane 3), neuraminidase (lane 4), or neuraminidase and O-glycosidase (lane 5). After the enzyme treatment, endomucin was electrophoresed on a 10% polyacrylamide gel, transferred to nitrocellulose, and detected either with peroxidase-conjugated streptavidin followed by enhanced chemoluminescence (A) or by immunoblotting with the anti endomucin MoAb V.1A7 (B), V.5C7 (C), and V.7C7 (D). Molecular mass markers (in kilodaltons) are indicated on the left.
Sensitivity of the antibody epitopes on endomucin to treatment with glycosidases. bEND.3 cells either surface biotinylated (A) or unlabeled (B, C, and D) were subjected to immunoprecipitations with the antiendomucin MoAb V.5C7 (A and C), V.1A7 (B), or V.7C7 (D). Immunoprecipitated endomucin was either mock-treated (lane 1) or treated with endoglycosidase F (lane 2), O-glycosidase (lane 3), neuraminidase (lane 4), or neuraminidase and O-glycosidase (lane 5). After the enzyme treatment, endomucin was electrophoresed on a 10% polyacrylamide gel, transferred to nitrocellulose, and detected either with peroxidase-conjugated streptavidin followed by enhanced chemoluminescence (A) or by immunoblotting with the anti endomucin MoAb V.1A7 (B), V.5C7 (C), and V.7C7 (D). Molecular mass markers (in kilodaltons) are indicated on the left.
Similar experiments were performed with unlabeled bEND.3 cells and immunoprecipitated and glycosidase-treated endomucin was subsequently examined in immunoblots with the three MoAbs V.1A7, V.5C7, and V.7C7 (Fig 9B, C, and D). Whereas the epitopes of all three antibodies were resistant to endoglycosidase F, they were highly sensitive to sialidase treatment of the antigen. The weak detectable signal with V.1A7 after sialidase treatment of the antigen is probably due to incomplete digestion. Thus, it is possible that the three antibodies recognize carbohydrate epitopes on endomucin. Alternatively, removal of sialic acid might affect the overall folding of the mucin and thereby might indirectly destroy the epitopes.
DISCUSSION
Searching for novel endothelial cell surface antigens, we have generated three MoAb against a novel membrane glycoprotein. All three antibodies specifically recognized endothelial cells in all mouse tissues examined. Cloning showed a sequence with no significant homology to any known glycoprotein. The high content of serine and threonine residues suggested strong O-glycosylation of the protein backbone, a characteristic feature of mucin-like glycoproteins. This could be verified by the large decrease in molecular weight upon O-glycosidase treatment as well as by the sensitivity of the glycoprotein to O-sialoglycoprotein endopeptidase, a protease with specificity for sialomucins. Thus, we conclude that the novel endothelial-specific cell surface glycoprotein is a sialomucin, which we suggest should be named endomucin.
The tissue distribution of endomucin suggests that it serves a specific function important in endothelial cells. Although endomucin is expressed on endothelial cells of all tissues examined, it is not expressed on all types of endothelial cells. Most importantly, it is not expressed on HEV of secondary lymphoid organs such as peripheral lymph nodes, mesenteric lymph nodes, and Peyer’s patches. These are the sites where lymphocytes leave the blood stream and enter into lymphatic tissue. Lymphocyte trafficking at these sites of the vascular system is very efficient. The lack of endomucin at these sites might lead us to speculate that endomucin is an antiadhesive molecule that suppresses leukocyte endothelial interactions in other vascular beds and whose absence in HEV might increase the probability of lymphocyte endothelial interactions at these sites. This would be in line with the antiadhesive effects of other sialomucins such as CD43,28epiglycanin,8 and the sialomucin complex formed of the mucins ASGP-1 and ASGP-2.9 Such a negative regulatory effect of endomucin on cell adhesion would also be in line with the absence of endomucin on arteriolar endothelium, because the higher shear forces in these vessels are already sufficient to suppress unwanted leukocyte endothelial interactions.
Although an antiadhesive effect of endomucin is still speculative, it is intriguing that this novel antigen is found in all venules but is absent from HEV. In contrast, several other antigens are known to be specifically expressed in endothelium of HEV but not other endothelia. Examples are the carbohydrate epitope MECA79 (also called PNAd),49 the vascular addressin MAdCAM-1,25 and the selectin-binding glycoforms of GlyCAM-120 and CD34.21,50 All of these molecules or epitopes are thought to be involved in lymphocyte trafficking. The secreted protein hevin, which is more prominent in HEV than in flat-walled vessel endothelium,51 52 was shown to modulate the binding of high endothelial cells to components of the basement membrane, a process that could possibly influence features of HEV endothelium that facilitate lymphocyte migration.
The sensitivity of endomucin to sialidase and O-glycosidase as well as to O-sialoglycoprotein endopeptidase indicates the high content of this antigen in sialic acid-carrying O-linked carbohydrate side chains, the common feature of sialomucins. Surprisingly, sialidase treatment of endomucin abolished the binding of each of the three MoAb against this antigen. This argues for a direct or indirect role of the carbohydrates in the generation of the antibody epitopes. Although it cannot be excluded, it is unlikely that the epitopes of all three antibodies are exclusively formed by carbohydrates, because each of the three antibodies only recognizes endomucin and no other glycoprotein. The effect of sialidase on the expression of the antibody epitopes could well be due to indirect effects on the conformation or accessibility of polypeptide epitopes. This was found for several antibody epitopes of antibodies against the sialomucin CD43.3 The binding of four antibodies against CD43 was abolished upon treatment of the antigen with neuraminidase, although three of these antibodies recognized bacterial, nonglycosylated fusion proteins of CD43 in Western blots. Thus, these antibodies, possibly similar to the three antiendomucin antibodies, recognize epitopes formed by the protein backbone, which are dependent on the sialylation of the antigen, but which are detectable in Western blots, ie, in the SDS-extended conformation of the protein.
Major differences in O-linked glycosylation structures have been described for leukocyte populations, with the complexity of the O-linked structures altering according to the activation states.4 It has been shown, for example, that, whereas simple O-linked oligosaccharide structures are found on CD43 on resting T cells, more complex structures are displayed on activated T cells.53,54 The larger forms of CD43 found on activated T lymphocytes that resulted from the modification of O-linked oligosaccharide could be resolved by SDS-PAGE.55 56 In contrast to these data on CD43, we did not observe any changes in molecular weight of endomucin upon activation of bEND.3 cells with the alarm cytokines IL-1β, IFN-γ, or TNF-α. This might indicate that substantial alterations in the O-glycan substitutions are not inducible by the treatment of bEND.3 cells with these cytokines.
Endomucin is a novel endothelial-specific sialomucin. It is the first constitutively expressed endothelial cell surface protein that is found on all venules but that is absent from HEV, the specialized site for most efficient lymphocyte trafficking. This could argue for an antiadhesive function of endomucin, as it has been demonstrated for other sialomucins. Alternatively, it is possible that endomucin is differently glycosylated on HEV-endothelia. Indeed, the binding of each of our antiendomucin antibodies is sensitive to changes in the glycosylation of endomucin. A different HEV-specific glycosylation might enable endomucin to bind to leukocyte lectins in a manner similar to that shown for the selectin ligands GlyCAM-1 or CD34. It is also possible that endomucin, like other sialomucins,11-13 might be involved in signal transduction. The presence of three putative protein kinase C phosphorylation sites in the cytoplasmic tail indicates that endomucin might have the capacity to be a signaling molecule. Further studies are necessary in the future to elucidate the physiological function of this novel endothelial sialomucin.
S.M.M. and U.S. contributed equally to the work.
Supported in part by a grant of the Interdisziplinäres Klinisches Forschungszentrum (IKF) Münster to D.V. and a grant from the Wellcome Trust (UK) to D.L.S.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Dietmar Vestweber, PhD, Institute of Cell Biology, ZMBE, University of Münster, Von-Esmarch-Str. 56, D-48149 Münster, Germany; e-mail:vestweb@uni-muenster.de.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal