Abstract
Although hepatitis C virus (HCV) mainly affects hepatocytes, infection is widespread and involves immunologically privileged sites. Whether lymphoid cells represent further targets of early HCV infection, or whether other cells in the hematopoietic microenvironment may serve as a potential virus reservoir, is still unclear. We studied whether pluripotent hematopoietic CD34+ cells support productive HCV infection and can be used to establish an in vitro infection system for HCV. Six patients were selected as part of a cohort of HCV chronic carriers who developed a neoplastic disease. Reverse transcriptase-polymerase chain reaction (RT-PCR) and branched DNA signal amplification assays were used to detect and quantitate HCV RNA in extracted nucleic acids from purified bone marrow and peripheral blood CD34+ cells. Direct in situ RT-PCR, flow cytometry analysis, and immunocytochemistry were applied to demonstrate specific viral genomic sequences and structural and nonstructural virus-related proteins in intact cells. Results indicated that both positive and negative HCV RNA strands and viral proteins were present in CD34+ cells from all HCV-positive patients and in none of the controls. Additional experiments showed that a complete viral cycle took place in CD34+ cells in vitro. Spontaneous increases in viral titers indicated that virions were produced by infected hematopoietic progenitor cells. To further define the cellular tropism, we attempted to infect CD34+ cells in vitro. We were unable to demonstrate viral uptake by cells. These findings suggest that HCV replication can occur in the early differentiation stages of hematopoietic progenitor cells, and that they may be an important source of virus production.
© 1998 by The American Society of Hematology.
THE CLINICAL COURSE of a viral infection reflects the fluctuating balance between the capacity of the virus to replicate and spread and the ability of the host to eliminate it. Persistence of a virus is evidence of its ability to escape the host’s immune surveillance through the exploitation of several strategies, including integration into the host genome,1 silencing of viral gene expression,2 inhibition of antigen processing and presentation,3,4 synthesis of proteins homologous to known immune regulatory molecules,5 mutations that either inhibit viral responsiveness to antiviral cytokines or preclude recognition by neutralizing antibodies3 or else modify residues that are critical for recognition by the major histocompatibility complex (MHC),6 or the T-cell receptor.7 A further defense mechanism is direct infection of immunologically priviliged sites that cannot be reached by virus-specific T cells or that do not express class I or costimulatory molecules.8
Hepatitis C virus (HCV) infection is characterized by its striking tendency to become chronic in a high proportion of patients. A 70% to 80% range of persistent infection has been documented by detection of HCV RNA.9 This persistence seems attributable to HCV’s ability to mutate rapidly and exist simultaneously as a series of related but immunologically distinct variants (quasi-speciesnature).10,11 Several studies have provided convincing evidence for the occurrence of HCV productive infection in both bone marrow (BM) and peripheral blood mononuclear cells (PBMC)12and in lymphoid organs,13 which may thus serve as major HCV reservoirs.
The susceptibility of B-, T-, and monocyte/macrophage cell lines to HCV infection has been demonstrated by the observation of polymerase chain reaction (PCR)-driven HCV specific sequences.12 These findings have been corroborated by in situ hybridization techniques showing both positive- and negative-polarity HCV genomic strands in circulating and/or BM-recruited mononuclear cells.14-16 However, it remains unclear whether PBMC are the principal targets of early HCV infection in hematologic tissue, or whether other cells within the hematopoietic microenvironment may also serve as potential virus sites. In particular, it would be of interest to determine whether early hematopoietic CD34+ progenitor cells17 are also susceptible to infection.
Data from the present study indicate that hematopoietic progenitor cells may indeed be an additional HCV infection site and provide a better understanding of the cellular tropism of HCV.
MATERIALS AND METHODS
Patients.
Six patients were recruited from a cohort of consecutive human immunodeficiency virus (HIV)-negative patients referred to the Department of Biomedical Sciences and Clinical Oncology of the University of Bari because of chronic HCV infection and a neoplastic disease. In four of them, non-Hodgkin’s lymphoma (NHL) was diagnosed from routinely stained sections prepared from formalin-fixed paraffin-embedded surgical biopsy samples and classified according to the International Working Formulation.18 Immunophenotypic analyses using routine procedures, including the study of antigens associated with B cells, T cells, plasma cells, natural killer cells, monocytes/macrophages, and granulocytes, were performed on snap-frozen tissues. Light chain restriction of surface IgG, IgA, IgM, and DNA and RNA extraction studies were performed by standard procedures. DNA analyses included Southern blotting and molecular hybridization to detect Ig gene rearrangement; assessment of B-cell clonality by VDJ PCR19; sensitive analysis for Epstein-Barr virus and human herpes virus-6 genomes by PCR.20 NHL patients underwent liver biopsy and iliac crest BM needle biopsy. In the remaining two patients, colon carcinoma and gastric carcinoma were diagnosed respectively, and liver biopsies were performed during laparotomy.
All were hepatitis B surface antigen (HBsAg) and hepatitis B virus (HBV)-DNA negative, although four patients had antibodies to hepatitis B surface (HBs) and hepatitis B core (HBc) and one had antibodies to HBc alone. Organ- and nonorgan-specific autoantibodies were not detected. Two patients had received a blood transfusion many years earlier.
In all patients, clinical evaluation and biochemical and hematologic testing was performed at 3-month intervals during the follow-up for liver disease (mean, 9.1 ± 1.86 years). The presence of anti-HCV antibodies was confirmed by recombinant immunoblot assay (RIBA-2; Ortho Diagnostic Systems, Raritan, NJ). At the time of diagnosis of NHL, two patients were receiving interferon treatment (9 MU/wk). The controls were four patients with NHL and negative serology for anti-HCV antibodies and HCV RNA. The main clinical, histologic, and virologic features of patients and controls are summarized in Table 1.
Clinical, Histological, and Virological Parameters in HCV Chronic Carriers and Controls
| Patients . | Age/ Sex . | Neoplastic Disease . | Liver Disease . | HCV Status . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antibody Pattern (5-1-1/c100-3/c33/c22) . | PCR-Driven HCV RNA . | ||||||||||||||
| Type . | Histology . | Working Formulation . | Staging . | Histology . | Duration (yr) . | Serum/ Genotypes . | Lymph Nodes . | Liver Tissue . | BM . | ||||||
| 1 | 66/F | NHL | Follicular small cleaved cells | B | IIB | CH | 12 | − | − | + | + | +/1b | + | + | + |
| 2 | 77/F | NHL | Large cells immunoblastic | H | IIA | CH | 17 | − | − | − | + | +/2a | + | + | + |
| 3 | 59/M | NHL | Mixed: Small cleaved/large cells | G + A | IA | CIR | 21 | + | + | + | + | +/1b | + | + | + |
| 4 | 65/M | NHL | Diffuse large cells | G | IIA | CH | 11 | − | − | + | + | +/1b | + | + | + |
| 5 | 56/M | Colon carcinoma | Adenocarcinoma | — | T2, N2, M0 | CIR | 25 | − | + | + | + | +/1b | ND | + | ND |
| 6 | 61/M | Gastric carcinoma | Adenocarcinoma | — | T2, N1, M0 | CIR | 22 | + | + | + | + | +/1b | ND | + | ND |
| Controls | |||||||||||||||
| 1C | 59/F | NHL | Diffuse large cells | G | IIB | NNL | — | − | − | − | − | − | − | − | − |
| 2C | 69/F | NHL | Small lymphocytic | A | IB | ST | — | − | − | − | − | − | − | − | − |
| 3C | 75/M | NHL | Diffuse large cells | G | IIA | ST | — | − | − | − | − | − | − | − | − |
| 4C | 68/M | NHL | Follicular small cleaved cells | B | IIA | NNL | — | − | − | − | − | − | − | − | − |
| Patients . | Age/ Sex . | Neoplastic Disease . | Liver Disease . | HCV Status . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antibody Pattern (5-1-1/c100-3/c33/c22) . | PCR-Driven HCV RNA . | ||||||||||||||
| Type . | Histology . | Working Formulation . | Staging . | Histology . | Duration (yr) . | Serum/ Genotypes . | Lymph Nodes . | Liver Tissue . | BM . | ||||||
| 1 | 66/F | NHL | Follicular small cleaved cells | B | IIB | CH | 12 | − | − | + | + | +/1b | + | + | + |
| 2 | 77/F | NHL | Large cells immunoblastic | H | IIA | CH | 17 | − | − | − | + | +/2a | + | + | + |
| 3 | 59/M | NHL | Mixed: Small cleaved/large cells | G + A | IA | CIR | 21 | + | + | + | + | +/1b | + | + | + |
| 4 | 65/M | NHL | Diffuse large cells | G | IIA | CH | 11 | − | − | + | + | +/1b | + | + | + |
| 5 | 56/M | Colon carcinoma | Adenocarcinoma | — | T2, N2, M0 | CIR | 25 | − | + | + | + | +/1b | ND | + | ND |
| 6 | 61/M | Gastric carcinoma | Adenocarcinoma | — | T2, N1, M0 | CIR | 22 | + | + | + | + | +/1b | ND | + | ND |
| Controls | |||||||||||||||
| 1C | 59/F | NHL | Diffuse large cells | G | IIB | NNL | — | − | − | − | − | − | − | − | − |
| 2C | 69/F | NHL | Small lymphocytic | A | IB | ST | — | − | − | − | − | − | − | − | − |
| 3C | 75/M | NHL | Diffuse large cells | G | IIA | ST | — | − | − | − | − | − | − | − | − |
| 4C | 68/M | NHL | Follicular small cleaved cells | B | IIA | NNL | — | − | − | − | − | − | − | − | − |
Abbreviations: CH, chronic hepatitis; NNL, near normal liver; CIR, cirrhosis; ST, steatosis; ND, not determined.
The study was approved by the ethical committee of the University of Bari Medical School and informed consent was obtained from all patients and controls.
CD34+ cell mobilization, isolation, and purification.
CD34+ cell mobilization was achieved by injecting patients with granulocyte colony-stimulating factor (G-CSF), given at a daily dose of 10 μg/kg. Injections were repeated for 6 to 7 days before the apheresis and BM aspiration. Leukocyte subsets were monitored frequently (every 1 to 3 days) until the end of the apheresis procedure. In all of the cases, leukapheresis was started when the absolute number of circulating CD34+ cells exceeded 10/μL. After volume reduction by centrifugation, each leukapheresis product was frozen in 1-3 DF 700 freezing bags (Gambro, Leuven, Belgium). The freezing medium consisted of 4% (wt/vol) human serum albumin and 7.5% (vol/vol) dimethylsulfoxide in 0.9% NaCl. All bags were frozen in a controlled rate freezer and stored in liquid nitrogen (−196°C).
For collection of peripheral blood CD34+ cells, an automated dual chamber computer-assisted continuous flow blood cell separator (Baxter CS 3000; Baxter Healthcare Co, Deerfield, IL) was used, using a preattached solution in a closed system kit. Common parameters included blood flow (60 mL/min), anticoagulation (ACD-A 500 to 700 mL), volume of processed blood (7.5 L), and rotating speed (1,400 rpm).
BM mononuclear cells and aliquots of the leukapheresis products were washed with RPMI 1640, separated by density gradient centrifugation over Ficoll-Hypaque (Pharmacia, Uppsala, Sweden), washed twice, suspended in Iscove’s modified Dulbecco’s medium (IMDM)/20% fetal calf serum (FCS), and cultured overnight to remove adherent cells.
The CD34+ mononuclear cell fraction was isolated by sequential immunomagnetic bead selection using the Dynal CD34 progenitor cell selection system (code 113.01/113.02 from Dynal A.S., Oslo, Norway). This reagent contains paramagnetic-polystyrene beads coated with monoclonal antibody (MoAb) 561 specifically directed against class III epitope on the CD34 antigen.21 Flow cytometry analysis showed that CD34 selection in the final product ranged from 81% to 96% with a mean of 92.3%.
CD34+ cell cultures and lymphocyte subpopulations.
Adherent BM stromal cells were recovered after enzymatic digestion, washed twice with RPMI 1640/10% FCS in medium, and seeded into flasks. They were then expanded by several passages. The medium was changed every week. CD34+ cells (0.5 × 106 per well) were added to the washed, stromal feeder layer and cultured for a further 2 to 4 weeks in IMDM/20% FCS. Control cultures included no stroma or stroma alone. Cultures were half-refed with complete medium twice weekly.
Positive selection of CD20+, CD4+, and CD8+ cell fractions was performed using the same type of magnetic beads (Dynal) already described, coated with the corresponding MoAbs.
Flow cytometry analysis.
CD34+ cell cultures were obtained by gentle aspiration, washed twice in phosphate-buffered saline (PBS), and stained with the following murine MoAbs against HCV: (1) anti-c22-3 antibody (clone 4,6 E7-F6), recognizing amino acids (AA) 29-43 of the core protein at a concentration of 0.9 μg/mL; (2) antienvelope glycoprotein 2 (E2)/nonstructural protein 1 (NS1) antibody (clone 15-B4-C7), recognizing a conformational antigen of E2/NS1-encoded protein of HCV-1 strain at a concentration of 1.5 μg/mL; (3) anti-c33c antibody (clone 1,4 G11-B4G10), recognizing a structural determinant in the NS3-encoded protein at a concentration of 1.0 μg/mL; (4) a mixture of three anti-c100-3 antibodies, namely clone 2C4G3 recognizing AA 1690-1696; clone 22A5B12 recognizing AA 1694-1711, and clone 20A6F3 recognizing alinker sequence at the junction of superoxide dismutase (SOD) and HCV sequences in the NS4-encoded protein. Protein concentration in the working mixture of each MoAb was 1.0 μg/mL; (5) anti-NS5 antibody (clone 1A6B7), recognizing AA 2278-2310 of NS5-encoded protein was used at a protein concentration of 1.2 μg/mL. Immunochemical characteristics and fine specificities of these reagents have been reported elsewhere.13 22
After a 1-hour incubation at 37°C, the cells were pelleted, washed with PBS, and reincubated with fluorescein isothiocyanate (FITC)-conjugated antimouse immunoglobulin F(ab′)2fragment (Boehringer Mannheim, Mannheim, Germany; code 1.214.616). A parallel number of cells fixed with 4% paraformaldehyde were analyzed for each unfixed sample.
CD34+ immunophenotype was detected by a pool of phycoerythrin (PE)-labeled anti-CD34 MoAbs (Immunotech, Marseille, France; code 1459), which contained optimized proportions of the following three MoAbs: QBend-10 directed to CD34 class II epitopes and Immu-33 and Immu-409 specific for different CD34 class I epitopes. Omission of the first antibody in indirect assay or PE-conjugated isotype-matched nonspecific mouse immunoglobulins were used as controls in each culture sample. CD34+ mononuclear cells recovered from HCV− patients acted as a further control. Flow cytometric analysis was performed with a FACScan cytometer (Becton Dickinson, Sunnyvale, CA).
For double-staining, cell suspensions were stained with the following antibody combinations: anti-CD34 conjugated with PE (Immunotech) and anti-HCV (E2/NS1 protein, clone 15-B4-C7) conjugated with FITC.
Immunocytochemical staining.
After harvesting, CD34+ cells were washed twice in PBS, resuspended at 1 × 105/mL, dried onto glass coverslips, and fixed for 15 minutes at −20°C with 100% acetone followed by 100% methanol. After extensive washings, adherent stromal cells were similarly fixed. After rehydration, cells were incubated with anti-HCV MoAbs (see above) and then with affinity-purified sheep IgG, F(ab′)2 fragment to mouse Ig conjugated to alkaline phosphatase (APh) (Boehringer, code 1.198.661). Unbound antibody was washed off by immersing the slides in 0.1 mol/L Tris-HCl buffer pH 7.4 containing 0.15 mol/L NaCl, followed by 0.1 mol/L Tris-HCl pH 9.5 containing 0.1 mol/L NaCl and 0.05 mol/L MgCl2. Levamisole (0.15 mg/mL) was added to the APh substrate (4-chloro-2-methylbenzenediazonium/3-hydroxy-2-naphthoic acid 2,4-dimethyl-anilide phosphate) at 1 mg/mL to block endogenous APh.
Specificity of HCV staining patterns was assessed on selected positive samples before and after absorption of the probe with recombinant proteins (1 mg/mL) derived from the equivalent structural and nonstructural regions of HCV genome. Recombinant SOD and HBV-associated antigens (HBsAg, HBcAg) and hepatitis A virus (HAV) antigen were included in these experiments as controls. Efficiencyof absorption was shown by testing the probes before and after absorption for antigen reactivities with commercially available RIBA. Replacement of primary antibody with an irrelevant antibody (mouse antihuman chorionic gonadotropin [HCG] MoAb) was also performed.
CD34 antigen was detected both directly by incubating the section with polystyrene beads coated with anti-CD34 MoAb (Dynal A.S.), and indirectly with an anti-CD34 MoAb (clone QBend-10, Immunotech) followed by antimouse Ig/APh (Boehringer Mannheim). Omission of the first antibody and mouse isotypic antibody recognizing an irrelevant epitope (IgG1 anti-HCG antibody) acted as the controls.
RT-PCR assay for HCV RNA and HCV genotyping.
RNA was extracted according to Chomczynsky and Sacchi.23The RNA pellet was washed in 75% ethanol and resuspended in 20 μL of diethyl-pyrocarbonate–treated autoclaved H2O. The total RNA yield was determined by spectrophotometry and processed for HCV RNA detection by RT-PCR assay, as described elsewhere.24Primers were selected from the 5′-noncoding (NC) region of the HCV genome.
To characterize the HCV genotypes, biotinylated universal primers referred to the 5′-NC region were used to amplify and hybridize to genotype-specific probes (Line Probe assay, LiPA HCV II; Innogenetics, Brussels, Belgium). Each sample was tested in triplicate, and appropriate positive and negative controls were always included.
Negative strand-specific RT-PCR.
Negative-polarity strand HCV RNA sequences, presumably representing replicative intermediates, were detected by RT-PCR with two sets of primers aimed at specifically priming and amplifying HCV negative strand RNA.25 One of them contained at the 5′ end atag sequence unrelated to HCV genome (TAG-based assay). The second was located in the nucleocapsid of viral genome outside the highly structured 5′-NC region (CAP-based assay). Conditions for the amplification of negative strand templates, their specificity, and sensitivity were those described for the “TAG-based” assay by Lanford et al26 and for the CAP-based assay by Lerat et al.27
HCV− strands were analyzed in CD34+ cells obtained from patients 5 and 6. Following the above-described Dynal protocol, CD34+ cell fractions were further purified with a microbeads system (MACS; Miltenyi Biotec Inc, Auburn, CA). After removal of magnetic beads from CD34+ cells using the detachment system (Detach-a-Bead CD34 No. 113.01/02; Dynal), cells were relabeled specifically with supermagnetic MACS microbeads coated with anti-CD34 MoAb (clone QBend-10) and passed through a separation column placed in a strong permanent magnet. Thus, only the magnetically labeled CD34+ cells were retained in the column and then recovered after removal of the column from the magnetic field. By sequentially applying these two methodologies, which combine the use of anti-CD34 antibodies directed against different epitopes on CD34 antigen, almost 100% pure CD34+ cells were obtained.
HCV RNA quantitation.
HCV RNA was quantitated by signal amplification using branched DNA (bDNA) in a sandwich hybridization assay,28 according to the manufacturer’s instructions (Quantiplex HCV RNA, Version 2.0; Chiron Corp, Emeryville, CA). Duplicate 50-μL samples were added to the wells in which lysis, hybridization, capture, and signal amplification occurred. Synthetic oligonucleotides, in a mixture which included probes that mediated capture and probes that bound to the bDNA amplifier molecule, hybridized equally well to the highly conserved 5′-NC and core regions of the HCV RNA of all known genotypes, thereby capturing the RNA molecules onto the surface of a microwell plate and linking to synthetic bDNA molecules added to the well. Multiple copies of an APh-linked synthetic probe hybridized to the immobilized complex, thereby amplifying the target signal. Detection was achieved by incubating the complex with a chemiluminescent substrate (dioxetane) and measuring light emission, which was proportional to the concentration of target nucleic acid in the specimen. The standard curve was based on a diluted sample from a patient with HCV infection whose serum had been quantitated by comparison with synthetic HCV RNA. Because the values assigned to the HCV RNA standards were based on comparison with highly purified RNA transcript covering the first 3,200 nucleotides from the 5′ end of the HCV genome, the results were expressed as genomic equivalents per mL (Eq/mL) rather than genomic copies. The lower limit of sensitivity of this assay was 0.2 million Eq/mL (MEq/mL). HCV genomic equivalents were divided by the number of the cells and results expressed as HCV Eq/cell when considering HCV RNA extracted from cultured cells.
Direct in situ RT-PCR (IS-RT-PCR).
CD34+ cells, washed with PBS to remove all traces of culture medium, were centrifuged onto silane-coated microscope slides (Perkin-Elmer, Foster City, CA; code: 804-0502). A mild acid hydrolysis was performed by incubating the slides in 0.02 mol/L HCl for 10 minutes. After repeated washings in PBS, cells were immersed in 0.01% Triton-X 100 in PBS for 2 minutes, transferred to a Coplin jar containing proteinase K (Boehringer Mannheim) in 0.1 mol/L Tris-HCl pH 7.5, 5 mmol/L EDTA, and placed in a microwave oven for 10 minutes. Microwaves were pulsed through the jar until boiling for 5 minutes. To further reduce the levels of endogenous APh, the slides were dipped in acetic acid at 4°C for 1 minute. They were then transferred to PBS and digested with RNase-free DNase (Boehringer Mannheim) at 37°C at 1 U/mL overnight. After washings in PBS and nuclease-free water, the slides were dehydrated through graded ethanols to 100%. RT-PCR was performed with one enzyme under a single set of buffer conditions using DNA polymerase from Thermus thermophilus (Tth) (Perkin-Elmer; code N808-0097), which possesses RT activity in the presence of manganese. RT-PCR mixture contained 200 mmol/L (each) deoxyribonucleoside triphosphates (deoxyadenosine triphosphate [dATP], deoxycytidine triphosphate [dCTP], deoxyguanosine triphosphate [dGTP], and deoxythymidine triphosphate [dTTP]), 2.5 mmol/L digoxigenin (DIG)-linked 11-deoxyuridine 5′-triphosphate (dUTP) (Boehringer Mannheim), 2.5 mmol/L manganese acetate [Mn(OAc)2], 5 U recombinant Tth DNA polymerase, 2.5 U RNasin (Promega, Madison, WI), 150 mmol/L of each primer (JHC 51, antisense: CCCAACACTACTCGGCTA; JHC93 B, sense: ACCATGAATCACTCCCCT) from the highly conserved 5′-NC of HCV genome,29 and 1× RT-PCR buffer (5× RT-PCR buffer consists of 250 mmol/L Bicine, 575 mmol/L potassium acetate, 40% glycerol, pH 8.2; Perkin-Elmer, code 808-0177). The reactions were performed in an Omnislide Thermal Cycler (Hybaid Limited, Middlesex, UK), and slides were assembled with amplicover clips (Perkin-Elmer; code N804-0501).
RT was allowed to proceed for 60 minutes at 70°C, followed by 1-minute incubation at 95°C to facilitate denaturation. PCR amplification was performed with two cycles at 95°C for 2 minutes followed by 38 cycles at 95°C for 1 minute, and a combined annealing and extension step for 1 minute at 60°C and 72°C. A final extension step of 7 minutes at 72°C followed the last PCR cycle. At the end of the cycling, the slides were washed in 2× SSC (1× SSC: 0.15 mol/L sodium chloride and 0.015 mol/L sodium citrate) at 60°C for 5 minutes and then in PBS for 2 minutes. Slides were dipped in 0.1 mol/L Tris-HCl, pH 7.5, 0.1 mol/L NaCl, 2 mmol/L MgCl2, 0.05% Triton-X 100 for 30 minutes. DIG-labeled PCR product was detected by incubating the slides with anti-DIG antibody conjugated with APh (Boehringer Mannheim) and the signal detected by nitroblue tetrazolium (NTB; 340 μg/mL) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP; 170 μg/mL) (Boehringer Mannheim) substrate.
IS-RT-PCR procedure included the following controls: (1) CD34+cells from HCV RNA-negative patients; (2) RT-PCR solution phase on nucleic acids extracted from CD34+ cells from each patient; (3) RT-PCR on cell suspensions containing a known number of CD34+ cells from HCV RNA-positive patients mixed with CD34+ cells from HCV RNA-negative patients; (4) addition of RNase to remove target RNA; (5) omission of specific primers, enzyme, nucleotides, Mn(OAc)2, or anti-DIG antibody; (6) reference control gene-specific primers to amplify 308-bp sequence complementary to interleukin-1α (IL-1α) site insert from pAW109-derived cDNA (Perkin-Elmer; code N0808-0037); (7) IS-RT-PCR efficiency, analyzed by targeting β-actin mRNA; sense: 5′ACACTGTGCCCATCTACCTAGGGG3′; antisense: 5′ATGATGGAGTTGAAGGTAGTTTCGTGGAT3′.
In vitro HCV infection.
Primary CD34+ hematopoietic precursors from either BM or peripheral blood were seeded at 1 × 105 cells per well and inoculated with undiluted serum #0715 containing 2 × 10 HCV MEq/mL (genotype 1b). This inoculum was selected because it contained a high genomic titer and was capable of establishing a productive HCV infection in MOLT-4 cells, a human T-cell line (American Type Culture Collection, Rockville, MD). A total of 1 mL of #0715 inoculum together with 3 mL of fresh culture medium was incubated for 24 hours at 37°C. Cells were then washed and fresh medium was added, followed by continued incubation with medium changes at 4-day intervals. At various times during the culture period, culture medium (0.25 mL) and cells (105 cells) were collected for HCV RNA detection and titration.
RESULTS
Experiments were designed to detect the presence of HCV RNA in CD34+ cells. Suspensions containing positively-selected CD34+ cells from BM and/or peripheral blood were collected during a single apheresis procedure, and extracted RNA was processed to demonstrate sequences of HCV genome.
Data reported in Table 2 provide evidence that all patients showed HCV genomic sequences in CD34+cells. Specifically, confirmation of negative-polarity strand HCV RNA likely representing viral replicative intermediates was examined in two highly purified CD34+ cell products obtained from patients 5 and 6, which were found to be 99.7% and 99.1% phenotypically pure, respectively. Nested PCR using capsid-derived primers detected a specific product at 1 × 103 HCV RNA Eq. Its sensitivity appeared slightly reduced at 5 × 103 HCV RNA Eq in the presence of 1 × 106 Eq of positive strand templates, whereas it was not significantly influenced by the addition of cellular RNA to the system. By the CAP-based assay, negative strand intermediates were demonstrated in CD34+cells of both patients (Fig 1). Strand-specific RT-PCR analysis was also performed with TAG-based derived primers. Negative strand HCV RNA was shown in one (patient 6) of the two CD34+ samples. As indicated in Table 1, products of PCR-driven amplification of 5′-NC region were obtained in the serum of all anti-HCV positive patients. Moreover, detection and distribution of HCV RNA in purified lymphocyte subsets of these patients did not show a consistent picture, in that CD20+and/or CD8+ or CD4+ subsets tested positive for HCV RNA (data not shown).
HCV RNA Titers Measured by Branched DNA Methodology and HCV RNA Negative-Polarity Strand
| Patients . | HCV RNA . | |||
|---|---|---|---|---|
| Serum . | CD34+Cells . | |||
| NegativePolarity Strand* . | Titer† (MEq/mL) . | NegativePolarity Strand* . | Titer† (Eq/cell) . | |
| 1 | −/− | 25.5 | ND | 0.74 |
| 2 | −/− | 5.34 | ND | 0.80 |
| 3 | −/− | 3.4 | ND | 0.31 |
| 4 | −/− | 1.43 | ND | 0.40 |
| 5 | −/− | 17.20 | +/+ | 1.06 |
| 6 | −/− | 11.71 | −/+ | 0.33 |
| Patients . | HCV RNA . | |||
|---|---|---|---|---|
| Serum . | CD34+Cells . | |||
| NegativePolarity Strand* . | Titer† (MEq/mL) . | NegativePolarity Strand* . | Titer† (Eq/cell) . | |
| 1 | −/− | 25.5 | ND | 0.74 |
| 2 | −/− | 5.34 | ND | 0.80 |
| 3 | −/− | 3.4 | ND | 0.31 |
| 4 | −/− | 1.43 | ND | 0.40 |
| 5 | −/− | 17.20 | +/+ | 1.06 |
| 6 | −/− | 11.71 | −/+ | 0.33 |
*TAG-/CAP-derived primers.
Branched DNA assay.
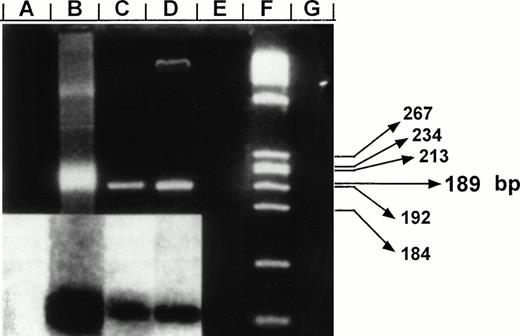
Negative-polarity strand HCV RNA and specificity of CAP-based RT-PCR assay. Products were fractionated on a 3% agarose gel and stained with ethidium bromide. Lane B: detection of synthetic HCV negative strand RNA generated from the vector pRTI HCV cDNA containing HCV capsid sequences cloned into pBluescript (Stratagene, San Diego, CA).24 Lanes A, E, and G: negative control reactions in which RNA templates were amplified in the absence of RT, omitting specific primers, or in nucleic acids extracted from CD34+ cells of a HCV-negative control, respectively. Lanes C and D: RT-PCR products for negative strand HCV RNA amplified from highly purified samples of CD34+ cells from patients no. 5 and 6, respectively. Results were confirmed after Southern blotting (inserted panel). A total of 1 μg of RNA extracts prepared from CD34+ cells was used along with 250 ng outer sense primer (5′CCAAAACCCCAAAGAAA3′, position: 750) for cDNA synthesis. Resulting cDNA was amplified after addition of 250 ng of the outer antisense primer (5′GTACCCCATGAGGTCGGCG3′, position: 355). A second PCR reaction was performed using a set of internal primers (sense-5′CAGATCGTTGGTGGAGTT3′, position: 427; antisense-5′CAAGCCCTCATTGCCAT3′, position: 616). The resulting product of 189 bp was probed after Southern blotting using a P32-labeled oligomer (5′GGTCGCAACCTCGAGGTAGACGTCAGCCT3′, position: 506). Lane F: molecular markers (HaeIII-digested ◊X174). Predicted size of amplified fragment is indicated in base pair (bp).
Negative-polarity strand HCV RNA and specificity of CAP-based RT-PCR assay. Products were fractionated on a 3% agarose gel and stained with ethidium bromide. Lane B: detection of synthetic HCV negative strand RNA generated from the vector pRTI HCV cDNA containing HCV capsid sequences cloned into pBluescript (Stratagene, San Diego, CA).24 Lanes A, E, and G: negative control reactions in which RNA templates were amplified in the absence of RT, omitting specific primers, or in nucleic acids extracted from CD34+ cells of a HCV-negative control, respectively. Lanes C and D: RT-PCR products for negative strand HCV RNA amplified from highly purified samples of CD34+ cells from patients no. 5 and 6, respectively. Results were confirmed after Southern blotting (inserted panel). A total of 1 μg of RNA extracts prepared from CD34+ cells was used along with 250 ng outer sense primer (5′CCAAAACCCCAAAGAAA3′, position: 750) for cDNA synthesis. Resulting cDNA was amplified after addition of 250 ng of the outer antisense primer (5′GTACCCCATGAGGTCGGCG3′, position: 355). A second PCR reaction was performed using a set of internal primers (sense-5′CAGATCGTTGGTGGAGTT3′, position: 427; antisense-5′CAAGCCCTCATTGCCAT3′, position: 616). The resulting product of 189 bp was probed after Southern blotting using a P32-labeled oligomer (5′GGTCGCAACCTCGAGGTAGACGTCAGCCT3′, position: 506). Lane F: molecular markers (HaeIII-digested ◊X174). Predicted size of amplified fragment is indicated in base pair (bp).
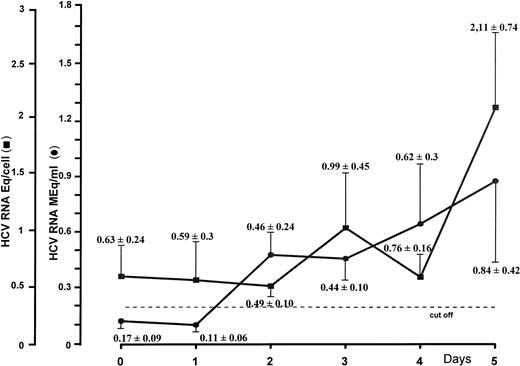
To gain insight as to whether HCV might sustain a productive infection in CD34+ cells, the latter were incubated and seeded in liquid culture at a concentration of 0.5 × 106 in the absence of growth factors. HCV RNA levels were titrated daily by the bDNA technique in cells and the corresponding supernatants. Results from separate experiments after 5 days of culture showed that increments of HCV genomic equivalents in the cells paralleled those found in the supernatants at corresponding intervals. Figure 2, which summarizes culture experiments, shows that 0.63 ± 0.24 (mean ± standard deviation [SD]) genomic HCV equivalents per cell could be detected on the starting day, whereas no viral RNA could be demonstrated in the supernatant. Significant progressive increases of viral RNA concentrations were shown during the culture period, and 5 days later, the number of viral Eq/cell was more than duplicated (2.11 ± 0.74). Remarkable changes were also shown in the supernatants, in that 0.84 ± 0.42 HCV RNA MEq/mL were found on day 5.
HCV RNA titration by branched DNA signal amplification assay of nucleic acids extracted from CD34+ cells and their corresponding supernatants. Peripheral CD34+ cells obtained from HCV-infected patients were cultured in RPMI 1640 medium supplemented with 10% FCS. Supernatants and cells were harvested daily for a period of 5 days.
HCV RNA titration by branched DNA signal amplification assay of nucleic acids extracted from CD34+ cells and their corresponding supernatants. Peripheral CD34+ cells obtained from HCV-infected patients were cultured in RPMI 1640 medium supplemented with 10% FCS. Supernatants and cells were harvested daily for a period of 5 days.
Data from cells and supernatants suggested that CD34+ cells contained viral particles and expressed a complete viral cycle. The time-dependent increase in viral titers strongly indicated that the virions present in the culture fluid were actually produced by infected cells. After 5 days, infected cultures compared with noninfected cultures differed neither in terms of number of CD34+ cells nor of viable cells. To assess whether cells in which HCV was suspected to replicate still expressed the CD34 antigen, cells were restained with anti-CD34 MoAbs and separated into CD34+ and CD34− cells. Molecular analysis showed that CD34+ and CD34− cells both expressed viral RNA in five of six patients. No difference in cell morphology and viability was found between the two fractions.
The presence of HCV RNA genomic sequences within CD34+cells was demonstrated morphologically by using a PCR-driven in situ hybridization technique. Intracellular RNA sequences were preserved in situ while the cell membrane was permeabilized with proteinase K. Specific intracellular viral RNA was amplified by a PCR protocol in which DIG-11-dUTP was used to obtain a DNA product that remained in situ. DIG-labeled dUTP incorporated into the PCR product was detected by antidigoxigenin-APh–labeled antibody and chromogenic substrate. Controls were carefully evaluated. Positive cells were mixed with uninfected CD34+ cells at different dilutions. Each experiment was performed in triplicate.
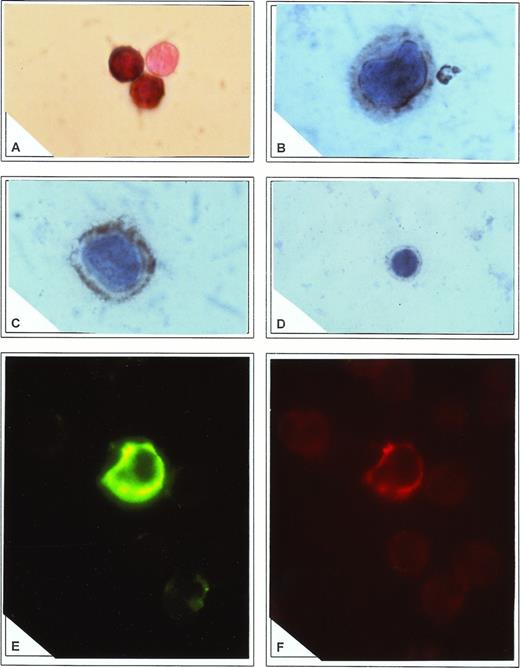
Amplified HCV RNA product was identified in a mixture containing at least 1 × 104 negative cells in excess. Cytocentrifuged samples showed that the cellular morphology was preserved, cellular debris was minimal, and intracellular localization of PCR product was maintained. Efficiency of the RT-PCR process was monitored by reverse transcription of cRNA transcribed from the plasmid pAW109 containing an insert of a synthetic linear array of primer sequences for the IL-1α site. Sensitivity was further evaluated by targeting in situ β-actin mRNA constitutively present in PBMC and showing that more than 95% of them were positive. Analysis of viral accumulation in the cell compartments showed that the signal corresponding to HCV-specific 240-bp amplicon was detected in the cytoplasm of CD34+ cells from all patients (Fig 3A) and in none of the controls. Albeit with different intensity, a large proportion of CD34+ cells were positive ranging from 36% to 65% (mean, 53.8 ± 25.1) of purified fractions.
(A) Direct in situ RT-PCR to amplify 5′-NC region of HCV genome in CD34+ cells. Note the complete absence of reaction in an adjoining cell. Immunocytochemistry to detect core (B) and E2/NS1 (C) antigens in CD34+ cells. In both cases a cytoplasmic appearance of immune reactants was found. Note adhesion of the core antigen to the nuclear membrane and accumulation of E2/NS1 antigen in cytoplasmic submembrane spaces. In (D) core reactivity was blocked by preadsorption of antibody with recombinant HCV core antigen. (E) Staining of CD34+ cells with FITC-conjugated anti-E2/NS1 protein. Specific signal outlining nonfluorescent nuclei is demonstrated. (F) The same cell as described in (E) was shown to coexpress CD34 antigen, stained with PE-labeled antibody.
(A) Direct in situ RT-PCR to amplify 5′-NC region of HCV genome in CD34+ cells. Note the complete absence of reaction in an adjoining cell. Immunocytochemistry to detect core (B) and E2/NS1 (C) antigens in CD34+ cells. In both cases a cytoplasmic appearance of immune reactants was found. Note adhesion of the core antigen to the nuclear membrane and accumulation of E2/NS1 antigen in cytoplasmic submembrane spaces. In (D) core reactivity was blocked by preadsorption of antibody with recombinant HCV core antigen. (E) Staining of CD34+ cells with FITC-conjugated anti-E2/NS1 protein. Specific signal outlining nonfluorescent nuclei is demonstrated. (F) The same cell as described in (E) was shown to coexpress CD34 antigen, stained with PE-labeled antibody.
Morphologic evidence for the cellular accumulation of HCV proteins was provided by immunocytochemistry on cultured CD34+ cells. Cells were tested with a large panel of MoAbs covering structural (core, E2/NS1 proteins) and nonstructural (NS3, NS4, NS5 proteins) encoding regions of the HCV genome. Results showed that antigens were detected as diffuse and homogeneous immune reactants within cytoplasmic compartments of CD34+ cells (Fig 3B and C). Immunoreactive cells rarely displayed granular submembrane deposits. Cell nuclei were persistently negative. Immunostaining was considered specific, as there was no reaction with CD34+ cells from HCV-unrelated controls, or with HCV-related samples after substitution of the primary antibody mixture. Preabsorption of the reagents with specific recombinant antigens abolished the signal (Fig 3D), with a parallel loss of reactivity in immunoblotting. The proportion of HCV-infected CD34+ cells ranged from 9% to 33% (mean, 19.2 ± 14.7) in the different purified preparations.
Morphologic studies on the coexpression of CD34 antigen and HCV structural protein (E2/NS1) using two different fluorochromes demonstrated a cytoplasmic colocalization of both reactivities (Fig 3E and F). With this methodology, however, the frequency of HCV-positivity in CD34+ cells was somewhat lower (9.2% ± 4.7%).
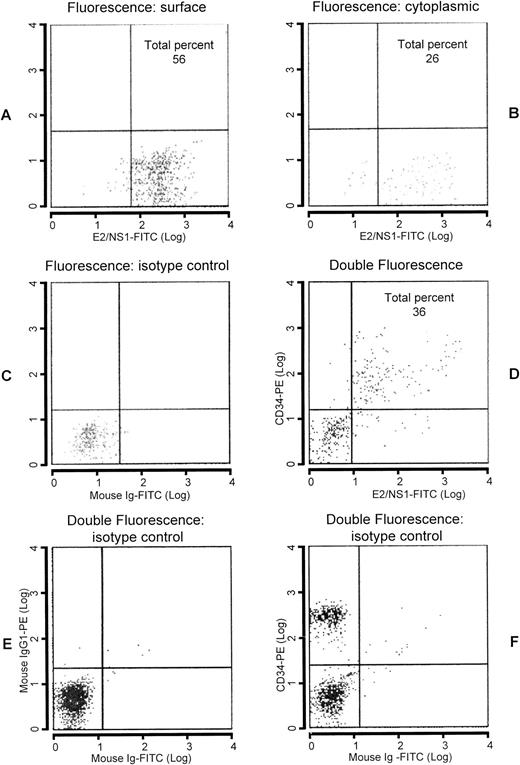
To determine whether viral protein translation was a regular and effective process following transcription of viral genes in CD34+ cells, core and E2/NS1 protein-related epitopes were analyzed by flow cytometry (Fig 4). CD34+ cells were enumerated by flow cytometry in the apheresis products with and without Ficoll-Hypaque centrifugation. The percentage of double-stained cells approximately reflected that found with immunohistochemistry in cytocentrifuged samples, in that almost one fourth of CD34+ cells (25.12% ± 17.88%) coexpressed HCV proteins. For cytoplasmic staining, CD34+cells were previously fixed in 4% formaldehyde solution and resuspended in acetone. The presence of intracytoplasmic core and E2/NS1 epitopes occurred less frequently than that found for surface staining (16.8 ± 15.2 v 48.6 ± 31.9), respectively.
Analysis by flow cytometry of Ficoll-Hypaque–purified leukapheresis products. Enumeration of CD34+ cells in light density cell suspension of peripheral blood labeled with anti-E2/NS1 whose labeling to the cells was defined by goat antimouse Ig conjugated with FITC. Surface (A) and cytoplasmic (B) stainings were performed. In (C), FITC-labeled isotypic control is shown. Double-staining analysis with PE-conjugated anti-CD34 antibody followed by FITC-labeled anti-E2/NS1 antibody is reported in (D). Isotypic controls are indicated in (E and F).
Analysis by flow cytometry of Ficoll-Hypaque–purified leukapheresis products. Enumeration of CD34+ cells in light density cell suspension of peripheral blood labeled with anti-E2/NS1 whose labeling to the cells was defined by goat antimouse Ig conjugated with FITC. Surface (A) and cytoplasmic (B) stainings were performed. In (C), FITC-labeled isotypic control is shown. Double-staining analysis with PE-conjugated anti-CD34 antibody followed by FITC-labeled anti-E2/NS1 antibody is reported in (D). Isotypic controls are indicated in (E and F).
These results led us to investigate whether CD34+ cells could be infected in vitro. CD34+ cells from anti-HCV, HCV RNA-negative patients were inoculated with undiluted serum #0750, which was known to contain a high titer HCV and to be extremely efficient in infecting the MOLT-4 cell line. Results were negative in all experiments, performed under different conditions, including the addition of feeder layers obtained from BM stromal cells and growth factors (IL-2, G-CSF, γ-interferon) (data not shown). Samples from cells and supernatants failed to show viral RNA genomic sequences after 4 weeks of culture.
DISCUSSION
Our results demonstrate that HCV-harboring CD34+ cells are a consistent biological feature in chronic HCV carriers. The following evidence points to productive infection of CD34+ cells by HCV: (1) detection of negative-polarity strand of viral RNA by RT-PCR amplification on nucleic acids extracted from highly purified CD34+ cell populations; (2) in situ demonstration of HCV RNA on intact cells; (3) detection of HCV-related structural and nonstructural proteins by flow cytometry analysis, immunocytochemistry, and immunofluorescence; and (4) an apparently complete extent of the viral cycle in cultured CD34+ cells.
Separation of CD34− from the CD34+ cell fraction showed that HCV infection occurs in both cells that do and do not express CD34 antigen, suggesting that HCV infection is not restricted to a CD34+ subset. In addition, it was demonstrated that other hematopoietic lineages including B- and T-cell subsets may be infected with HCV, as already reported.12 14-16
The presence or absence of HCV RNA within PBMC did not correlate with any specific profile of serum or histologic markers. During the purification of CD34+ cells and lymphocyte subsets, the last washing was tested for HCV RNA. In every instance, no PCR product was detected, suggesting that the HCV RNA signal detected in the cells was not due to plasma contamination. The presence of both positive- and negative-polarity HCV RNA strands in CD34+ cells and only the positive HCV RNA strand detected in the corresponding serum reflects the specificity of the RT-PCR products. Furthermore, the uneven distribution of HCV RNA in the blood mononuclear cell subsets in the same patient is consistent with the specific nature of the signal.
In vitro experiments indicated that HCV-infected BM and peripheral blood hematopoietic progenitors did not show obvious morphologic abnormalities when compared with noninfected CD34+ cells. Furthermore, the percentages of viable cells after many days of liquid culture were not significantly different between infected and noninfected CD34+ cells. These data support the contention that HCV infection has little or no effect on the proliferation and differentiation of CD34+ cells. They also suggest that, although HCV infection occurs in the early steps of blood progenitor differentiation, it does not result in viral-induced myelosuppression. These considerations are in good accordance with clinical studies showing that HCV is not directly responsible for hepatitis-associated aplastic anemia.30 However, the biological significance of these findings is still uncertain, and their impact on the clonogenic differentiation of infected CD34+ cells should be directly investigated.
Our experiments showed that CD34+ cells are not susceptible to HCV infection in vitro. None of the variations in the culture conditions tested, including different concentrations of the virus and the presence of a variety of growth factors or different accessory cells, resulted in susceptibility to HCV infection. These observations raise the possibility that several mechanisms, acting alone or in combination, could mediate HCV infection of CD34+ cells in vivo, namely infection of accessory cells necessary for maintenance and regulation of normal hematopoiesis through the production of growth factors and infection of stromal cells in the microenvironment interfering with cytokine(s) production or disrupting normal stromal cell-hematopoietic cell interactions.
As compared with HCV RNA-containing CD34+ cells, those expressing HCV-related antigens were significantly fewer. It is clear from our experiments that differing sensitivities in the detection of CD34+ cells infected with HCV are related to the techniques used. Immunofluorescence on cytocentrifuged samples appears to be less sensitive as compared with immunocytochemistry and flow cytometry analysis, which gave roughly the same frequency. These differences were particularly striking with in situ RT-PCR, in that mispriming and incorrect priming were carefully excluded. Indeed, a pattern of viral latency with silencing of viral genes expression can be suggested, as already described in hepatitis B virus (HBV) infection in which not all cells that contain HBV DNA display HBV-related proteins.31Because CD34+ cells include self-renewing stem cells, they can be an initial site of infection, a continuous source of virus, and possibly a mode of dissemination.
Previous studies on liver characterization and distribution of CD34 reactivity have indicated that anti-CD34 antibodies are more specific and sensitive than anti-von Willebrand factor or Ulex europeusagglutinin 1 for labeling endothelial cells.32 Moreover, it has been shown that CD34 antigen is expressed by endothelial cells in the sinusoid-like vessels of primary liver tumor and not by the endothelial cells in the normal sinusoids. CD34 staining, as detected by QBend-10 clone product, in normal liver was confined to vessels of the portal tracts and in a few sinusoids of the periportal areas in patients with chronic hepatitis and cirrhosis.33Experimental studies have indeed shown that expression of CD34 transcripts fluctuate and decrease after the birth, but increase during processes such as wound healing or tumor growth.34 In any case, the demonstration of CD34 reactivity in the liver suggests that this organ may retain CD34+ cells and perhaps act as a hematopoietic microenvironment from the fetal period, or as a reservoir of circulating CD34+ cells still remaining in the adult organ, as already suggested.35 Further studies are needed to recognize and characterize the distribution of CD34+reactivity in various tissues, in view of the evidence which indicate that it may be involved in leukocyte adhesion and “homing” during inflammatory processes, as well as in the localization of progenitor cells in the BM.17 Although the precise function of CD34 protein remains unknown, its widespread detection is consistent with the multiple sites of demonstration of HCV infection, namely BM,12,16 liver,36 and endothelium.22
Whether HCV infection of CD34+ cells is also related to lymphomagenesis is an intriguing possibility37 suggested by the fact that HCV infection in our patients preceded the appearance of lymphoma by many years and by the demonstration of HCV proteins in the pathologic lymph nodes.13 Indeed, HCV might sustain indolent stages of B-cell lymphoproliferation (putatively antigen-dependent), as recently demonstrated by PCR analysis of Ig-VH gene rearrangements using genomic DNA derived from intrahepatic B cells of chronically HCV-infected patients with type II mixed cryoglobulinemia.38 A common origin from B cells selected by the triggering antigen may be inferred in both low-grade and high-grade NHLs.39
Supported in part by the Finalized Project “Clinical Applications of Oncologic Research,” National Research Council, Rome (Contract No. 92.02269.PF39), by “Associazione Italiana per la Ricerca sul Cancro” (AIRC), and by a grant from Italian Ministry of University and Scientific and Technological Research, group “Liver Cirrhosis and Viral Hepatitis.” V.C. is the recipient of a fellowship from AIRC.
Address reprint requests to Franco Dammacco, MD, Department of Biomedical Sciences and Human Oncology, University of Bari Medical School, Policlinico, P.zza G. Cesare, 11, 70124 Bari, Italy.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal