To the Editor:
Fibroblast growth factor receptor 3 (FGFR3) is one of four distinct tyrosine-kinase receptors (FGFR1-4) that are capable of binding a repertoire of at least nine related mitogenic fibroblast growth factors (FGFs). FGFRs encode proteins that all contain three glycosylated extracellular Ig-like domains, a transmembrane domain (TM), and a split cytoplasmic tyrosine-kinase domain. Point mutations in distinct domains of the FGFR3 gene are associated with autosomal dominant human skeletal disorders, such as achondroplasia, thanatophoric dysplasia types I and II, and hypochondroplasia.1,2 Recent reports indicate that the point mutations associated with these disorders produce constitutively activated FGFR3, which shows autophosphorylation in the absence of ligand and is no longer regulated by FGF binding.3-6
We and others have recently provided the first evidence ofFGFR3 gene involvement in human cancer.7,8 In particular, the FGFR3 gene located at 4p16.3 is translocated to chromosome 14q32 as a result of a novel and karyotypically undetectable t(4;14)(p16.3;q32) chromosomal translocation in multiple myeloma (MM), a malignant proliferation of plasma cells. Molecular studies have shown this lesion in five MM-derived cell lines and in four primary tumors. Although the breakpoints on 4p16.3 are located approximately 50 to 120 kb centromeric to FGFR3, the gene is overexpressed in these cases, but absent or barely detectable in cell lines without the translocation. Interestingly, FGFR3 gene mutations associated with distinct human skeletal disorders2 have also been identified in some MM tumors carrying the t(4;14)(p16.3;q32): in particular, the Y373C mutation in the KMS-11 cell line,7,8the K650E mutation in the OPM2 cell line,7 and the K650M mutation in a primary MM tumor.7
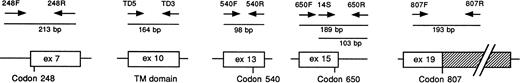
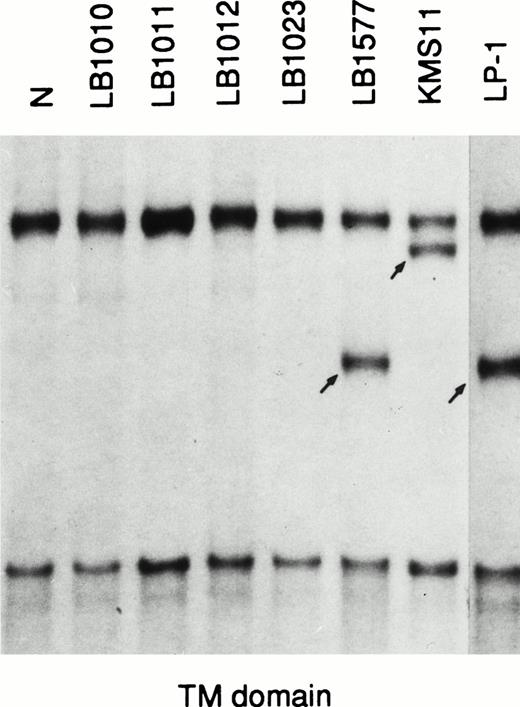
These findings prompted us to look for FGFR3 mutations known to be associated with skeletal disorders in a representative panel of MM, including 80 primary cases (60 patients at first diagnosis, 12 at relapse, and 8 affected by plasma cell leukemia) and 10 MM-derived cell lines (including the KMS-11 and OPM2 cell lines). The analysis was performed by means of the polymerase chain reaction–single-strand conformation polymorphism (PCR-SSCP) direct sequencing of genomic DNA. We amplified five distinct genomicFGFR3 fragments containing codons affected by mutations: codon 248, the entire TM domain (codons 371, 373, 375, and 380), codon 540, codon 650, and codon 807 (Fig 1). The mutations at codon 650 were also investigated by means of a restriction enzyme analysis of the PCR-amplified fragment using Mbo II andBbs I enzymes, as previously described.9 We detected allelic variations of the FGFR3 gene only in the fragment specific for the TM domain. An abnormal fragment with the same pattern of migration was observed in 2 cases (the LP-1 cell line and a primary tumor; Fig 2); in both cases, a novel single basepair mutation involving codon 384 in the form of a T to C transition (TTC-CTC) led to a conservative Phe → Leu amino acid substitution (data not shown). Interestingly, this mutation abrogates aMbo II restriction site and creates a new Mnl I site that allows restriction enzyme analysis of the PCR-amplified fragment. The apparently similar intensity of the normal and mutated bands in both cases, as well as the detection of the mutation in 2 of 100 normal individuals by means of restriction enzyme analysis, suggest that it may represent a rare genetic polymorphism. Finally, the FGFR3gene was apparently not expressed in the LP-1 cell line; it remains to be seen whether this particular variant may affect FGFR3 biological activity.
Schematic representation of the primers from the humanFGFR3 gene used in the study. The FGFR3 exons are indicated by white boxes, and the introns are indicated by lines. The 3′ untranslated region is indicated by the dashed box. The approximate locations of the primers, the length of the amplified fragments, and the approximate positions of codons 248, 540, 650, and 807 are indicated. The nucleotide sequence of FGFR3 cDNA and the intron-exon organization of the gene have been previously reported.12,13 The sequences of the primers are as follows: 248F (intron 6), 5′-CCTGAGCGTCATCTGCC-3′, and 248R (exon 7), 5′-CCATTGCATCCCACACGG-3′; TD5 (exon 10), 5′-AGGAGCTGGTGGAGGCTGA-3′, and TD3 (exon 10), 5′-GGAGATCTTGTGCACGGTGG-3′14; 540F (exon 13), 5′-ACTGACAAGGACCTGTCGGAC-3′, and 540R (exon 13), 5′-GCCCTGCGTGCAGGCGCC-3′; 650F (exon 15), 5′-GCATCCACAGGGACCTGG-3′, and 650R (intron 15), 5′-AGGCGGTGTTGGCGCCAG-3′; 14S (exon 15), 5′-GTGCACAACCTCGACTAC-3′ (this primer was used with 650R to obtain a DNA fragment suitable for the restriction enzyme analysis of codon 650); 807F (exon 19), 5′-CCTGTCGGCGCCTTTCGAGCAGTAC-3′, and 807R (exon 19), 5′-CACCAGCAGCAGGGTGGGCTGCTAG-3′.15
Schematic representation of the primers from the humanFGFR3 gene used in the study. The FGFR3 exons are indicated by white boxes, and the introns are indicated by lines. The 3′ untranslated region is indicated by the dashed box. The approximate locations of the primers, the length of the amplified fragments, and the approximate positions of codons 248, 540, 650, and 807 are indicated. The nucleotide sequence of FGFR3 cDNA and the intron-exon organization of the gene have been previously reported.12,13 The sequences of the primers are as follows: 248F (intron 6), 5′-CCTGAGCGTCATCTGCC-3′, and 248R (exon 7), 5′-CCATTGCATCCCACACGG-3′; TD5 (exon 10), 5′-AGGAGCTGGTGGAGGCTGA-3′, and TD3 (exon 10), 5′-GGAGATCTTGTGCACGGTGG-3′14; 540F (exon 13), 5′-ACTGACAAGGACCTGTCGGAC-3′, and 540R (exon 13), 5′-GCCCTGCGTGCAGGCGCC-3′; 650F (exon 15), 5′-GCATCCACAGGGACCTGG-3′, and 650R (intron 15), 5′-AGGCGGTGTTGGCGCCAG-3′; 14S (exon 15), 5′-GTGCACAACCTCGACTAC-3′ (this primer was used with 650R to obtain a DNA fragment suitable for the restriction enzyme analysis of codon 650); 807F (exon 19), 5′-CCTGTCGGCGCCTTTCGAGCAGTAC-3′, and 807R (exon 19), 5′-CACCAGCAGCAGGGTGGGCTGCTAG-3′.15
PCR-SSCP analysis of the FGFR3 gene. N, normal control; migrating fragments different from the normal control are indicated by arrows.
PCR-SSCP analysis of the FGFR3 gene. N, normal control; migrating fragments different from the normal control are indicated by arrows.
Although no specific genetic lesions have been found to be associated with MM (unlike other types of lymphoid neoplasms), cytogenetic and more recent molecular analyses suggest that chromosomal translocations involving the Ig locus on chromosome 14q32 may play an important role in gene deregulation.7,8 In this context, the recent identification of the t(4;14)(p16.3;q32) in MM, associated with an apparent deregulation of the FGFR3 gene, may provide some insights into the pathogenesis of this neoplasia. Although more work is needed to assess the role and frequency of the t(4;14) in MM, it can be suggested that deregulation of FGFR3 gene expression may lead to a constitutive oncogenic signal for the growth and/or survival of malignant plasma cells. This possibility is supported by the evidence that the bone marrow environment and, in particular, the stromal cells with which the plasma cells interact10are able to produce FGFs.11 The FGFR3 mutations reported in MM probably represent somatic events, suggesting thatFGFR3 gene may be deregulated by different mechanisms. However, we were unable to detect FGFR3 mutations associated with skeletal disorders in our series of samples, except in the cell lines previously reported.7 8 This finding suggests that such mutations represent rare events in MM and support the hypothesis that they may occur after the translocation and deregulation of theFGFR3 gene, thus contributing to tumor progression by means of ligand-independent activation.
ACKNOWLEDGMENT
We are grateful to Dr T. Otsuki, Dr F. Malavasi, and Dr A. Solomon for providing us with the some of the MM-derived cell lines (KMM1, KMS-11, KMS-12, LP-1, and UTMC-2) used in this study and to G Ciceri for technical assistance. The cell lines U266, Sultan, ARH-77, and RPMI 8226 were obtained from ATCC and the OPM2 cell line was obtained from DSMZ. This work was supported by a grant from the Associazione Italiana Ricerca sul Cancro (AIRC) to A.N. and a grant “Ricerca Corrente 1994” from the Ministero Italiano della Sanità to Ospedale Maggiore IRCCS.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal