Abstract
Hodgkin’s disease (HD) represents a malignant lymphoma in which the putative malignant Hodgkin and Reed-Sternberg (H-RS) cells are rare and surrounded by abundant reactive cells. Single-cell analyses showed that H-RS cells regularly bear clonal Ig gene rearrangements. However, there is little information on the clinical evolution of HD in a given patient. In this study, we used the single-cell polymerase chain reaction (PCR) to identify H-RS cells with clonal Ig gene rearrangements in biopsy specimens of patients with relapsed HD. The obtained clonal variable region heavy-chain (VH) gene rearrangements were used to construct tumor-clone-specific oligonucleotides spanning the complementarity determining region (CDR) III and somatically mutated areas in the rearranged VHgene. A number of biopsies were obtained during a period of 3 years from two HD patients. H-RS cells with identical VHrearrangements were detected in two separate infiltrated lymph nodes from one patient with nodular sclerosis HD. In a second patient with mixed cellularity HD subtype, clonal VH rearrangements with identical sequences were detected in infiltrated spleen and two lymph node biopsies. Despite the high sensitivity of the PCR method used (one clonal cell in 105 mononuclear cells), residual H-RS cells were not found in peripheral blood, leukapheresis material, purified CD34+ stem cells or bone marrow. The results show that different specimens from relapsed patients suffering from classical HD carry the same clonotypic IgH rearrangements with identical somatic mutations, demonstrating the persistence and the dissemination of a clonal tumor cell population. Thus, PCR assays with CDRIII-specific probes derived from clonal H-RS cells are of clinical importance in monitoring the dissemination of HD and tumor progression and could be useful for analysis of minimal residual disease after autologous stem cell transplantation.
© 1998 by The American Society of Hematology.
IN HODGKIN’S DISEASE (HD), the mononuclear Hodgkin and multinucleated Reed-Sternberg (H-RS) cells represent only a minority of 0.1% to 1% of the cells in the infiltrated tissue and are surrounded by a mixture of reactive cells.1,2 Most of the H-RS cells in classical HD, including nodular sclerosis (NS), mixed cellularity (MC), and lymphocyte depleted (LD) subtype, coexpress CD30 and CD15.3 H-RS cells of the lymphocyte predominant (LP) subform express in the majority of cases a panel of B-lineage–associated antigens.4
It has been shown by amplification of rearranged Ig genes that H-RS cells in classical as well as in LP HD represent a clonal population of mature B cells.5-12 The presence and pattern of somatic mutations within rearranged Ig genes carried by H-RS cells identified germinal center B cells as the precursor of H-RS cells in both types of HD.9-12
Little is known about the persistence of a clonal tumor cell population and the migration of this tumor clone throughout the body of the patient at different stages of the disease. Reports of the persistence of clonal Epstein-Barr virus (EBV) episomes in multiple HD-affected lesions indicate that clonal H-RS cells can disseminate in the body of an HD patient.13,14 It was recently shown that the rearranged Ig gene of the cell line L1236 is detectable in lymph node sections from the HD patient at primary diagnosis and in the relapse, suggesting the dissemination and persistence of clonal tumor cells.7,15 16
Polymerase chain reaction (PCR) assays based on the amplification of rearranged Ig heavy chains are useful tools for detecting clonal residual tumor cells. Based on the highly variable complementarity determining region (CDR) III sequence of the rearranged heavy chain, clone-specific oligonucleotides can be chosen to detect the presence of one malignant tumor cell in a background of 105 reactive cells.17-21 Supporting evidence for the role of residual tumor cells as a potential source of relapse comes from reports on leukemia,22 lymphoma,23,24neuroblastoma,25 and breast cancer,26 showing that the rate of relapse correlates with the presence of occult tumor cells in bone marrow or in peripheral blood.
Thus, the detection of residual tumor cells in HD may have clinical importance and has to our knowledge not yet been analyzed with PCR-based techniques.
We describe here the PCR analysis of blood and tissue samples from two patients with relapsed HD using tumor-specific primers derived from clonal Ig heavy-chain gene rearrangements of single H-RS cells. H-RS cells with identical IgH gene rearrangements were detected in different infiltrated lesions obtained at various time points, indicating the persistence and dissemination of an H-RS tumor clone in both patients. However, tumor cells were not detected in peripheral blood, apheresis material, purified CD34+ stem cells, or bone marrow even at the time of overt relapse.
MATERIALS AND METHODS
Patient Samples
Patient no. 1.
In December 1986, a 22-year-old woman presented with HD of the nodular sclerosis subtype. Clinical staging showed stage IIA with a bulky mediastinal tumor. The patient was treated according to the HD1 protocol of the German Hodgkin Study Group with two cycles of COPP/ABVD (cyclophosphamide, vincristine, procarbazine, prednisone, adriamycin, bleomycin, vinblastine, dacarbazine) followed by consolidating radiotherapy. The patient went into complete remission. In June 1992, a first relapse was diagnosed with involvement of bone marrow and enlarged mediastinal, inguinal, axillary, and iliac lymph nodes. The treatment included four cycles of salvage chemotherapy, under which the patient achieved complete remission. In February 1995, a second relapse occurred with enlarged retroperitoneal and mediastinal lymph nodes. Histological examination confirmed the relapse in an inguinal lymph node. The patient was again treated with salvage chemotherapy and went into partial remission. In January 1996, a peripheral stem cell apheresis was performed. In February 1996, the patient relapsed again with enlarged cervical lymph nodes and suspicious liver and bone marrow infiltration. Histological examination showed HD infiltration in a cervical lymph node. The patient was again treated with polychemotherapy and went into partial remission.
Patient no. 2.
In March 1990, a 28-year-old woman was diagnosed with HD mixed cellularity subtype, stage IVB. The patient presented with enlarged supraclavicular, axillary, mediastinal and inguinal lymph nodes, and pulmonary lesions. The patient was treated with three cycles of MOPP/ABVD (nitrogen mustard, vincristine, procarbazine, prednisone, adriamycin, bleomycin, vinblastine, dacarbazine), one cycle of COPP/ABVD, and consolidating radiotherapy and went into complete remission. In September 1993, a first relapse occurred with mesenteric and paraaortal lymph nodes. The patient was treated with four cycles of salvage chemotherapy, followed by high-dose chemotherapy and autologous stem cell transplantation. In July 1995, a second relapse was detected with mesenteric and axillary lymph nodes and spleen involvement. A laparotomy with splenectomy was performed and histological examination confirmed the infiltration of the spleen and the mesenteric lymph node with H-RS cells. The patient was subsequently treated with involved field radiotherapy. In June 1996, progressive lymphomas were detected in the mediastinum, the axilla, and the paraaortal region, but not in the bone marrow. After treatment with polychemotherapy, the patient achieved partial remission.
Cell and Tissue Samples
Granulocyte colony-stimulating factor (G-CSF)–mobilized leukapheresis material, heparinized peripheral blood mononuclear cells (PBMCs), and bone marrow aspirates were collected from the patients. Frozen and paraffin-embedded tissue sections were obtained from the Department of Pathology of the University of Cologne (Cologne, Germany). Peripheral blood was also obtained from healthy volunteers, and PBMCs were isolated by separation over Ficoll-Paque (Pharmacia Biotech, Freiburg, Germany). High gradient magnetic cell sorting (MACS) of CD34+ cells from G-CSF–mobilized leukapheresis material was performed using the CD34 progenitor cell isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany). Immunophenotyping of cells was performed before and after the separation procedure using anti-CD34 phycoerythrin (PE; HPCA2, mouse IgG1) or isotype control (mouse IgG1) (Becton Dickinson, Mountain View, CA) for immunofluorescence staining.
Immunostaining and Micromanipulation
Single-Cell PCR
Rearranged Ig genes were amplified from single cells using V-gene family-specific primers together with JH and Jκ primers as described.27,28 In some experiments, the VH gene family-specific framework region (FR) I primers were exchanged by a set of VH FRII primers (5 nmol/L of each primer) using an annealing temperature of 68°C in the first cycle and 59°C in the following 34 cycles and 2.5 mmol/L MgCl2. The sequences of the FRII primers were as follows: VH1FRII, 5′ GACAAGGGCTTGAGTGGATGGGA 3′; VH2FRII, 5′ GAAGGCCCTGGAGTGGCTTGC 3′; VH3FRII, 5′ CAGGGAAGGGGCTGGAGTGGGT 3′; VH4FRII, 5′ GAAGGGRCTGGAGTGGATTGGG 3′; VH5FRII, 5′ CGCCAGAGTCCCGGGAAAGGC 3′; VH6FRII, 5′ GGATCAGGCAGTCCCCATCGAG 3′. The second round of amplification was performed as described,27 28 using 2.0 mmol/L MgCl2. and an annealing temperature of 63°C. An aliquot of 5 μL of the reaction mixture was analyzed on a 2% agarose gel (Biozym, Oldendorf, Germany).
Sequence Analysis
PCR products were gel-purified and directly sequenced using the Ready Reaction DyeDeoxy Terminator cycle sequencing kit (Perkin Elmer, Weiterstadt, Germany). Sequence products were analyzed on an automatic sequencing system (ABI377; Applied Biosystems, Weiterstadt, Germany) and compared with the Genbank data library using DNASIS software (Version V3.6; Pharmacia).
DNA Preparation
High molecular weight DNA was isolated using a DNA extraction kit (Qiamp Blood/Tissue kit; Qiagen, Hilden, Germany). For paraffin-embedded tissue, sections were first incubated with xylol to remove paraffin and incubated overnight with proteinase K. As a control for cross-contamination of the analyzed samples with PCR products, DNA extraction of mononuclear cells from healthy volunteers or tissue sections from irrelevant donors was performed in parallel to all patient samples.29
Estimation of the Assay Sensitivity
Cells of the HD-derived cell line L428 were mixed with 107PBMCs of a healthy volunteer and serial dilutions of lymphoma cells in 107 PBMCs were performed. DNA was isolated from the cell mixtures containing 10 to 104 L428 cells as described above.
Clone-Specific PCR and Southern Blot
For each VH gene, one forward primer, spanning a somatically mutated area of the sequenced VH gene (CDRI for patient no. 1 and CDRII for patient no. 2), and two reverse CDRIII-specific primers were designed. For L428 cells, the VH5FRI was used for amplification. The primer sequences and annealing temperatures were as follows: L428rev1, 5′ CCGGGAGACAACTCCCCCCATCAT 3′; and L428rev2, 5′ AACTCCCCCCATCATCTGACTATG 3′ (1.5 mmol/L MgCl2; annealing temperature, 63°C); NSfor, 5′ TACACCTTCAACACCCATGGTC 3′; NSrev1, 5′ GAAGTCAGGTGAGTACATTCCATA 3′; and NSrev2, 5′ ATTCCATAATTACAACGAAGTGCC 3′ (2 mmol/L MgCl2; annealing temperature, 61°C); MCfor, 5′ GAGACTTCAGGACAGACTCACCAT 3′; MCrev1, 5′ ATGTCGAAGGGTGGAACAGGTTTG 3′; and MCrev2, 5′ GGTGGAACAGGTTTGAGTATCGCA 3′ (1.5 mmol/L MgCl2; annealing temperature, 63°C). The first and second round of amplification was performed as described,27,28 using 1 μg of DNA and 0.125 μmol/L of the primers mentioned above. After DNA amplification for 40 cycles, 1 μL of the PCR product was reamplified for 40 cycles using the same forward primer and the corresponding nested CDRIII reverse primer. The reactions were performed in five replicates to avoid false-negative results in cases when the amount of target molecules was at the detection limit of 1 tumor cell in 105 mononuclear cells. All PCR products were separated on a 2% agarose gel and then transferred to a nylon membrane (Hybond-N+; Amersham Buchler, Braunschweig, Germany) using an alkali transfer buffer (0.4 mol/L NaOH/1.0 mol/L NaCl).30 Hybridization with a fluorescein isothiocyanate (FITC)-labeled oligonucleotide was performed at 42°C overnight. The following oligonucleotides were used for hybridization: L428intern, 5′ CACCAACTATGGGTCGTCCTTCGG 3′; NSintern, 5′ AAATACTCACAGACGTTTAAAGAC 3′; and MCintern, 5′ GTGCGGACGATTTCATGGGGACA 3′. The washing steps were performed at 42°C with 5× SSC/0.1% sodium dodecyl sulfate (SDS) for 5 minutes twice. The detection of specific PCR products was performed following the protocol for the ECL detection system (Amersham Buchler). Filters were exposed to x-ray film (AR, Kodak, Integra Bioscience, Fernwald, Germany) for 10 seconds to 10 minutes.
RESULTS
Estimation of PCR Sensitivity
The detection limit of the PCR approach was estimated by mixing PBMCs of healthy volunteers with different amounts of cells from the HD cell line L428. This cell line harbors a rearranged VH5 gene (M.V., unpublished data).
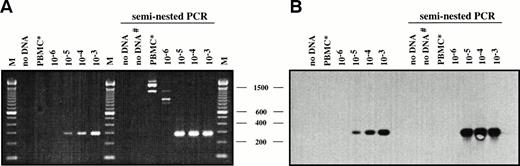
DNA samples of 1 μg containing the equivalent of 0.2 to 200 L428 cells in 2 × 105 PBMCs were analyzed by seminested PCR with a VH5 FRI primer and two nested CDRIII-specific primers (L428rev1/rev2). Reactions containing water or PBMC DNA of a healthy volunteer served as negative controls. Southern blot hybridization with a specific internal oligonucleotide (L428intern) and ECL visualization was performed with all reactions to show specificity of the amplificates. As shown in Fig 1, specific amplification products of 296 bp were visible on ethidium bromide-stained agarose gels and the corresponding Southern blot at a dilution corresponding to 1 tumor cell in 105 mononuclear cells after the first round of amplification with the primers VH5 and L428rev1. The second amplification cycle with the nested CDRIII-specific primer L428rev2 led to an increase of the amount of specific PCR products corresponding to the dilution steps of 10−3 to 10−5 but did not increase the sensitivity to the dilution step of 10−6, because the dilution of 10−6 did not contain any target molecules for amplification. The same results were found for five replicates (not shown).
Estimation of PCR and Southern blot sensitivity for detection of L428 cells admixed with normal PBMCs. (A and B) DNAs isolated from serial dilutions of L428 cells in normal PBMCs were amplified using VH5FRI and L428rev1 (296 bp)/rev2 primers (287 bp). PCR products were separated on a 2% agarose gel and stained with ethidium bromide (A) and subjected to Southern blot hybridization using an internal FITC-labeled oligonucleotide (L428intern) and visualized by ECL (B). M, 100-bp DNA ladder. Negative controls: DNA from a healthy volunteer (PBMC*), water control after first and second (#) PCR.
Estimation of PCR and Southern blot sensitivity for detection of L428 cells admixed with normal PBMCs. (A and B) DNAs isolated from serial dilutions of L428 cells in normal PBMCs were amplified using VH5FRI and L428rev1 (296 bp)/rev2 primers (287 bp). PCR products were separated on a 2% agarose gel and stained with ethidium bromide (A) and subjected to Southern blot hybridization using an internal FITC-labeled oligonucleotide (L428intern) and visualized by ECL (B). M, 100-bp DNA ladder. Negative controls: DNA from a healthy volunteer (PBMC*), water control after first and second (#) PCR.
Identification of Tumor-Specific VH Rearrangements in Single H-RS Cells of HD Patients
Two independent micromanipulations and single-cell PCRs (experiment no. 1a/b) were performed of single CD30+ H-RS cells and control CD3+ T cells picked from a frozen section of an inguinal lymph node that was biopsied in March 1995 (LN 3/95) from patient no. 1 (Table 1). Overall, 20 of 31 H-RS cells yielded PCR products with VH1 primers. Sequence analysis showed an identical sequence for all H-RS cells (Fig 2A). The heavy-chain rearrangement (NSVH1) is potentially functional and uses a JH4 gene.31 Comparison to the most homologous germline gene DP1432 shows that the rearranged gene is highly mutated. A DH gene could not be identified. A second micromanipulation was necessary, because clonally unrelated IgH rearrangements were obtained from six of eight T cells in experiment no. 1a. We speculate that a specific antibody for B-cell antigens was erroneously used for the “T-cell staining.” In experiment no. 1b, all control CD3+ T cells and buffer controls were negative for VH gene rearrangements.
Summary of Single-Cell Analysis of Two Cases for V Gene Rearrangements
| Patient No. . | Experiment No. . | Cells Positive for V Rearrangements After PCR With Different Forward Primers . | PCR Products . | Rearrangements . | |||||
|---|---|---|---|---|---|---|---|---|---|
| Vκ . | VHFRI . | VHFRII . | Clone- Specific . | Total . | Sequenced . | Repeated . | Unique . | ||
| 1 | 1a | ND | 13/17 | —-150 | ND | 13 | 13 | 13 VH1 | |
| 1b | ND | 7/14 | —-150 | 7/14 | 7 | 7 | 7 VH1 | ||
| 2 | 2′LN | 0/13 | 0/13 | —-150 | 3/13 | 3 | 3 | 3 VH1 | |
| 2"LN | 0/7 | —-150 | 1/7 | 3/7 | 3 | 3 | 3 VH1 | ||
| 2"spleen | 0/10 | —-150 | 1/10 | 2/10 | 2 | 2 | 2 VH1 | ||
| Control | 1a-151 | ND | 6/8 T cells | —-150 | ND | 8 | 8 | 5 VH3,3 VH4 | |
| 1b | ND | 0/8 T cells | —-150 | 0/8 T cells | |||||
| ND | 0/10 buffer | —-150 | 0/10 buffer | ||||||
| Control | 2′LN | 0/4 T cells | 0/4 T cells | —-150 | 0/4 T cells | ||||
| 2"LN | 0/6 T cells | —-150 | 0/6 T cells | 0/6 T cells | |||||
| 2"spleen | 0/5 T cells | —-150 | 0/5 T cells | 0/5 T cells | |||||
| Patient No. . | Experiment No. . | Cells Positive for V Rearrangements After PCR With Different Forward Primers . | PCR Products . | Rearrangements . | |||||
|---|---|---|---|---|---|---|---|---|---|
| Vκ . | VHFRI . | VHFRII . | Clone- Specific . | Total . | Sequenced . | Repeated . | Unique . | ||
| 1 | 1a | ND | 13/17 | —-150 | ND | 13 | 13 | 13 VH1 | |
| 1b | ND | 7/14 | —-150 | 7/14 | 7 | 7 | 7 VH1 | ||
| 2 | 2′LN | 0/13 | 0/13 | —-150 | 3/13 | 3 | 3 | 3 VH1 | |
| 2"LN | 0/7 | —-150 | 1/7 | 3/7 | 3 | 3 | 3 VH1 | ||
| 2"spleen | 0/10 | —-150 | 1/10 | 2/10 | 2 | 2 | 2 VH1 | ||
| Control | 1a-151 | ND | 6/8 T cells | —-150 | ND | 8 | 8 | 5 VH3,3 VH4 | |
| 1b | ND | 0/8 T cells | —-150 | 0/8 T cells | |||||
| ND | 0/10 buffer | —-150 | 0/10 buffer | ||||||
| Control | 2′LN | 0/4 T cells | 0/4 T cells | —-150 | 0/4 T cells | ||||
| 2"LN | 0/6 T cells | —-150 | 0/6 T cells | 0/6 T cells | |||||
| 2"spleen | 0/5 T cells | —-150 | 0/5 T cells | 0/5 T cells | |||||
H-RS cells were analyzed together with B and T cells coded in a blinded fashion from two patients with HD. The number of PCR products obtained and sequenced from H-RS and T cells for the first and the second experiment is indicated. Clonally independent VHgene rearrangements of various VH gene families were obtained from control B cells (not shown). Repeated rearrangements indicate clonally related rearrangements; unique rearrangements indicate rearrangements amplified once and not related to any other rearrangement. The sequences obtained from H-RS cells of both patients were deposited in the Genbank under the accession numbers AJ007389 and AJ0007390.
Abbreviations: LN, lymph node; ND, not determined.
PCR was performed either with VHFRI or with VHFRII primers.
In this experiment, a B-cell–specific antibody was eventually used for the “T-cell staining.”
Comparative sequence analysis of rearranged VH gene sequences from H-RS cells. Dashes indicate sequence identity. CDRs I-III are marked. Corresponding amino acids are shown above each codon. (A) PCR products of the VH1 rearrangement amplified from 20 H-RS cells of a patient with NS-HD are compared with the germline VH1 gene DP1432 and the JH4 gene.31 The upper sequences were obtained from single H-RS cells (NSVH1), the lower sequences were obtained from lymph node tissues (LN 3/95 and LN 2/96). (B) VH1 rearrangement amplified from eight H-RS cells from a patient with MC-HD, compared with the germline VH1 gene DP1031 and the JH3 gene.30 The upper sequences were obtained from single H-RS cells (MCVH1); the lower sequences were obtained from spleen (SP 7/95) and lymph node tissue (LN 7/96).
Comparative sequence analysis of rearranged VH gene sequences from H-RS cells. Dashes indicate sequence identity. CDRs I-III are marked. Corresponding amino acids are shown above each codon. (A) PCR products of the VH1 rearrangement amplified from 20 H-RS cells of a patient with NS-HD are compared with the germline VH1 gene DP1432 and the JH4 gene.31 The upper sequences were obtained from single H-RS cells (NSVH1), the lower sequences were obtained from lymph node tissues (LN 3/95 and LN 2/96). (B) VH1 rearrangement amplified from eight H-RS cells from a patient with MC-HD, compared with the germline VH1 gene DP1031 and the JH3 gene.30 The upper sequences were obtained from single H-RS cells (MCVH1); the lower sequences were obtained from spleen (SP 7/95) and lymph node tissue (LN 7/96).
As shown in Table 1, 13 CD15+ H-RS cells were analyzed by single-cell PCR picked from a frozen section of a mediastinal lymph node (LN 7/95) that was biopsied in July 1995 from patient no. 2 (Table1, experiment no. 2′). Because the use of VH and Vκ family-specific FRI primers did not lead to any amplification product after the second round of PCR, another set of single cells, which was picked at the same day from the HD-infiltrated frozen lymph node section (LN 7/95) and from a frozen HD-infiltrated spleen section (SP 7/95), was analyzed by single-cell PCR with VH family-specific FRII primers (Table 1, experiment no. 2′′). One of seven micromanipulated CD15+ H-RS cells from the lymph node H-RS cell and 1 of 10 CD30+ H-RS cells from the spleen section led to an amplified product with the VH1 FRII primer. Comparison to the most homologous germline gene (DP-10)32 showed that this highly mutated rearrangement (MCVH1) is rearranged out of frame to the JH3 gene31 (Fig 2B). We assume that the MCVH1 gene from other H-RS cells was either not amplified or amplified only with reduced efficiency because of mutations at the primer binding sites. Therefore, we screened all micromanipulated PCR product-negative cells again using a clone-specific primer in combination with the 5′ JH-Mix for the second round of PCR. Three PCR products were each obtained from experiment no. 2′ and experiment no. 2′′ with identical DNA sequences.
Detection of Clonal H-RS Cells in Different HD Infiltrated Tissues
Twenty-two DNA samples of two HD patients obtained from different tissue and blood samples and different time points of the disease were analyzed using a seminested PCR approach with a clone-specific forward and two nested CDRIII-specific reverse primers followed by Southern blot hybridization with an internal oligonucleotide. In Fig 3, the analyzed DNA samples are depicted schematically. In Fig 4, representative agarose gel electrophoreses and the corresponding Southern blot hybridization of PCR products from the two patients are shown.
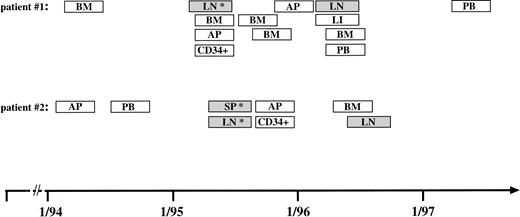
PCR-based monitoring of clonal H-RS cells in different blood and tissue samples from two patients with relapsed HD for 3 years from March 1994 to May 1997. Collection of samples analyzed from a patient with NS-HD (patient no. 1) and from a patient with MC-HD (patient no. 2). (▧) Specific PCR products were amplified from these samples with clone-specific primers. (□) No PCR products were detected in the DNA samples. (*) Single-cell PCR was performed from a frozen tissue section. Abbreviations: BM, bone marrow aspirate; LN, lymph node tissue; AP, apheresis material; LI, liver tissue; SP, spleen cells; PB, peripheral blood; CD34+, MACS-separated CD34+ cells.
PCR-based monitoring of clonal H-RS cells in different blood and tissue samples from two patients with relapsed HD for 3 years from March 1994 to May 1997. Collection of samples analyzed from a patient with NS-HD (patient no. 1) and from a patient with MC-HD (patient no. 2). (▧) Specific PCR products were amplified from these samples with clone-specific primers. (□) No PCR products were detected in the DNA samples. (*) Single-cell PCR was performed from a frozen tissue section. Abbreviations: BM, bone marrow aspirate; LN, lymph node tissue; AP, apheresis material; LI, liver tissue; SP, spleen cells; PB, peripheral blood; CD34+, MACS-separated CD34+ cells.
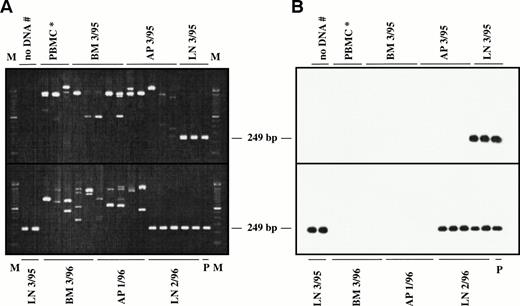
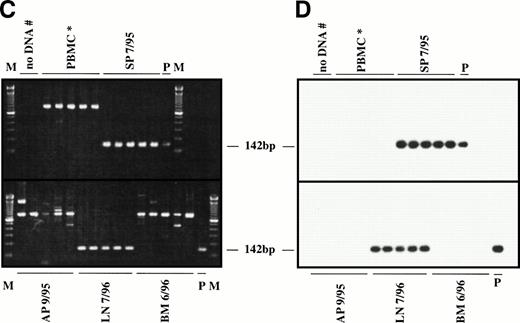
Representative seminested PCR analysis of H-RS cell-specific DNA in apheresis material, bone marrow, spleen, and lymph node samples from a patient with NS-HD (patient no. 1; A and B) and a patient with MC-HD (patient no. 2; C and D). DNA samples (1 μg) were amplified in five replicates (except control PBMCs for patient no. 1) using NSfor and NSrev1/rev2 primers (patient no. 1) and MCfor and MCrev1/rev2 primers (patient no. 2). PCR products were fractionated by agarose gel electrophoresis (2%) and stained with ethidium bromide (A and C). After Southern blot, the DNA was hybridized to the internal FITC-labeled oligonucleotide NSintern (B) or MCintern (D) and visualized by ECL. M, 100-bp DNA ladder; P, PCR product amplified from single H-RS cells served as the positive controls (NSVH1, 249 bp; MCVH1, 142 bp). Negative controls: DNA from a healthy volunteer (PBMC*), water control after first and second (#) PCR.
Representative seminested PCR analysis of H-RS cell-specific DNA in apheresis material, bone marrow, spleen, and lymph node samples from a patient with NS-HD (patient no. 1; A and B) and a patient with MC-HD (patient no. 2; C and D). DNA samples (1 μg) were amplified in five replicates (except control PBMCs for patient no. 1) using NSfor and NSrev1/rev2 primers (patient no. 1) and MCfor and MCrev1/rev2 primers (patient no. 2). PCR products were fractionated by agarose gel electrophoresis (2%) and stained with ethidium bromide (A and C). After Southern blot, the DNA was hybridized to the internal FITC-labeled oligonucleotide NSintern (B) or MCintern (D) and visualized by ECL. M, 100-bp DNA ladder; P, PCR product amplified from single H-RS cells served as the positive controls (NSVH1, 249 bp; MCVH1, 142 bp). Negative controls: DNA from a healthy volunteer (PBMC*), water control after first and second (#) PCR.
Patient no. 1.
Fourteen DNA samples (from March 1994 to May 1997) were analyzed. The specific DNA fragment of 249 bp was detected in the DNA sample of the HD-infiltrated lymph node tissue (LN 3/95) and in cervical lymph node taken in February 1996 (LN 2/96) (Fig 4A and B). Sequence analysis of the two obtained PCR products showed identical sequences as compared with the H-RS cells (NSVH1) analyzed by single-cell PCR (Fig 2A). The amplification of DNA obtained from the bone marrow aspirate (BM 3/95) and apheresis material (AP 3/95) taken during the second relapse led to unspecific amplification products of different size (Fig 4A and B). No specific PCR products were detected in apheresis material collected in January 1996 (AP 1/96) and in the bone marrow aspirate (BM 3/96). All bone marrow and blood samples taken at different time intervals remained negative in the PCR analysis.
Patient no. 2.
Eight DNA samples (from February 1994 to July 1996) were analyzed. PCR analysis of the DNA sample from fresh spleen cells (SP 7/95) and an axillary lymph node (LN 7/96) showed the presence of the specific tumor-specific amplification product (Fig 4C and D). Sequence comparison presented in Fig 2B showed identical sequences for the DNA sequences obtained from the spleen (SP 7/95), the lymph node tissue (LN 7/96), and micromanipulated single H-RS cells from spleen and lymph node cells (MCVH1). No specific amplification products were found in DNA extracted from apheresis material (AP 9/95), bone marrow aspirate (BM 6/96), and various blood samples obtained at different times of the disease.
DISCUSSION
There is now a general agreement that the putative malignant H-RS cell belongs in most cases of HD to a clonal population of mature B cells.33 However, little is known about the clinical evolution of the tumor clone in patients with HD in the course of the disease.
We analyzed whether CDRIII-specific DNA probes derived from clonal H-RS cells could be used as a diagnostic marker to follow-up the dissemination of HD in different biopsy specimens over a time period of several years. Our results show that independently derived specimen from relapsed patients suffering from classical HD carry the same clonotypic IgH rearrangements with identical somatic mutations, demonstrating the persistence and the dissemination of a clonal tumor cell population.
To establish H-RS cell-specific DNA probes, single H-RS cells were micromanipulated from frozen lymph node sections and a spleen section and single-cell PCR was performed to detect rearranged V genes. In both cases, clonally related, highly mutated VH region genes were amplified from single H-RS cells. The detection of rearranged Ig genes in H-RS cells is consistent with earlier studies, where clonally related, highly mutated VH rearrangements were found.5,7 9-12
We find it noteworthy that the clonal rearrangement of single H-RS cells of patient no. 2 was amplified from CD15+ H-RS cells of a mediastinal lymph node as well as from CD30+ H-RS cells of the spleen, demonstrating an extensive dissemination of this tumor clone. The VH rearrangement of patient no. 2 could only be amplified by the application of the VH1 FRII primer or a clone-specific primer. This indicates that the low number of H-RS cells with rearranged Ig genes detected primarily was due to the insufficient binding efficiency of the used FRII primer because of mutations at the binding site. The inability of primer binding might also partly explain why several groups failed to amplify V-region genes from single H-RS cells.34 35 Because the MCVH1 rearrangement represents an out-of-frame rearrangement and because H-RS cells appear to be derived from mature B cells that (before malignant transformation) expressed antigen receptors, it is likely that the H-RS cells harbor an additional, potentially functional heavy chain rearrangement. This rearrangement was most likely not detected by PCR because of mutations at primer binding sites.
The detection limit of our PCR and Southern blot approach was estimated in a dilution assay with the HD-derived cell line L428. Our assay could detect one tumor cell in 105 mononuclear cells. One microgram of DNA was analyzed in every reaction, which corresponds to approximately 2 × 105 cells. Thus, a PCR that contains DNA from the dilution step of 10−5 contains on average two target cells. This is the reason why a seminested PCR with a nested CDRIII-specific primer and the Southern blot hybridization did not lead to further enhancement of the detection limit. We did not attempt to use a greater amount of DNA in our assay, because only a few micrograms of DNA were available from the HD patients. The sensitivity of 1 tumor cell in 105mononuclear cells was also achieved in a similar dilution assay with the Burkitt lymphoma cell line BL41 (not shown), which harbors a rearranged VH3 gene.36 Therefore, we assume that this sensitivity might be representative for PCR assays with CDRIII-specific primers. The second round of clone-specific PCR led sometimes to unspecific amplification products when no target molecules were present. This is probably due to the fact that the VHsegment primers may cross-hybridize to V-region genes of the B cells present in the samples.
The results of the present report show that the same tumor clone is present at different time points and in different lesions. Sequence analyses showed identical sequences amplified from DNA samples of tissue sections as in single H-RS cells in both patients. This confirms the expansion of clonal cells, which are present in the analyzed tissues with minimum of 1 tumor cell surrounded by 105 nonmalignant cells. There was no indication for ongoing mutations in these VH genes in the dominant tumor clones as described for LP-HD.10-12 Thus, the chemotherapy after the second relapse neither eliminated this tumor clone and prevented the dissemination to other compartments of the lymphatic system nor was the DNA of the analyzed part of the V-region genes altered by point mutations, deletions, or insertions.
The absence of additional mutations within V-region genes under chemotherapy shows that the clone-specific PCR of V region genes is a suitable assay to analyze dissemination and persistence as well as tumor progression in HD patients. Strikingly, in our study, PCR positivity was found in lymph node or spleen samples, whereas at the same time cells of the tumor clone were undetectable in bone marrow and apheresis material of both patients. The absence of tumor cells in these samples even at the point of relapse demonstrates that H-RS cells may home primarily to lymphatic organs. It still remains unclear whether the spreading of H-RS cells throughout the body takes place in the blood or in the lymphatic system. The studies of Wolf et al have provided strong evidence that clonogenic cells can circulate in peripheral blood, because they established the cell line L1236 from peripheral blood of an HD patient.15 However, the dissemination of H-RS cells in bone marrow and peripheral blood seems to be a rare event in advanced stages of tumor progression.
Upon relapse, both patients were scheduled for high-dose chemotherapy and autologous stem cell transplantation, which may improve the rates of complete remission and disease-free survival.37-39 A concern with the treatment protocols used is that residual tumor cells may be transferred through the stem cell transplant and can contribute to a later relapse. Some investigators have demonstrated that survival of non-Hodgkin’s lymphoma (NHL) patients was significantly better in the groups of patients who received PCR-negative transplants.40 41 It is unknown whether stem cell harvests from the peripheral blood of HD patients might contain lymphoma cells that can lead to a relapse. In the current study, we analyzed four apheresis materials and two preparations of MACS-purified CD34+ cells by PCR and found no contaminating tumor cells, indicating that the tumor burden in the transplants was below the detection limit of our PCR assay of 1 malignant cell in 105reactive cells. The amplified CDRIII of clonal H-RS cells could now be used as an individual tumor marker to screen stem cell harvests for residual H-RS cells.
ACKNOWLEDGMENT
The authors thank Andrea Jox and Martin Kornacker for providing us with several patient samples and for critical reading of the manuscript.
Supported by a grant from the Bundesministerium für Forschung (01KS 9052) and the Deutsche Forschungsgemeinschaft (Sonderforschungsbereich 502).
Address reprint requests to Martina Vockerodt, Klinik I für Innere Medizin, LFI, E4, R.508, Joseph-Stelzmann Str.9, D-50924 Köln, Germany.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal