To the Editor:Human herpesvirus 6 (HHV-6) is a recently discovered member of the human herpesvirus family.1 Although primary infection with variant B HHV-6 causes exanthem subitum,2 the clinical features of variant A HHV-6 infection remains unclear. The virus probably latently infects the body after the primary infection and then reactivates in an immunosuppressed state like other human herpesviruses. HHV-6 has recently been recognized as an opportunistic pathogen in transplant recipients.3-7 It has been shown that HHV-6 might be associated with fever and skin rash resembling acute graft-versus-host disease (GVHD),3 interstitial pneumonitis,4encephalitis,6 and bone marrow suppression7after bone marrow transplantation (BMT). Since infection with the virus after BMT could be fatal,6 it is important to prevent the infection. Therefore, if we are able to predict HHV-6 infection after BMT, it should prove invaluable in helping to prevent the virus infection.
There are two likely sources for HHV-6 infection after BMT: one is reactivation from the recipient body and another is infection via the donor marrow from a seropositive donor. Therefore, virus genome latently infected in peripheral blood mononuclear cells (PBMCs) of donors and recipients could be an important source of the virus infection after transplantation. The aim of this study is to determine whether the presence of HHV-6 genome in PBMCs before BMT is a valuable predictor of virus infection after BMT. We also analyzed whether HHV-6 antibody titers of donors and recipients at the time of transplantation were associated with virus infection.
Thirty recipients (20 male and 10 female), who received allogeneic BMT at the Children's Medical Center of the Japanese Red Cross Nagoya First Hospital, and their donors, were employed in this study. All guardians of these patients consented to be in this study. Patient characteristics relating to age, sex, and underlying disease are summarized in Table 1. The median age of these recipients was 5.9 years old (ranging from 1 year to 15 years old) at the time of transplantation. EDTA peripheral blood samples were collected from donor and recipient pairs at the time of transplantation. In addition, EDTA blood samples were collected at 2 weeks before transplantation and biweekly after transplantation until 2 months after transplantation from recipients. We attempted to isolate HHV-6 from PBMCs and measure antibody titers to HHV-6 by indirect immunofluorescence assay. We also analyze for the presence of HHV-6 DNA in PBMCs obtained from the donor at the time of BMT and from the recipient at 2 weeks before BMT.
Patient Characteristics
| Age (yr) | 5.9 (range: 1-15) |
| Sex (M/F) | 20/10 |
| Underlying disease | |
| Severe aplastic anemia | 5 |
| Acute lymphoblastic leukemia | 8 |
| Acute myelogenous leukemia | 8 |
| Chronic myelocytic leukemia | 3 |
| Neuroblastoma | 2 |
| Myelodysplastic syndrome | 1 |
| Malignant lymphoma | 2 |
| Adrenoleukodystrophy | 1 |
| Donor | |
| HLA-matched sibling | 22 |
| HLA-mismatched related donor | 3 |
| HLA-matched unrelated donor | 5 |
| GVHD prophylaxis | |
| MTX | 12 |
| MTX + CyA | 18 |
| Conditioning regimen | |
| TBI + L-PAM + others | 13 |
| TBI + CY + others | 5 |
| BU + L-PAM | 7 |
| Others | 5 |
| Age (yr) | 5.9 (range: 1-15) |
| Sex (M/F) | 20/10 |
| Underlying disease | |
| Severe aplastic anemia | 5 |
| Acute lymphoblastic leukemia | 8 |
| Acute myelogenous leukemia | 8 |
| Chronic myelocytic leukemia | 3 |
| Neuroblastoma | 2 |
| Myelodysplastic syndrome | 1 |
| Malignant lymphoma | 2 |
| Adrenoleukodystrophy | 1 |
| Donor | |
| HLA-matched sibling | 22 |
| HLA-mismatched related donor | 3 |
| HLA-matched unrelated donor | 5 |
| GVHD prophylaxis | |
| MTX | 12 |
| MTX + CyA | 18 |
| Conditioning regimen | |
| TBI + L-PAM + others | 13 |
| TBI + CY + others | 5 |
| BU + L-PAM | 7 |
| Others | 5 |
Abbreviations: MTX, methotrexate; CyA, cyclosporin A; TBI, total body irradiation; L-PAM, melphalan; CY, cyclophosphamide; BU, busulfan.
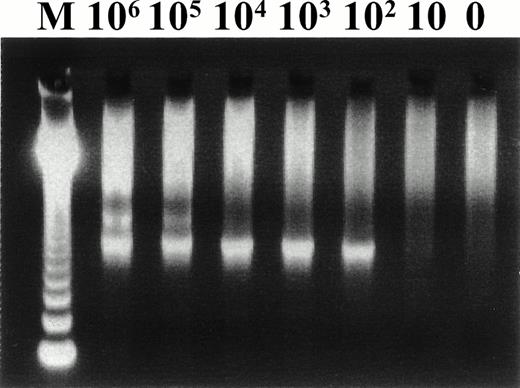
Five hundred nanograms of DNA extracted from PBMCs obtained from recipients approximately 2 weeks before transplantation and from donors at the time of transplantation was used for nested polymerase chain reaction (PCR) amplification. Nested PCR was performed for amplification of HHV-6 DNA by using two primer sets (A/C, HS6AE/HS6AF) as previously described.8 The PCR resulted in the amplification of a 751-bp DNA fragment encoding a putative large tegument protein gene. The type of HHV-6 was determined by the presence of an HindIII site in each second PCR product. The sensitivity of the PCR assay was determined with the use of serial dilutions of the plasmid, pSTY-05 (kindly provided by Dr K. Yamanishi, Department of Bacteriology, Osaka University, Osaka, Japan). As shown in Fig 1, we could routinely detect 100 copies of the virus genome. To ensure the accuracy of each PCR assay, dilutions containing 100 copies and 10 copies of the plasmid were coamplified with each of the samples in the following analyses. Statistical analyses were performed by using Fisher's exact test and Student's t-test.
Representative second PCR products from serial dilution of plasmid (pSTY-05) for determining the sensitivity of our PCR assay. Numbers on the top of the panel indicate the copy number of the pSTY-05 plasmid.
Representative second PCR products from serial dilution of plasmid (pSTY-05) for determining the sensitivity of our PCR assay. Numbers on the top of the panel indicate the copy number of the pSTY-05 plasmid.
If HHV-6 was isolated from PBMCs or a significant increase of HHV-6 antibody was observed, we defined this as a virus infection. HHV-6 infection was confirmed in 17 (57%) of the 30 recipients. The virus was isolated from 10 (33%) of the 30 recipients between 7 and 29 days after transplantation. Seven additional recipients had a significant increase in their HHV-6 antibody titer during the observation period. All of the donors and the recipients were seropositive to HHV-6 at the time of BMT. Seven of the 10 isolates were analyzed to determine the variant of the virus. All 7 isolates were variant B HHV-6 on the basis of restriction fragment length polymorphism analysis of the second PCR products (data not shown).
HHV-6 DNA was detected in 10 (33%) of the 30 donors at the time of BMT, and in 15 (50%) of the 30 recipients before BMT. Details of the presence of the virus genome in PBMC and the results of virological examinations after transplantation are shown in Table2. HHV-6 infection occurred in 3 of 4 donor-positive/recipient-negative patients, in 7 of 9 donor-negative/recipient-positive patients, in all of the donor-positive/recipient-positive patients, and in 1 of 11 donor-negative/recipient-negative patients. Moreover, all of the second PCR products detected in this study were digested withHindIII indicating variant B HHV-6 (data not shown). Details relating to the presence of the virus genome and the antibody titers of the donor and the recipient at risk for HHV-6 infection after allogeneic BMT are shown in Table 3. If HHV-6 genome was detected in a donor's or recipient's PBMCs, we defined this as a positive for the virus genome. Sixteen (84%) of the 19 patients who were positive for the virus genome before BMT were infected with the virus after transplantation. However, only 1 (9%) of the 11 patients who were negative had HHV-6 infection. Thus, the presence of the virus genome in the donor or the recipient before transplantation was a good predictor of HHV-6 infection after allogeneic BMT (P < .001). The geometric mean titers (GMT) (log10) of HHV-6 antibody in donors with HHV-6 infection (1.3) and donors without infection (1.4) were not significantly different. Moreover, GMT of the antibody to HHV-6 in recipients with infection (1.5) and without infection (1.5) were not significantly different.
Summary of the Virological Examinations
| Before BMT | After BMT | |||
| HHV-6 Infection | ||||
| Positive | Negative | |||
| Status of Virus Genome in PBMC | No. of Cases | With Viremia | Without Viremia | |
| D: Positive | 4 | 3 | 0 | 1 |
| R: Negative | ||||
| D: Negative | 9 | 3 | 4 | 2 |
| R: Positive | ||||
| D: Positive | 6 | 4 | 2 | 0 |
| R: Positive | ||||
| D: Negative | 11 | 0 | 1 | 10 |
| R: Negative | ||||
| Before BMT | After BMT | |||
| HHV-6 Infection | ||||
| Positive | Negative | |||
| Status of Virus Genome in PBMC | No. of Cases | With Viremia | Without Viremia | |
| D: Positive | 4 | 3 | 0 | 1 |
| R: Negative | ||||
| D: Negative | 9 | 3 | 4 | 2 |
| R: Positive | ||||
| D: Positive | 6 | 4 | 2 | 0 |
| R: Positive | ||||
| D: Negative | 11 | 0 | 1 | 10 |
| R: Negative | ||||
Abbreviations: D, donor; R, recipient.
Comparison of Patients at Risk of HHV-6 Infection After Transplantation
| Category . | HHV-6 Infection . | |
|---|---|---|
| Yes (n = 17) . | No (n = 13) . | |
| HHV-6 DNA in PBMC* | ||
| Positive | 16 | 3† |
| Negative | 1 | 10† |
| HHV-6 antibody titers in donor (GMT ± SD) | 1.3 ± 0.4 | 1.4 ± 0.3 |
| HHV-6 antibody titers in recipient (GMT ± SD) | 1.5 ± 0.3 | 1.5 ± 0.3 |
| Category . | HHV-6 Infection . | |
|---|---|---|
| Yes (n = 17) . | No (n = 13) . | |
| HHV-6 DNA in PBMC* | ||
| Positive | 16 | 3† |
| Negative | 1 | 10† |
| HHV-6 antibody titers in donor (GMT ± SD) | 1.3 ± 0.4 | 1.4 ± 0.3 |
| HHV-6 antibody titers in recipient (GMT ± SD) | 1.5 ± 0.3 | 1.5 ± 0.3 |
Abbreviation: GMT, geometric mean titer (log10).
If HHV-6 DNA is detected in donor's or recipient's PBMC, we defined it as positive.
P < .001 by Fisher's exact test.
As we expected, the presence of HHV-6 DNA in a recipient's or donor's PBMCs before transplantation correlated with the virus infection after allogeneic BMT. Although many studies have been done to elucidate the occurrence, time course, and clinical features of HHV-6 infection after BMT, this is the first demonstration that the presence of the virus genome before transplantation is predictive of virus infection after allogeneic BMT. It is likely that infection with HHV-6 after BMT is affected by a number of factors, such as the degree of immunosuppression, HLA mismatch between donor and recipient, and the occurrence of acute GVHD. We have to evaluate other factors on prediction of the virus infection after allogeneic BMT in the future. A large number of prospective studies is needed to perform multivariate analysis.
Although HHV-6 DNA was not detected in 20 donors and 15 recipients, all of them were seropositive to HHV-6 at the time of BMT. This indicates that the amount of latently infected HHV-6 genome was below the level of detection in our PCR assay. Hence, the sensitivity of the PCR assay is a critical issue for this study. Therefore, all of the PCR assays were performed carefully to eliminate contamination as previously described. The detection limit of our nested PCR was 100 copies of HHV-6 genome. We used standard solutions containing 10 copies and 100 copies of the plasmid pSTY-05 as negative and positive controls, respectively, to maintain the accuracy of the PCR. Five hundred nanograms of viral DNA was used for each PCR analysis in this study and a good correlation was found between the results of the PCR and the virus infection. Some of the DNA samples which gave positive PCR results in this assay using 500 ng gave negative results when only 50 ng of DNA was used (data not shown). Therefore, it is likely that the quantity of DNA used for PCR is another important factor to consider for obtaining good results. We collected PBMC from the recipients 2 weeks before BMT to analyze for the presence of the virus genome. This is because the recipients generally had a severe bone marrow suppression at the time of BMT, and it might be impossible to obtain sufficient PBMCs at that time.
As we expected, all of the donors and the recipients had HHV-6 antibody at the time of BMT. It is impossible to predict virus infection on the basis of the sero-status of the donor or the recipient, as suggested by studies on cytomegalovirus infection. Moreover, no clear correlation between antibody titers of either the recipients or the donors and the virus infection was shown. Although Willborn et al9 used a PCR assay for determining HHV-6 infection after BMT, they reported a similar finding to what we have reported here. This indicates that antibody titers of the donor or the recipient do not have a predictive value for HHV-6 infection.
In this study, we demonstrated that the presence of the virus genome in a donor's or recipient's PBMCs is a good predictor of HHV-6 infection after allogeneic BMT. Prediction of the clinical features caused by the virus infection will be an important issue for the future study. Moreover, measurement of the amount of the virus genome in blood products transfused to the recipient will be another important problem to resolve in future studies. Prospective study consisting of a large number of cases is now proceeding to resolve such issue.
ACKNOWLEDGMENT
Supported in part by grants from Fujita Health University and a Grant-in-Aid for Scientific Research, The Ministry of Education, Science and Culture, Japan. We thank Michael Conlon, PhD, for revision of the language.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal