Abstract
The Rh polypeptides and the glycoproteins Rh50, CD47, LW, and glycophorin B, which interact in the red blood cell membrane to form a multisubunit complex, are lacking or are severely reduced in the Rh-deficiency syndrome. We previously reported that in several Rhnull patients the RH50 gene was altered at the coding sequence level, resulting in either a single amino acid substitution or the synthesis of a truncated polypeptide. In the present report, we have detected two mutations in the intronic region of the RH50 gene that identify a new molecular mechanism involved in Rh-deficiency. The first mutation affected the invariant G residue of the 3′ acceptor splice-site of intron 6, causing the skipping of the downstream exon and the premature termination of translation. The second mutation occurred at the first base of the 5′ donor splice-site of intron 1. Both these mutations were found in homozygote state. RNase protection assays demonstrated that the Rh50 mRNA level was strongly reduced or undetectable in the 3′ and 5′ splice mutants, respectively. The different mutations affecting the RH50 gene are indicative of an heterogeneous mutational pattern, which further supports the hypothesis that the lack of the Rh50 protein may prevent the assembly or transport of the Rh membrane complex to the red blood cell surface.
THE Rh (Rhesus) ANTIGENS are defined by a multisubunit complex composed of the unglycosylated Rh polypeptides and of several glycoproteins (Rh50, CD47, LW, and glycophorin B) held together by noncovalent linkages. These proteins are collectively absent or severely reduced on erythrocytes from Rhnullpatients who suffer from a chronic hemolytic anemia known as the Rh deficiency syndrome.1-3 Among the components of the Rh membrane complex, Rh, Rh50, and CD47 are of special interest, because there is no selective deficiency of these proteins presently identified and, therefore, each of them may play a crucial role in the transport and assembly of the Rh membrane complex to the red blood cell (RBC) membrane. In contrast, deficiency of LW and GPB without alteration of Rh antigen expression is well known,4 suggesting that these proteins represent dispensable accessory chains of the Rh membrane complex.
The Rh50 glycoprotein (50 kD) and the RhD/RhCE proteins (30 kD) are erythroid specific. They share about 36% amino acid identity and a similar topology of their 12 transmembrane domains.5 These proteins are encoded by the RH50 and RH genes, respectively, that belong to the same protein family. Both genes are composed of 10 exons whose size and exon/intron junctions are highly conserved.6,7 CD47 has a wide tissue distribution and is identical to the integrin-associated calcium channel of endothelial cells.8-11
Rh deficiency may arise from two distinct genetic backgrounds called amorph (or silent) and regulator.4,12 The amorph type is very rare and arises by homozygosity of a silent allele at the RH locus. Recent studies in our laboratory have shown that, in 2 of these patients, the RHCE gene was altered by different point mutations, but there was no sequence alteration of either the Rh50 or CD47 transcripts.13 In the regulator type, the most common form of Rhnull, which is caused by the homozygosity of a rare autosomal suppressor gene (X°r) unlinked to the RH locus, all of the genes and transcripts noted above were normal, except for RH50. The Rh50 protein was absent or severely reduced because of frameshift, missense mutations, or absence of detectable transcript.14,15 Because Rh and Rh50 are thought to interact with each other in the cell membrane,16 17these findings suggested that, when the Rh50 glycoprotein is lacking, the Rh membrane complex is not assembled or transported to the cell surface. Therefore, mutant alleles of the RH50 gene are likely candidates as suppressors of the RH locus in the Rhnull of regulator type.
Very few mutations of the RH50 gene have been described so far.14 15 We describe here two splice-site mutations in theRH50 gene from 2 unrelated patients that identify a new molecular mechanism leading to the Rhnull phenotype. Moreover, we trace the inheritance of the mutations by family studies and provide additional information on the mapping of Rh50 mutations causing Rh deficiency.
MATERIALS AND METHODS
Blood samples.
Rhnull samples of the regulator type and family members were collected on heparin (10 U/mL) and immediately shipped to Paris, France. Sample AL was provided by L. Lebeck (Loma Linda, CA) and sample AC, from Bilbao (Spain), was provided by one of us. Both patients exhibited the typical Rhnull syndrome. Blood samples from common RhD-positive and RhD-negative phenotypes were obtained from the Institut National de la Transfusion Sanguine (Paris, France).
Flow cytometry and Western blot analysis.
Immunostaining of intact RBCs was performed as described,18using murine monoclonal antibodies (MoAbs) directed against CD47 (MoAb 6H9; gift of Dr M. Telen, Durham, NC) and the Rh50 glycoprotein (2D10; gift of Dr A. von dem Borne, Amsterdam, The Netherlands). Fluorescein-conjugated F(ab′)2 fragments of goat antimouse IgG (Immunotech, Marseille, France) were used and the mean fluorescence intensity was determined with a FACScan flow cytometer (Becton Dickinson, San Jose, CA). Determination of antigen site densities was performed with QIFIKIT microbeads coated with variable numbers of mouse Ig molecules (Biocytex, Marseille, France).
RBC membrane proteins (60 μg) were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 10% acrylamide), transferred onto nitrocellulose sheets, and incubated with the A23 rabbit polyclonal antibody (1:200 dilution) directed against the N-terminus of the Rh50 glycoprotein (residues 2 through 16) and with a rabbit anti-p55 antibody13 (1:10,000) used as control. Bound antibodies were detected with alkaline phosphatase-labeled goat antirabbit IgG (1:800), followed by revelation with the alkaline-phosphatase conjugate substrate kit (Biorad Laboratories, Rockville Centre, NY).
Reverse transcription coupled with polymerase chain reaction (PCR) amplification.
Reticulocyte RNAs from 30 mL of peripheral blood were extracted by selectively lysing RBCs with the Orskov reaction.19 One microgram of RNA was reverse transcribed in a total volume of 33 μL using the First Strand cDNA synthesis kit (Pharmacia, Uppsala, Sweden) following the manufacturer's instructions. Five microliters of cDNA products was then subjected to PCR in 50 mmol/L KCl, 10 mmol/L Tris (pH 8.3), 0.01% gelatin, 0.2 mmol/L of the four dNTPs, 50 pmoles of each primer, and 2.5 U of Taq polymerase (Perkin-Elmer Cetus, Norwalk, CT). PCR conditions were as follows: denaturation for 1 minute at 94°C, annealing for 1 minute at 55°C, and extension for 1.30 minutes at 72°C for 30 cycles. Relevant PCR fragments were purified on a 1% low melting agarose gel followed by a Wizard PCR preps minicolumns (Promega, Madison, WI) and subcloned into a pCR II vector using the TA cloning kit (Invitrogen, Leek, The Netherlands). Primers used to amplify Rh (sense, nt −19 to −2; antisense, nt 1,350 to 1,330), Rh50 (sense, nt −22 to −5; antisense, nt 1,262 to 1,243), and CD47 (sense, nt −2 to +14; antisense, nt 1,007 to 989) cDNAs were deduced from published sequences.5,8,20 21
Genomic DNA analysis and DNA sequencing.
DNA was prepared from peripheral leukocytes by the SDS/proteinase K extraction procedure.22 A 316-bp fragment, encompassing the exon 1 to intron 1 junction of the RH50 gene, was obtained using an appropriate primer pair (sense, nt −2 to +15; antisense intronic, 5′ccaggtaagccgtggaggttatgg3′) after 30 cycles each, including 1 minute at 94°C, 30 seconds at 50°C, and 30 seconds at 72°C. A 1.3-kb fragment encompassing the exon6/exon7 junctions was also amplified between specific primers (sense, A3, nt 857 to 880; antisense, A4, nt 1,013 to 990) with 2.5 U of enzyme mix (Taq & Pwo DNA polymerases) using the Expand long template PCR system (Boehringer Mannheim, Mannheim, Germany) under the following conditions: 2 minutes at 94°C (1 cycle); 10 seconds at 94°C, 30 seconds at 65 °C, and 4 minutes at 68°C (10 cycles); 10 seconds at 94°C, 30 seconds at 65 °C, and 4 minutes at 68°C plus cycle elongation of 20 seconds for each cycle (15 cycles); and an elongation time up to 7 minutes at 68°C. PCR products were purified through a microcon ultrafiltration membrane (Amicon, Beverly, MA). Sequencing was performed on an automated ALF express sequencer (Pharmacia) using either the Cy5 Autoread sequencing kit (Pharmacia) with plasmid DNA or the Thermo Sequenase sequencing kit (Amersham, Bucks, UK) for direct sequencing of PCR products, following the manufacturer's instructions.
Ribonuclease protection assay.
Two DNA fragments of 270 bp between nucleotides (nt) 162 and 431 of the Rh50 cDNA5 and of 207 bp between nt 616 and 822 of the XK cDNA encoding the Kx antigen23 were amplified and cloned into a pCR II vector using the TA cloning kit (Invitrogen). From the linearized recombinant plasmids, antisense RNA probes were synthesized using [α-32P]-UTP (800 Ci/mmol; NEN, Boston, MA) in an in vitro transcription system (Riboprobe Core System; Promega). The full-length transcripts were purified from a 5% acrylamide, 8 mol/L urea gel with 0.5 mol/L ammonium acetate, 1 mmol/L EDTA, and 1% SDS. Rh50 and XK RNA probes (8 × 104 and 1 × 104 cpm, respectively) were hybridized with 12.5 μg of total reticulocyte RNAs overnight at 52°C and digested with RNase A and T1, following the manufacturer's instructions (RPAII; Ambion, Austin, TX). The protected RNA fragments were separated through a 5% denaturing polyacrylamide gel. Hybridizing signals on the autoradiograph were quantified using the NIH-Image software.
RESULTS
Cell surface expression of the antigens and proteins from the Rh membrane complex.
RBC samples from the Rhnull propositus and members of two families (C and L) were analyzed by flow cytometry with MoAbs specific for Rh (D, C, c, E, and e) and LW antigens as well as MoAbs directed against Rh50 and CD47. RBC samples from patients AC and AL did not react with anti-Rh, anti-LW, and anti-Rh50 reagents, but reacted only weakly with CD47 (2 to 3 × 103v 35 to 39 × 103 molecules/cell) as compared with controls. In contrast, RBC samples from the family members reacted normally (data not shown).
Analysis of the Rh and Rh50 transcripts in Rhnullpatients.
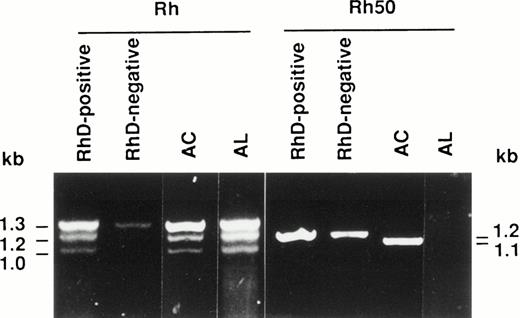
Reticulocyte RNAs isolated from patients AC and AL were reverse transcribed and the Rh and Rh50 cDNAs were amplified using appropriate primer pairs (see Materials and Methods). As expected for Rh transcripts, three PCR products were detected in the RhD-positive control (Fig 1). The 1.3-kb fragment corresponds to both RHCE and RHD full-length cDNAs. The 1.2- and 1.0-kb products are derived from RHD spliceoforms, as previously described.24-26 In the RhD-negative sample, the 1.3-kb and the faint 1.2-kb amplified products correspond to the full-length RHCE cDNA and to the Rh4 spliceoform of theRHCE gene,27 respectively. The amplification pattern distinctive of the RhD-positive phenotype was obtained in AC and AL samples, indicating that both RH genes are present and transcribed in these patients (Fig 1). Moreover, sequence analysis of the full length Rh transcripts (1.3 kb) from AC and AL showed no alteration (data not shown).
Reverse transcription-PCR (RT-PCR) amplification of reticulocyte RNAs from AC and AL Rhnullpatients. The Rh and Rh50 cDNAs were amplified by RT-PCR from reticulocyte mRNAs as described in Materials and Methods. The Rh50 amplification products showed a smaller size in patient AC as compared with RhD-negative or RhD-positive controls, and no product is present in AL.
Reverse transcription-PCR (RT-PCR) amplification of reticulocyte RNAs from AC and AL Rhnullpatients. The Rh and Rh50 cDNAs were amplified by RT-PCR from reticulocyte mRNAs as described in Materials and Methods. The Rh50 amplification products showed a smaller size in patient AC as compared with RhD-negative or RhD-positive controls, and no product is present in AL.
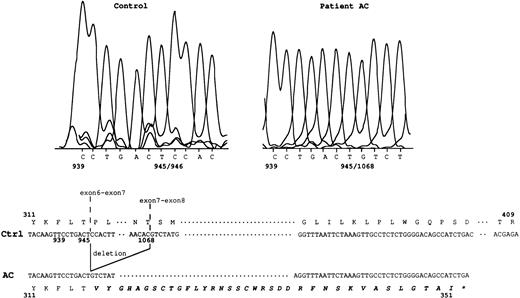
The amplification product from the Rh50 cDNA was smaller in size in the AC sample (1.1 kb) with respect to the controls (1.2 kb), whereas no product was detected in AL (Fig 1), as previously suggested.14 Sequencing of the Rh50 cDNA amplified from AC mRNAs showed a loss of 122 bp (positions 946 to 1067) corresponding to the entire transcribed region of exon 7, as compared with the normalRH50 sequence5 (Fig 2). This deletion introduced a frameshift after the threonine codon at position 315 and resulted in a premature stop codon, 108 bp downstream, located in the exon 9 region of the gene. The predicted truncated protein is 351 amino acids long (instead of 409) and includes 36 novel residues at the C-terminus. This result raised the hypothesis that the lack of Rh50 expression in AC erythrocytes, determined by flow cytometry analysis (see above), may result from the failure of the anti-Rh50 antibody (2D10) to bind to the mutated region of the protein. This possibility was ruled out by Western blot analysis, using a polyclonal antibody directed against the Rh50 N-terminus (see Materials and Methods), which confirmed the absence of the Rh50 glycoprotein in the RBC membrane (Fig 3). In addition, the CD47 cDNA from AC and AL patients were amplified and their sequences were found to be identical to that already published.8
Rh50 transcript analysis in patient AC. In the control sample, nt 945/946 correspond to the exon 6-exon 7 junction. In patient AC, a 122-bp deletion (nt 946 to 1067, ie, the entire transcribed exon 7 region) results in a new junction, nt 945/1068. This deletion introduced a frameshift resulting in 36 novel amino acids (in bold italics) at the C-terminal region, after the threonine at position 315. Ctrl, normal Rh50 cDNA.5 The star identifies the stop codon located in exon 9. Junctions of exons 6-7 and exons 7-8 are shown.
Rh50 transcript analysis in patient AC. In the control sample, nt 945/946 correspond to the exon 6-exon 7 junction. In patient AC, a 122-bp deletion (nt 946 to 1067, ie, the entire transcribed exon 7 region) results in a new junction, nt 945/1068. This deletion introduced a frameshift resulting in 36 novel amino acids (in bold italics) at the C-terminal region, after the threonine at position 315. Ctrl, normal Rh50 cDNA.5 The star identifies the stop codon located in exon 9. Junctions of exons 6-7 and exons 7-8 are shown.
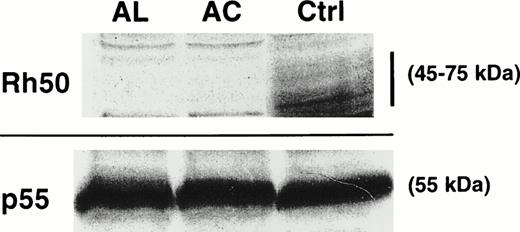
Immunostaining of RBC membrane proteins from AC and AL erythrocytes. The Rh50 glycoprotein carries a N-linked polylactosylaminyl carbohydrate chain and is shown as a diffuse band of 45 to 75 kD in the RhD-positive control (Ctrl) using an antibody directed against the Rh50 N-terminus. No signal was detected in both AC and AL. All samples exhibited equivalent amounts of p55.
Immunostaining of RBC membrane proteins from AC and AL erythrocytes. The Rh50 glycoprotein carries a N-linked polylactosylaminyl carbohydrate chain and is shown as a diffuse band of 45 to 75 kD in the RhD-positive control (Ctrl) using an antibody directed against the Rh50 N-terminus. No signal was detected in both AC and AL. All samples exhibited equivalent amounts of p55.
RH50 gene analysis.
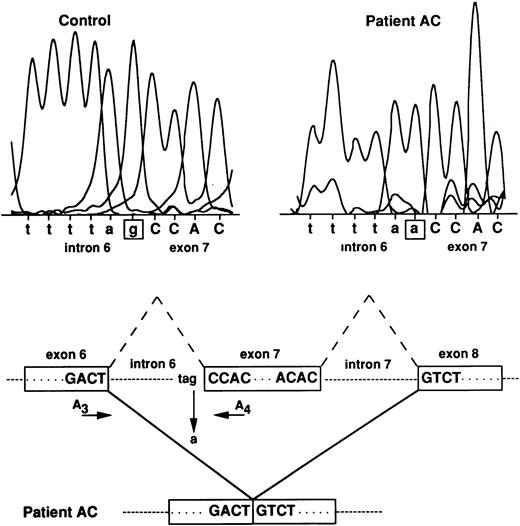
No obvious abnormality of the RH50 locus was observed by Southern blot analysis on genomic DNA extracted from AC and AL blood samples (not shown). Therefore, the 122 nt deletion of the Rh50 cDNA in patient AC may be attributed to an exon skipping during the processing of the pre-mRNA that may well be the result of a 3′ acceptor splice-site mutation localized upstream from nucleotide position 946. To test this hypothesis, the intron 6 regions in AC and RhD-positive genomic DNAs were amplified using primers located in exons 6 and 7 (A3 and A4, see Materials and Methods) and the 1.3-kb PCR genomic product was directly sequenced (Fig 4). Patient AC was found to be homozygous for a G→A point mutation at the invariant G residue of the 3′ acceptor splice-site of intron 6. All members of the family C, sequenced at the genomic level, were found to be heterozygous (G or A) for the same mutation (not shown).
RH50 gene analysis in patient AC. The G residue at the 3′ acceptor splice-site of intron 6 is mutated to A in patient AC, thus resulting in the skipping of exon 7. Normal splicings of introns 6 and 7 are indicated by the broken lines. The approximate position of primers A3 and A4, used to amplify intron 6, is indicated. Intron and exon sequences are in lowercase and uppercase, respectively.
RH50 gene analysis in patient AC. The G residue at the 3′ acceptor splice-site of intron 6 is mutated to A in patient AC, thus resulting in the skipping of exon 7. Normal splicings of introns 6 and 7 are indicated by the broken lines. The approximate position of primers A3 and A4, used to amplify intron 6, is indicated. Intron and exon sequences are in lowercase and uppercase, respectively.
To characterize the mutation affecting the RH50 gene in AL, a 316-bp genomic fragment, encompassing exon 1 as well as 157 bp of intron 1, was amplified (see Materials and Methods) and cloned into the PCR II vector. Sequence analysis showed a G→A transition at the invariant first base of the 5′ donor splice-site of intron 1 (Fig 5A). This point mutation created a novel Rca I restriction site polymorphism in genomic DNA, which was used to test the inheritance of the RH50 mutation in family L by a PCR-restriction fragment length polymorphism (RFLP) assay (Fig 5B). As expected, the 316-bp PCR product remained uncut in BL (wife of the propositus) as well as in control DNAs, whereas it was cleaved into two fragments of 158 bp each in AL (homozygous for the mutation). The daughter LL exhibited a heterozygous pattern in which both uncleaved and cleaved amplification products are present.
Mutation of the RH50 gene in patient AL and its inheritance in the family. (A) The G nucleotide of the 5′ donor splice-site of intron 1 in the control sequence is replaced by an A in AL, thus creating a new Rca I site. (B) Tree of family L andRca I restriction pattern of the genomic PCR products are shown. The 316-bp PCR product (see Materials and Methods) encompassing the splice-site mutation shown in (A) was cleaved into two 158-bp fragments in AL, demonstrating the homozygosity of the mutation in this patient. As expected, the product from a control DNA was uncut. The arrow shows the Rhnull proposita AL. The initials of each family member are indicated. Intron and exon sequences are in lowercase and uppercase, respectively.
Mutation of the RH50 gene in patient AL and its inheritance in the family. (A) The G nucleotide of the 5′ donor splice-site of intron 1 in the control sequence is replaced by an A in AL, thus creating a new Rca I site. (B) Tree of family L andRca I restriction pattern of the genomic PCR products are shown. The 316-bp PCR product (see Materials and Methods) encompassing the splice-site mutation shown in (A) was cleaved into two 158-bp fragments in AL, demonstrating the homozygosity of the mutation in this patient. As expected, the product from a control DNA was uncut. The arrow shows the Rhnull proposita AL. The initials of each family member are indicated. Intron and exon sequences are in lowercase and uppercase, respectively.
Quantification of Rh50 mRNA in Rhnull patients.
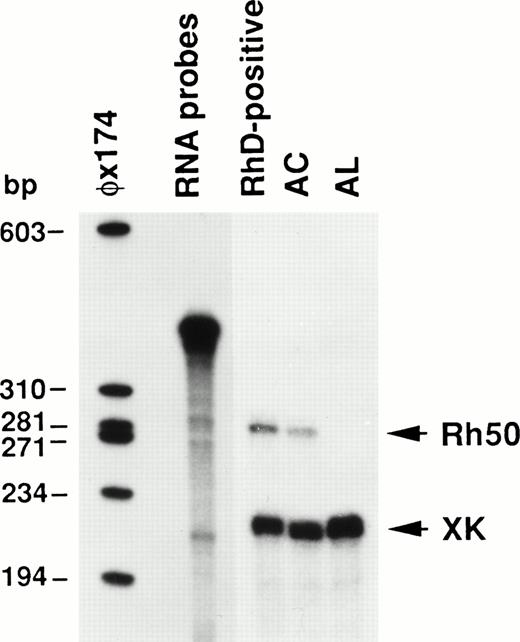
To elucidate the functional significance of the mutations affecting theRH50 gene in AL and AC, the Rh50 mRNAs from these patients were quantified by ribonucleotide protection assay (RPA) using the XK mRNA as an internal standard. The results, shown in Fig 6, definitely demonstrate that no Rh50 mRNA was detectable in the AL patient. Moreover, the amount of Rh50 mRNA in the AC sample was found to be reduced to about 54% of the level detected in the RhD-positive control.
Ribonuclease protection analysis of Rh50 mRNAs. Total reticulocyte RNAs (12.5 μg) protected a 270 nt Rh50 fragment and a 207 nt XK fragment. Rh50 mRNA levels in AC, compared after normalizing them to the XK internal standard, were about 54% of those detected in the RhD-positive control. No Rh50 mRNA was detected in AL. RNA probes of 396 nt and 333 nt for Rh50 and XK, respectively, migrate very close to each other in the gel. A radiolabeled HaeIII digest of phage ◊X174 was used as a size marker.
Ribonuclease protection analysis of Rh50 mRNAs. Total reticulocyte RNAs (12.5 μg) protected a 270 nt Rh50 fragment and a 207 nt XK fragment. Rh50 mRNA levels in AC, compared after normalizing them to the XK internal standard, were about 54% of those detected in the RhD-positive control. No Rh50 mRNA was detected in AL. RNA probes of 396 nt and 333 nt for Rh50 and XK, respectively, migrate very close to each other in the gel. A radiolabeled HaeIII digest of phage ◊X174 was used as a size marker.
DISCUSSION
In the Rhnull patients of the regulator type studied so far, the RH50 gene is altered by mutations that most likely account for the lack or the severe reduction of RBC surface expression of the Rh50 glycoprotein.14,15 Therefore, this protein may be essential for the correct expression of the Rh antigens on RBCs.5 14 In the present study, we have identified new mutations affecting different splice-sites of the RH50 gene in 2 unrelated Rhnull individuals.
Patient AC was found to be homozygous for a G→A transition at the invariant base of a 3′ acceptor splice-site, which leads to the skipping of the downstream exon 7 (Fig 4). Interestingly, the RPA analysis showed that AC Rh50 mRNA was reduced to about half of the amount found in the RhD-positive phenotype (Fig 6). This could be due to either the lower transcriptional activity of the mRNA or its lower stability. The abnormality of the Rh50 mRNA, disrupted by a 122 nt deletion, seems more likely to favor the second hypothesis. The presence of potential crytpic splice-sites in the intron 6 or exon 7 regions is ruled out by the fact that no PCR products were detected in AC in amplification reactions encompassing the cDNA region corresponding to exon 6 and exon 7 (not shown).
The premature termination of translation is expected to alter the structure and conformation of the protein. Indeed, the predicted truncated protein would be 351 amino acids long (v 409 residues for the normal Rh50 protein), including a C-terminus region containing 36 new amino acids. Moreover, hydropathy analysis of the deduced protein sequence predicted a reverse orientation of the C-terminal region, which may become exposed extracellularly (not shown). However, the mutated protein was not found on the RBC surface, because no reactivity of both the 2D10 and the anti–N-terminus antibodies could be detected by flow cytometry and Western blot analyses, respectively. These results suggest that the mutant protein is either unstable (degradation) or that it is not transported to the cell surface. In the latter case, one may speculate that the C-terminal region of the Rh50 protein is critical for the assembly or transport of the Rh membrane complex to the plasma membrane. Experimental confirmation of this hypothesis will be necessary when an efficient system for the coexpression of both Rh and Rh50 proteins will became available.
The mutation described in patient AL occurs at a critical position of the 5′ splice-site of intron 1 of the RH50 gene and therefore completely inactivates splicing. Indeed, mutations at the canonical GT dinucleotide at the donor splice-site alter RNA processing and can lead to several splicing defects.28-30 As clearly demonstrated by RPA analysis (Fig 6), the mutation affecting AL results in the lack of Rh50 mRNA. This may be accounted for by the instability and the consequent degradation of the misspliced RNA in the nucleus due to either the inclusion of intron 1 (about 17.7 kb)7and/or the appearance of a premature stop codon. Reduction of the levels of mRNA associated with splice mutations and premature termination codon has been reported for several mutant alleles31-33 (and references therein).
In conclusion, we have described a novel type of mutation affecting two different splice-sites of the RH50 gene, which identify an additional molecular mechanism responsible for Rh deficiency. These findings further substantiate the hypothesis that the RH50mutants represent distinct alleles of the X°r gene, the so-called Rh suppressor gene,34 which most likely function by preventing the assembly and transport of the Rh complex to the RBC membrane. Clearly, these mutants do not act as a transcriptional regulators of the RH locus, because in all Rhnull regulator individuals analyzed so far, the Rh transcripts are normally present and their sequence is not altered. Mapping the Rh50 mutations in Rhnull patients, therefore, might help to define critical positions or domains of the Rh50 protein that are involved in the assembly of the Rh membrane complex and the expression of Rh antigens.
ACKNOWLEDGMENT
The authors thank L. Lebeck for the gift of blood sample AL and P. Bailly for the gift of the A23 antibody and helpful discussion.
Supported in part by a EC Grant No. BIO2-CT93-0348. G.M. thanks ORTHO Clinical Diagnostics for financial support.
Address reprint requests to Jean-Pierre Cartron, PhD, INSERM U76, Institut National de la Transfusion Sanguine, 6 rue Alexandre Cabanel, 75015 Paris, France; e-mail: cartron@infobiogen.fr.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal