Abstract
The Duffy (FY) blood group antigens are carried by the DARC glycoprotein, a widely expressed chemokine receptor. The molecular basis of the Fya/Fyb and Fy(a-b-) polymorphisms has been clarified, but little is known about the Fyxantigen and the FY*X allele associated with weak expression of Fyb, Fy3, Fy5, and Fy6 antigens. We analyzed here the structure and expression of the FY gene in 4 Fy(a-bweak) individuals. As compared with Fy(a-b+) controls, the Fy(a-bweak) red blood cell membranes contained residual amount of DARC polypeptide and these cells were poorly bound by anti-Fy antibodies and chemokines. The FY gene from Fy(a-b+) and Fy(a-bweak) individuals differed by one substitution, C286T. The resulting Arg89Cys amino acid change reduced the binding of anti-Fy antibodies and chemokines to DARC transfectants. We concluded that the Fybweak donors carried theFY*X allele at the FY locus and that the Fyxantigen corresponds to highly reduced expression of a grossly normal Fyb polypeptide caused by the Arg89Cys substitution. Because FY is a single copy gene, this defect should also affect DARC expression in nonerythroid cells. Because the Fyx phenotype is not associated with apparent clinical consequences, we discussed these findings in the light of the putative roles of DARC in various tissues. Finally, we developed a Fyx DNA typing assay that should be useful for genetic studies and clinical transfusion medicine.
© 1998 by The American Society of Hematology.
THE DUFFY BLOOD group antigens are of wide interest in clinical medicine because of their involvement in transfusion incompatibilities and hemolityc disease of the newborn (HDN).1 The Duffy blood group system should represent one of the best illustrations of the link between genetics and biology. Indeed, before their molecular cloning, the Duffy antigens were already recognized as the erythrocyte receptor for malaria parasites and for chemokines because Duffy-positive but not Duffy-negative erythrocytes can be invaded by Plasmodium vivax2 andPlasmodium knowlesi3 and can bind interleukin-8 (IL-8).4,5 The Duffy antigens are carried by a 336 amino acid glycoprotein, originally named gpD,6 that exhibits significant protein sequence homology with the human and rabbit IL-8 receptors and that is most likely organized into seven transmembrane domains, like all members of the G-protein-coupled chemokine receptors.7 gpD is now referred to as the promiscuous chemokine receptor or the Duffy antigen/receptor for chemokine (DARC), because it expresses all Fya/b, Fy3, and Fy6 antigens7-9 and can bind chemokines of both the CC (RANTES, MCP-1) and CXC (IL-8, MGSA) classes.7,8 In addition to erythroid cells, DARC is expressed on endothelial cells lining postcapillary venules throughout the body,10,11 on vascular endothelial and epithelial cells in some nonerythroid organs,12 and on Purkinje cells in the cerebellum.13 The same DARC polypeptide isoform is expressed in all tissues studied so far,10,12,14 including the brain, where a larger FY mRNA (7.5 kb v 1.35 kb) is produced by the use of a specific promoter.14 The function(s) of the Duffy antigens as a widely expressed promiscuous chemokine receptor is not elucidated. However, it has been postulated that DARC on red blood cells (RBCs) could act as a sink or scavenger to inactivate excess chemokines released into the circulation.4 Accordingly, it has been demonstrated that IL-8 released after myocardial infarction is mainly bound to erythrocytes.15 There is no evidence that DARC on nonerythroid cells could transduce a signal across the membrane upon chemokine binding.7 However, transfectant cell analysis indicated that endothelial DARC could internalize ligands,11 and a recent study suggested that DARC might participate to transcytosis and surface presentation of IL-8 by venular endothelial cells,16 an important site for chemokine-induced leukocyte emigration during inflammatory process.
The receptor properties of DARC for malaria parasites and chemokines, together with the existence of phenotypes associated with quantitative and/or qualitative alteration of Fy antigen expression, provided new interest in the elucidation of the molecular genetic basis of the Duffy blood group system. The single copy FY gene is composed of two exons,17 and the two common alleles in Caucasians, FY*A and FY*B, differ by a single G to A nucleotide substitution resulting in the amino acid change Gly42Asp in the NH2 extracellular domain.9,18,19,20 In Caucasian populations, the three phenotypes Fy(a+b-), Fy(a-b+), and Fy(a+b+) given by these codominant alleles have a frequency of 0.195, 0.33, and 0.475, respectively. The phenotype Fy(a-b-), conveyed by homozygosy for the FY*Fy allele, is extremely rare in Caucasians but represents the major phenotype in blacks. The FY*Fy allele corresponds to a normal FY*B coding sequence but exhibits a mutation in the promoter region, T-46C, that, by disrupting a binding site for the h-GATA-1 erythroid transcription factor, abolishes expression of the FY gene in erythroid but not in nonerythroid cells.21,22 Thus, Fy(a-b-) individuals resistPlasmodium vivax infection because they lack DARC on their RBCs18 but have no obvious abnormality in the regulation of inflamation most likely because they express DARC normally on their nonerythroid tissues.11 12
In 1965, Chown et al23 reported that the very weak and variable reaction of some RBCs with anti-Fyb antibodies accounts for many seeming anomalies of inheritance in Caucasian families. These investigators postulated that the Fybweakphenotype is conveyed by a fourth allele at the FY locus with a frequency of 2%, which they named FY*X because the total gene product was not certain. Further serological studies indicated that expression of Fy3, Fy5, and Fy6 antigens is also depressed in Fybweak RBCs from donors with the putative genotypeFY*X/*X.24- 28 Although it has been proposed that Fyx should correspond to a quantitative rather than a qualitative variant of Fyb, the molecular nature, the biological properties, and the genetic background of the Fyx antigen and of the FY*X allele, constituted one of the dark holes in the study of the Duffy blood group system. To clarify these issues, we analyzed the structure and expression of the FY locus from 4 Fy(a-bweak) donors. We identified a mutation that accounts for very low DARC expression and chemokine binding to RBCs and transfected eukaryotic cells. Our results, together with the previous characterization of FY*A, FY*B, andFY*Fy, provide the definitive proof that the Duffy blood group system is controlled by four alleles.
MATERIALS AND METHODS
Blood samples.
Blood samples from healthy donors were collected on EDTA and the Fy phenotypes were determined by agglutination studies using the antiglobulin gel test (Diamed SA, Morat, Switzerland) and by subsequent adsorption/elution studies, when necessary. The Fy(a-bweak) donors BE.T (and family members) and donors SEV and BAR were obtained from the Etablissement de Transfusion Sanguine of Hopital PitiéSalepêtrière (Paris, France) and the Centre National de Réference pour les Groupes Sanguins (CNRGS; Paris, France), respectively. The Fy(a-bweak) blood sample TAR was collected at the Centre Regional de Transfusion Sanguine of Toulouse (Toulouse, France).
Materials.
125I-labeled IL-8, MGSA, and RANTES (specific activity, 2,200 Ci/mmol) were obtained from DuPont NEN (Boston, MA). Unlabeled and fluorescent fluorescein isothiocyanate (FITC)-conjugated recombinant IL-829 were donated by Dr A. Proudfoot (Glaxo Wellcome Research and Development, Geneva, Switzerland). The anti-Fy6 (i3A), anti-Fy3 (CRC-512), and anti-Fya MoAbs were kindly provided by Dr D. Blanchard (CRTS, Nantes, France), Dr Makato Uchikawa (Japanese Red Cross, Tokyo, Japan), and Dr F. Buffiere (ETS, Bordeaux, France), respectively. Anti-Fya and anti-Fyb human polyclonal antibodies were from Ortho Diagnostic Systems (Raritan, NJ). The anti-p55 rabbit polyclonal antibody, prepared by immunizing rabbit with synthetic peptide residues 28 to 47 of the human p55 protein,30 was donated by Dr P. Bailly (INTS, Paris, France).
Polymerase chain reaction (PCR) amplification of FY gene and transcripts.
Total RNAs were extracted from 400 μL of whole peripheral blood by the miniscale acid-phenol-guanidium method.31 The pellet was resuspended in 50 μL H2O, and 10 μL was used to produce first cDNA strands using the first-strand cDNA synthesis kit (Pharmacia, Uppsala, Sweden). One sixth of the cDNA products were enzymatically amplified between primers FY100, 5′-GAACCAAACGGTGCCATGGGGAACTGTCTG-3′ (sense, positions −15 to +15), and FY99, 5′- GGGAAGAGAACTAGGATTTGCTTCCAAGGG-3′ (antisense, positions +1021 to +992). Genomic DNA (200 ng) isolated from whole blood was amplified between primers FY94, 5′-AACAGCGTCCCCTAACCAG-3′ (sense, positions −1253 to −1236), and FY99. Thirty cycles of PCR were performed as follows: 1 minute at 94°C, 1 minute at 58°C, and 2 minutes at 72°C. PCR products were subcloned in plasmid vector and sequenced by the dideoxy chain termination method using an ALFexpress automatic DNA sequencer (Pharmacia).
Restriction analysis of PCR-amplified genomic sequences.
For the FY*X restriction fragment length polymorphism (RFLP) assay, the PCR reactions were performed with 200 ng of leukocyte DNA between primers FY7, 5′-ACTCTGCACTGCCCTTCTTC-3′ (sense, positions +176 to +195), and FY57, 5′-TGGGCAAAGGCTGAGCCA-3′ (antisense, positions +428 to +411), under the following conditions: 30 cycles of 40 seconds at 94°C, 1 minute at 58°C, and 45 seconds at 72°C. PCR products were purified on Ultrafree-MC (30,000) filter units (Millipore, Bedford, MA) and one third was digested for 2 hours with 20 U of the Aci I restriction enzyme. Restriction fragments were directly analyzed in 12% acrylamide minigels.FY*A/FY*B genotyping was performed as decribed.9FY*Fy typing was performed essentially as described,21 except that the reverse primer P39 was changed to FY97, 5′ TGTGGCAGACAGTTCCCCATGG-3′ (position +40 to +27), to better separate the FY*Fy specific Sty I restriction fragment (64 bp) generated by the T-46C mutation from other fragments.
Construction of Fy expression plasmid and mutagenesis.
pcDNA-3 expression vector (Invitrogen BV, Leek, The Netherlands) carrying the coding sequence of the major isoform (spliced) of Fyb was described elsewhere.32Mutagenesis was subsequently performed on the recombinant plasmids by the use of PCR primers carrying appropriate nucleotide substitutions and the Quick Change Site Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Inserts of the mutated plasmids were sequenced as described above.
Cell culture and transfection.
COS-7 cells were obtained from the American Type Culture Collection (Rockville, MD) and were grown in Iscove medium supplemented with 10% fetal calf serum and 50 μg/mL penicillin and streptomycin. Cells (3 × 106/assay) were transfected with 10 μg of recombinant plasmid using Lipofectin reagent (Life Technologies, SARL, Cergy-Pontoise, France). Transiently transfected COS-7 cells were analyzed after 36 hours of culture.
Flow cytometry analysis.
Expression of Duffy antigens on RBCs or transfectant cell lines was measured on a FACScan flow cytometer (Becton Dickinson, San Jose, CA) using anti-Fya, anti-Fy6, and anti-Fy3 MoAbs and anti-Fyb PoAb. Cells (3 × 105) were incubated for 60 minutes at 22°C with appropriate dilution of antibody in 0.15 mol/L phosphate-buffered saline (PBS). After washing with PBS supplemented with 0.5% bovine serum albumin (BSA), the cell suspension was incubated with fluorescein-conjugated antimouse or antihuman IgG (H+L) (Immunotech, Marseille, France). After another washing step, 0.1 ng of propidium iodide (PI) was finally added to 1 mL of cell suspension. PI-positive cells (dead cells) were excluded from analysis. Fy(a-b-) RBCs or mock-transfected cells and irrelevant mouse and human MoAbs were used as negative controls.
Receptor binding assay.
RBCs (108) and COS-7 cell transfectants (106) were analyzed for their ability to bind 125I-chemokines as previously described.32 Cells transfected with the pcDNA3 vector alone were used as negative controls.
The binding of FITC-conjugated IL-8 to RBCs was directly measured by flow cytometry analysis.
Protein chemistry.
RBCs membranes from Fy-typed donors were prepared by hypotonic lysis.33 For Western blot analysis, RBC membrane proteins (50 μg) were separated by discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)34using a Novex apparatus (San Francisco, CA) and transferred to nitrocellulose sheets. The blot was incubated with an appropriate dilution of the i3A anti-Fy 6 MoAb and then with antimouse IgG peroxydase-tagged antibody (Biosis, Compiègne, France). Finally, the immunoblot was stained with the ECL chemiluminescent system from Amersham (Bucks, UK) and exposed to x-ray film.
RESULTS
Duffy antigen expression on Fy(a-bweak) RBCs.
In agglutination studies with polyclonal anti-Fy reagents, RBCs from donors BAR and SEV were unreactive with the anti-Fyaantibodies and gave positive reactions with some anti-Fybantisera, but with an agglutination titer fivefold lower than those given by Fy(a−b+) and Fy(a+b+) controls. RBCs from donors TAR and BE.T were unreactive with all anti-Fya and anti-Fyb reagents and thus were initially considered as Fy(a−b−) (data not shown). However, in subsequent adsorption-elution experiments, these two RBC samples yielded an antibody with Fyb specificity. From these analysis, donors BAR, SEV, TAR, and BE.T were phenotyped Fy(a−bweak). The altered expression of Fyb on these RBC samples was better shown by flow cytometry analysis (Table 1). When compared with the Fy(a−b+), Fy(a+b+), Fy(a+b−), and Fy(a−b−) controls, a faint fluorescence signal was detected on the Fybweak RBCs, ranging from 3% to 7.7% of that obtained with Fy(a−b+) (FY*B/*B) control RBCs. Similarly, the use of anti-Fy3 and anti-Fy6 monoclonal antibodies (MoAbs) also showed a drastic alteration of Fy3 and Fy6 antigen expression at the surface of the Fy(a−bweak) RBCs. The decrease of anti-Fy3 and anti-Fy6 antibody binding capacity were strictly correlated between samples and the RBCs from donor BAR were stained approximatively twice as much as the RBCs from the 3 other Fy(a−bweak) donors with all anti-Fy antibodies (Table 1). The apparent Duffy site numbers per cell, as estimated from the anti-Fy3 and anti-Fy6 MoAb binding capacity, were 2,200 to 3,400 for the Fy(a−b+) controls; 150 to 230 for SEV, TAR, and BE.T; and 250 to 470 for BAR. As expected, anti-Fya MoAb bound neither to Fy(a−bweak) nor to the Fya-negative control RBCs. The Fybweak, Fy3weak, and Fy6weakphenotype of the 4 Fy(a−bweak) donors strongly suggested that they belong to the Fyx phenotype originally described and therefore should carry the FY*X allele of the Duffy blood group system.
Serological Characteristics of the Duffy Samples as Determined by Flow Cytometry Analysis
| . | Antigens . | |||
|---|---|---|---|---|
| Fya * . | Fyb * . | Fy3-151 . | Fy6-151 . | |
| Samples | ||||
| BAR | 0 | 140 | 470 | 250 |
| SEV | 0 | 33 | 220 | 150 |
| TAR | 0 | 80 | 230 | 150 |
| BE.T | 0 | 50 | 275 | 190 |
| BE.L | 1,600 | 0 | 2,300 | 1,560 |
| BE.C | 1,800 | 110 | 2,950 | 1,990 |
| Controls (n = 3) | ||||
| Fy a−b− | 0 | 0 | 0 | 0 |
| Fy a−b+ | 0 | 1,500-1,800 | 3,200-3,400 | 2,200-2,400 |
| Fy a+b− | 3,400-3,700 | 0 | 4,100-4,500 | 3,100-3,300 |
| Fy a+b+ | 1,500-1,600 | 800-820 | 3,500-3,800 | 2,200-2,600 |
| . | Antigens . | |||
|---|---|---|---|---|
| Fya * . | Fyb * . | Fy3-151 . | Fy6-151 . | |
| Samples | ||||
| BAR | 0 | 140 | 470 | 250 |
| SEV | 0 | 33 | 220 | 150 |
| TAR | 0 | 80 | 230 | 150 |
| BE.T | 0 | 50 | 275 | 190 |
| BE.L | 1,600 | 0 | 2,300 | 1,560 |
| BE.C | 1,800 | 110 | 2,950 | 1,990 |
| Controls (n = 3) | ||||
| Fy a−b− | 0 | 0 | 0 | 0 |
| Fy a−b+ | 0 | 1,500-1,800 | 3,200-3,400 | 2,200-2,400 |
| Fy a+b− | 3,400-3,700 | 0 | 4,100-4,500 | 3,100-3,300 |
| Fy a+b+ | 1,500-1,600 | 800-820 | 3,500-3,800 | 2,200-2,600 |
*FITC-equivalent/cell.
Site number/cell.
Chemokine binding to Fy(a−bweak) RBCs.
The Duffy antigens expressed on the Fy(a−bweak) RBCs were further characterized by analyzing their chemokine receptor properties. In a first set of experiments, the binding of IL-8 to the four Fy(a−bweak) samples was measured by flow cytometry using increasing concentrations of fluorescent FITC-conjugated IL-8 (Fig 1). As expected, IL-8 bound strongly to Fy(a−b+) RBCs but did not bind significantly to Fy(a−b−) RBCs that are deficient in DARC glycoprotein.6Fy(a−bweak) RBCs exhibited little but reproducible binding to IL-8 as compared with Fy(a−b−) RBCs, with the staining of BAR RBCs being twice as strong as compared with the other Fy(a−bweak) samples. In a second set of experiments, the binding of 125I-labeled chemokines to Fy(a−bweak) samples BAR and SEV was examined. Quantitative analysis indicated that IL-8 binding to these cells represented 24% and 12%, respectively, of the binding to the positive controls (Table 2). When tested for binding of 125I-labeled MGSA and RANTES, the RBCs from BAR also exhibited a 60% and 80% reduction of chemokine binding, respectively, as compared with Fyb-positive controls.
Flow cytometry analysis of IL-8 binding to RBCs. RBCs (5 × 104) from the Fy(a−bweak) donors SEV and BAR and from Fy(a−b+) and Fy(a−b−) controls with the determined genotypes FY*B/*B and FY*Fy/*Fy, respectively, were incubated with increasing concentration of fluorescent FITC-conjugated IL-8. Chemokine binding was analyzed by flow cytometry. Results are expressed as the specific IL-8 binding (arbitrary units) versus ligand concentration. Nonspecific signal was determined when incubation with the FITC-labeled IL-8 was performed in presence of a 100-fold excess of unlabeled IL-8.
Flow cytometry analysis of IL-8 binding to RBCs. RBCs (5 × 104) from the Fy(a−bweak) donors SEV and BAR and from Fy(a−b+) and Fy(a−b−) controls with the determined genotypes FY*B/*B and FY*Fy/*Fy, respectively, were incubated with increasing concentration of fluorescent FITC-conjugated IL-8. Chemokine binding was analyzed by flow cytometry. Results are expressed as the specific IL-8 binding (arbitrary units) versus ligand concentration. Nonspecific signal was determined when incubation with the FITC-labeled IL-8 was performed in presence of a 100-fold excess of unlabeled IL-8.
Chemokine Binding to RBCs
| . | IL-8 . | MGSA . | RANTES . | |||
|---|---|---|---|---|---|---|
| A . | B . | A . | B . | A . | B . | |
| Samples | ||||||
| BAR | 23.5 | 5.7 | 176 | 7.5 | 37 | 15 |
| SEV | 16 | 5 | NT | NT | NT | NT |
| Controls (n = 3) | ||||||
| Fy a−b+ | 84-86 | 11-12 | 380-390 | 28.5-30 | 95-100 | 12-15 |
| Fy a−b− | 4-4.5 | 3.6-3.7 | 3.5-3.8 | 2.8-3 | 6-8 | 10-11 |
| . | IL-8 . | MGSA . | RANTES . | |||
|---|---|---|---|---|---|---|
| A . | B . | A . | B . | A . | B . | |
| Samples | ||||||
| BAR | 23.5 | 5.7 | 176 | 7.5 | 37 | 15 |
| SEV | 16 | 5 | NT | NT | NT | NT |
| Controls (n = 3) | ||||||
| Fy a−b+ | 84-86 | 11-12 | 380-390 | 28.5-30 | 95-100 | 12-15 |
| Fy a−b− | 4-4.5 | 3.6-3.7 | 3.5-3.8 | 2.8-3 | 6-8 | 10-11 |
(A) 108 RBCs were incubated with 0.5 nmol/L125I-labeled chemokine. Results are expressed as bound cpm (×103). (B) Same as (A), but 200 nmol/L of cold ligand was added.
Abbreviation: NT, not tested.
Western blot analysis.
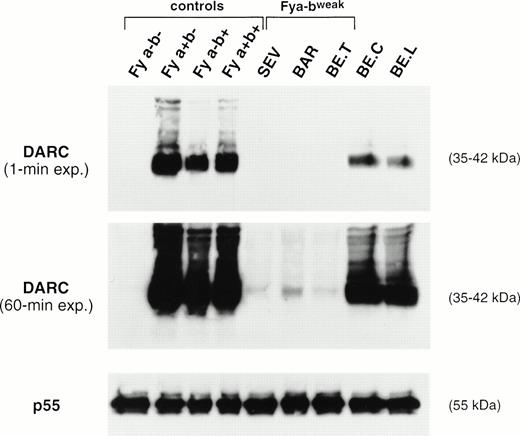
The amount of DARC polypeptide in erythrocyte membranes from donors with different Fy phenotypes was compared by Western blot analysis, using the iA3 MoAb directed against the Fy6 antigen (Fig 2). As expected, no signal was observed in the Fy(a−b−) negative controls. DARC was consistently detected in membranes from Fy(a+b−), Fy(a+b+), and Fy(a−b+) controls with the FY*A/*A, FY*A/*B, and FY*B/*Bgenotypes, respectively, whereas a signal 50% lower was observed in membranes from donors with the FY*A/*Bweak(BE.C) and FY*Fy/*A (BE.L) genotypes, respectively (see below). Nevertheless, DARC was not detected in membranes from SEV, BE.T, and BAR Fy(a−bweak) donors (Fig 2A). However, prolonged radiographic exposure showed that these membranes had a barely detectable signal, equivalent to less than 5% of the normal intensity, with the signal in SEV and BE.T samples being 50% of that in BAR membranes (Fig 2B). As a control, all samples exhibited normal amounts of the p55 peripheral protein (Fig 2C).
Immunoblot analysis of RBC membrane proteins. Total membrane proteins (60 μg) separated on SDS-PAGE were blotted on nitrocellulose sheets and incubated with the murine anti-Fy6 MoAb i3A and with a rabbit anti-p55 antibody. The DARC glycoproteins and p55 proteins were visualized by chemioluminescence autoradiography using goat antimouse or goat antirabbit IgG conjugated to horseradish peroxidase, as second antibodies, respectively. The 35- to 45-kD DARC glycoprotein forms aggregates migrating as bands of 90 and 200 kD.45
Immunoblot analysis of RBC membrane proteins. Total membrane proteins (60 μg) separated on SDS-PAGE were blotted on nitrocellulose sheets and incubated with the murine anti-Fy6 MoAb i3A and with a rabbit anti-p55 antibody. The DARC glycoproteins and p55 proteins were visualized by chemioluminescence autoradiography using goat antimouse or goat antirabbit IgG conjugated to horseradish peroxidase, as second antibodies, respectively. The 35- to 45-kD DARC glycoprotein forms aggregates migrating as bands of 90 and 200 kD.45
FY gene and transcript analysis.
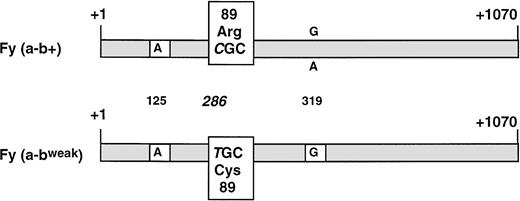
RNAs extracted from whole blood of the 4 Fy(a−bweak) donors and from a Fy(a−b+) control were reverse transcribed to cDNA and used as template to amplify the entire coding region for the major (spliced) isoform of DARC between primers FY100 and FY99. A PCR product of the expected size (1,036 bp) was obtained in all samples (data not shown) and subcloned in plasmid vector. Sequence analysis of several clones from each Fy(a−bweak) sample showed one kind of cDNA that differed from the common FY*B allele by only one substitution, C286T (+1 taken as the erythroid cap site21; Fig 3). A survey of the literature indicated that T286 has never been found in theFY*A, FY*B, or FY*Fy alleles of donors with common Duffy phenoypes. The C286T nucleotide change resulted in the amino-acid substitution Arg to Cys at position 89 of the DARC polypeptide. It is noteworthy that all Fybweak cDNA clones carried a G at nucleotide 319, whereas G or A, resulting in Ala100Thr substitution, was identified at this position by sequencing theFY*B or FY*A alleles from Fy(a−b+) or Fy(a+b+) donors7 20 (and our unpublished data).
Schematic comparative structure of the DARC cDNAs isolated from whole blood of Fy(a−b+) and Fy(a−bweak) donors. The full-length major isoform (spliced) of the FY transcripts was isolated by RT-PCR. The nucleotide sequences of Fy(a−b+) and Fy(a−bweak) cDNA clones were identical, except for the C286T substitution (+1 taken as the erythroid cap site) resulting in the Arg89Cys polymorphism on the DARC protein. G was found at nucleotide position 319 in all clones from four unrelated Fy(a−bweak) donors, whereas G or A, resulting in Ala100Thr amino acid change, could be found in the FY*B allele from Fy(a+b+) and Fy(a-b+) donors. The nucleotide residue found at the Fya/Fyb-associated polymorphic position (G125A) is indicated.
Schematic comparative structure of the DARC cDNAs isolated from whole blood of Fy(a−b+) and Fy(a−bweak) donors. The full-length major isoform (spliced) of the FY transcripts was isolated by RT-PCR. The nucleotide sequences of Fy(a−b+) and Fy(a−bweak) cDNA clones were identical, except for the C286T substitution (+1 taken as the erythroid cap site) resulting in the Arg89Cys polymorphism on the DARC protein. G was found at nucleotide position 319 in all clones from four unrelated Fy(a−bweak) donors, whereas G or A, resulting in Ala100Thr amino acid change, could be found in the FY*B allele from Fy(a+b+) and Fy(a-b+) donors. The nucleotide residue found at the Fya/Fyb-associated polymorphic position (G125A) is indicated.
The C286T nucleotide substitution was confirmed at the genomic level after amplification of the FY gene, including intron 1 and 1,253 nucleotides in 5′ of the erythroid cap site. However, in contrast to the cDNA analysis, sequence analysis of several genomic clones suggested homozygosity for donor BAR and showed the heterozygosity of donors SEV, BE.T, and TAR. One species of clones from SEV, BE.T, and TAR samples carried a normal Fyb coding sequence and exhibited the T-46C mutation in the promoter region typical ofFY*Fy and was previously shown to abolish the erythroid expression of this bone marrow silent FY*Ballele.21 Accordingly, this allele could not be shown by cDNA analysis of erythroid cells. Conversely, the second species of clones exhibited the C286T substitution but no additional polymorphism compared with the noncoding and 5′ flanking sequences ofFY*B gene. These results indicated that the genotype of the Fy(a-bweak) donors SEV, TAR, and BE.T was FY*Fy/*Xand suggested that the genotype of donor BAR was FY*X/*X.
Fyx DNA typing by PCR-RFLP.
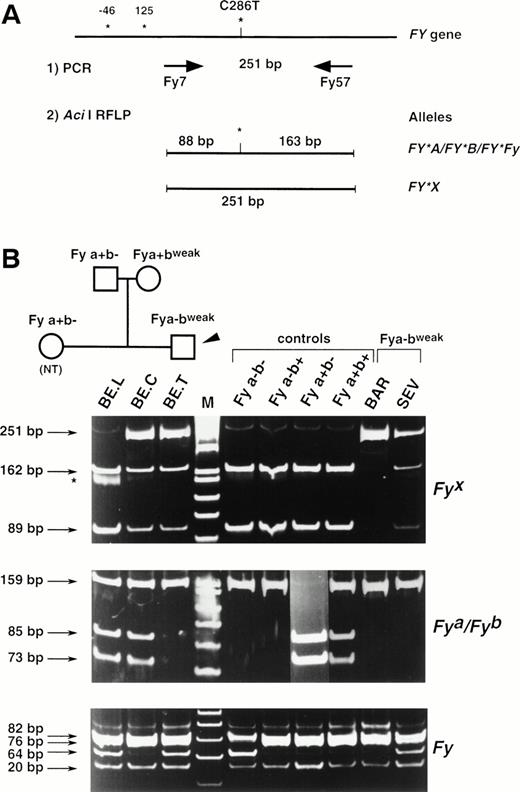
The substitution identified at nucleotide 286 of the FY mRNAs was correlated with the presence or the absence of an Aci I restriction site (CCGC→CTGC) on the Fy(a−b+) and Fy(a−bweak) clones, respectively. To confirm that the C286T substitution is associated with the Fybweakphenotype, a PCR-RFLP assay was developed. Primers FY7 and FY57 (see Materials and Methods) were designed to amplify a 251-bp fragment encompassing this polymorphic position on the FY gene. PCR-RFLP of genomic DNAs from Fy typed donors have been performed and typical results are shown in Fig 4. AfterAci I digestion, the 251-bp PCR product was cleaved in two fragments of 88 and 163 bp in all Fy samples except those with the Fy(a−bweak) or Fy(a+bweak) phenotypes. Only the uncleaved 251-bp fragment was observed in donor BAR, whereas the 251-, 163-, and 88-bp fragments were all detected in the heterozygous Fy(a+bweak) sample (BE.C) as well as in samples BE.T, SEV, and TAR.
Full Duffy DNA typing. (A) Strategy of the PCR-RFLP for the detection of the C286T substitution specific of FY*X allele. Primers Fy7 and Fy57 were designed to amplify a 251-bp FYfragment encompassing the single base substitution identified between the Fy(a−b+) and Fy(a−bweak) clones and that was correlated with an allele-specific Aci I restriction site. Nucleotide numbers are as in Fig 3 and do not take into account the intronic sequence of the FY gene. (B) DNA from donors with the indicated Duffy phenotype was used as templates in PCR-RFLP assays. Ten donors with each control phenotypes were analyzed and typical results are shown. The detection of the FY*A-, FY*B-, andFY*Fy-associated polymorphisms (G125A and C-46T, respectively) were based on Ban I and Sty I RFLP, as previously described, with some modifications for FY*Fy typing (see Materials and Methods). The TAR sample gave the same RFLP pattern as SEV and BE.T. The tree of the BE. family is shown to follow the inheritance of the FY*X mutation and to demonstrate that the presence at the heterozygous state of the silent allele FY*Fyin BE. father accounts for the apparent exclusion of paternity (BE.T being Fya-negative, with both parents being Fya-positive).
Full Duffy DNA typing. (A) Strategy of the PCR-RFLP for the detection of the C286T substitution specific of FY*X allele. Primers Fy7 and Fy57 were designed to amplify a 251-bp FYfragment encompassing the single base substitution identified between the Fy(a−b+) and Fy(a−bweak) clones and that was correlated with an allele-specific Aci I restriction site. Nucleotide numbers are as in Fig 3 and do not take into account the intronic sequence of the FY gene. (B) DNA from donors with the indicated Duffy phenotype was used as templates in PCR-RFLP assays. Ten donors with each control phenotypes were analyzed and typical results are shown. The detection of the FY*A-, FY*B-, andFY*Fy-associated polymorphisms (G125A and C-46T, respectively) were based on Ban I and Sty I RFLP, as previously described, with some modifications for FY*Fy typing (see Materials and Methods). The TAR sample gave the same RFLP pattern as SEV and BE.T. The tree of the BE. family is shown to follow the inheritance of the FY*X mutation and to demonstrate that the presence at the heterozygous state of the silent allele FY*Fyin BE. father accounts for the apparent exclusion of paternity (BE.T being Fya-negative, with both parents being Fya-positive).
Together with DNA typing of the FY*A-, FY*B-, andFY*Fy-associated polymorphisms by previously published PCR- RFLP methods (Fig 5B), these results indicated that the C286T substitution is associated with the Fybweak phenotypes [Fy(a−bweak) and Fy(a+bweak)] and confirmed the FY*X/*X genotype of donor BAR and the FY*Fy/*X genotype of donors SEV, BE.T, and TAR.
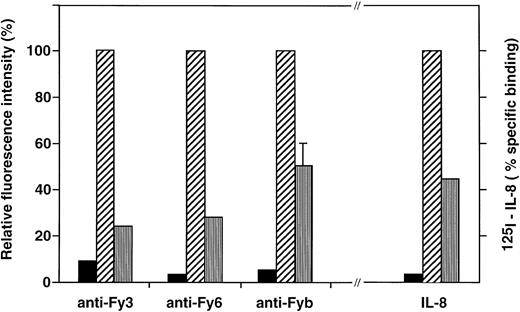
Effect of DARC Arg89Cys substitution on the binding of anti-Fy antibodies and chemokine to COS-7 cell transfectants.COS-7 cells were transiently transfected by the pcDNA3 expression vector alone (▪) and the recombinant vectors containing the cDNA encoding the Fyb (Arg89) (▨), or Fyx (Cys89) (□) DARC protein. Transfectant cells were analyzed for Duffy antigen expression by flow cytometry with anti-Fyb PoAb and anti-Fy3 and anti-Fy6 MoAbs and for chemokine binding with 125I-IL-8, as described in Materials and Methods. Specific binding of 125I-IL-8 yielded 100,000, 45,000, and 2,500 cpm with Fyb, Fyx, and mock transfectants, respectively.
Effect of DARC Arg89Cys substitution on the binding of anti-Fy antibodies and chemokine to COS-7 cell transfectants.COS-7 cells were transiently transfected by the pcDNA3 expression vector alone (▪) and the recombinant vectors containing the cDNA encoding the Fyb (Arg89) (▨), or Fyx (Cys89) (□) DARC protein. Transfectant cells were analyzed for Duffy antigen expression by flow cytometry with anti-Fyb PoAb and anti-Fy3 and anti-Fy6 MoAbs and for chemokine binding with 125I-IL-8, as described in Materials and Methods. Specific binding of 125I-IL-8 yielded 100,000, 45,000, and 2,500 cpm with Fyb, Fyx, and mock transfectants, respectively.
Furthermore, complete FY genotyping of BE.T family showed the inheritance of the C286T substitution with that of weak Fybexpression and demonstrated that the apparent paternity exclusion, suggested by the expression of Fya on RBCs from both parents but not from the propositus, resulted from the presence of the silent FY*B allele, FY*Fy., in the father (BE.L).
Binding of anti-Fy antibodies and chemokine to transfected COS-7 cells.
The C286T substitution identified in the FY*X allele, as defined above, was introduced by site-directed mutagenesis in an expression vector carrying the Fyb cDNA,32 and simian COS-7 cells were transiently transfected by the two recombinant plasmids, pcDNA-Fyx and pcDNA-Fyb. Several independent experiments were performed, and transfectants were further analyzed only when transfection efficiency by the two constructs were similar (4 experiments). Flow cytometry analysis showed that the Fyx transfectants stained poorly with the anti-Fy3 (75% reduction) and anti- Fy6 (70% reduction) MoAbs compared with the Fyb transfectants (Fig 5). Reactivity of anti-Fyb was also significantly lower on Fyxtransfectants, but this polyclonal reagent showed variable reduction of Fyb expression between the different experiments, ranging from 35% to 60%. Calculation of the anti-Fy3 and anti-Fy6 apparent binding site numbers suggested that about 200,000 and 60,000 copies of DARC were expressed at the surface of Fyb and Fyx transfectants, respectively. Binding of125I-labeled IL-8 to transfected cells, performed in parallel to Fy antigen expression analysis, showed a 55% reduction in the chemokine binding capacity of Fyx transfectant compared with Fyb transfectants (Fig 4).
DISCUSSION
The Fyx antigen.
Whereas the previous analyses of the Fyx phenotype were based on barely quantitative agglutination studies, we have performed flow cytometry and chemokine binding analysis to accurately estimate the variation of Fy antigen expression on RBCs from different phenotypes. Using HD50 agglutination assay and human polyclonal antisera, Habibi et al27 previously found a full correlation between Fyb and Fy3 depression on Fyx RBCs. We confirmed these results with the four Fy(a−bweak) samples, which indicated that these cells exhibited a typical Fyx phenotype, as previously defined.23 27 Fy5 expression could not be analyzed because anti-Fy5 was not available. However, we demonstrated that there is also a full correlation between Fy3 and Fy6 expression on RBCs of different phenotypes, including Fyx cells (R = .98, data not shown). Furthermore, by the use of murine MoAbs that allow antigen site density to be precisely estimated, we calculated that, as compared withFY*B/*B control cells, FY*X/*X RBCs expressed only 12% of Fy3 and Fy6 antigens. The decrease in Fyb expression, as measured with human anti-Fy b antisera, was slightly higher, but the polyclonal nature of the reagent did not allow an accurate calculation of Fyb site numbers.
Apart from its biological relevance (see below), chemokine binding to RBCs also represents a powerful approach in the comparison of DARC expression on RBCs with different phenotypes. It has been previously shown that chemokines from both C-C and C-X-C classes bind to RBCs from the three common Fy-positive phenotypes, but not to Fy-negative RBCs.7 We show here that these chemokines bind also to Fyx RBCs, but that the FY*X/*X RBCs could only bind 20% to 30% of the amount of chemokines that bind to FY*B/*BRBCs. It is noteworthy that both flow cytometry and chemokine binding analysis allowed us to discriminate between FY*X/*X andFY*Fy/*X RBCs and that cells mistyped as Fy-negative could be clearly reclassified as Fy-positive.
Recent structure-function analysis of DARC showed that the binding of the various ligands used in this study involved different domains of DARC. The Fy6 epitope has been precisely mapped by synthetic peptide/pins technology and mutagenesis analysis to the heptapeptide comprising residues Q19-25 of the NH-2 terminal extracellular domain.32,35 The epitopes recognized by anti-Fyb and anti-Fy3 have not been fully characterized. However, the Fyb epitope is thought to encompasse the polymorphic G42D position in the NH-2 terminal region associated with the Fya/Fyb antigenic polymorphism9,18-20 and analysis of chimeric receptors indicated that the third extracellular loop of DARC is necessary for the binding of anti-Fy3.36 We have recently demonstrated that the Fy6, Fya/b, and Fy3 epitopes are all involved in the binding of chemokines and that the close association of the first and fourth extracellular domains of DARC by a disulfide bond is required for ligand binding, because it may create a chemokine binding pocket.32 Thus, the ability of Fyx RBCs to bind all the anti-Fy antibodies and chemokines tested indicated that the overall structure of the DARC polypeptide expressed at the membrane of these cells is most likely not altered. Conversely, the parallel decrease of the binding capacity for all ligands together with the highly reduced amount of DARC polypeptide detected in FyxRBC membrane by Western blot analysis strongly suggested that the Fyx antigen represents a poorly expressed but grossly normal Fyb antigen.
The FY*X allele.
Not only has the structure of the FY*X allele not been previously characterized, but whether “Fyb weak reactions lies with Fyx being the tail-end expression of Fyb37 or with it being the expression of a fourth allele,”23 also remained an unresolved matter of controversy. Sequence analysis of the FY transcripts showed that the 4 Fy(a−bweak) donors investigated in this study carry the same allele, the coding sequence of which differs from that of a normalFY*B allele by a single substitution, C286T. Further sequence analysis of the FY gene did not show mutation in the intronic sequence or in the promoter region that could account for transcriptional or posttranscriptional alteration of FYexpression. Conversely, the C286T mutation is most likely causative of low expression of DARC in Fy(a−bweak) erythroid cells, because expression of the FY gene linked to an heterologous promoter demonstrated that the construct with the Fybweaksequence was expressed about twofold to threefold less in COS-7 cells compared with the Fyb construct.
These results provide the definitive proof that the Fybweakphenotype is conveyed by a fourth allele at the FY locus, FY*X(GenBank accession no. AF055992), which is as distinct from theFY*B allele as is the FY*Fy allele in Africans and Afro-Americans. However, the present characterization of FY*X, together with that of FY*A, FY*B,9,18-20and FY*Fy in blacks21,22 and Caucasians20 does not complete the elucidation of the molecular genetic basis the Duffy blood group system. Indeed, the lack of mutation in the sequence encoding for the minor, unspliced, isoform of DARC in 2 unrelated Fy(a−bweak) donors investigated by Mallinson et al20 suggested that the Fybweakphenotype might arise from at least two different genetic mechanisms. However, the sequences of the promoter, exon 1, and intron 1 ofFY was not elucidated when these investigators performed their study. Hence, it remains to be determined whether mutation in the 5′ part of FY accounts for quantitative or qualitative alteration of Fyb expression in the 2 Fybweak donors studied. A further complexity of the Duffy blood group system was recently suggested by Shimizu et al38 in the course of a serotyping study of 434 individuals from several Thai ethnic groups. The investigators emphasized the presence in 8 individuals of a weak-Fya antigen that has never been described before. It is anticipated that these studies could lead to the characterization of a fifth allele at the FY locus, FY*Aweak, which should be to FY*A what FY*X is to FY*B.
FY*X DNA typing in clinical and transfusion medicine.
A FY*X DNA typing based on the C286T substitution has been developed that, together with the previously published FY*A,FY*B, and FY*Fy genotyping, can discriminate between alleles associated with normal (FY*B), bone marrow silent (FY*Fy), and weak (FY*X) expression of the Fyb antigen and made a full FY genotyping of a large series of donors easy to perform. Because FY*B/*B and FY*B/*XRBCs are generally indistinguishable and because Fyx is often undetetected in FY*A/*X and FY*Fy/*X RBCs by standard serological methods, the frequency of FY*X will certainly appear to be much higher than previously reported (0.015)39 when population studies are performed using DNA typing. Interestingly, by using the FY*B genotyping test, Murphy et al40 have already reported that weakened expression of the Fyb antigen is responsible for 12% of discrepancies between the genotypically (FY*A/*B) and serologically [Fy(a+b−)] determined Fyb status of 109 Caucasian donors. It is assumed that complete FY genotyping could show the presence of FY*X in most of these discrepancies.
In the present study, FY genotyping was used to demonstrate that the inheritance of the silent FY*Fy allele accounts for apparent paternity exclusion in family BE. Similarly, FY*X DNA typing will be useful to investigate apparent anomalous inheritance within the Duffy system in Caucasian families, which in most cases is due to weakened expression of Fyb.23
Anti-Fya and, to a lesser extent, anti-Fybantibodies can cause HDN, with some of them being fatal,1and a recent study has pointed out the clinical value of antenal Fya/Fyb genotyping in pregnancies at risk of HDN due to anti-Fya.41 It is expected that the Fyx genotyping test will also prove to be useful in the management of pregnancies at risk of Fy hemolytic disease by discriminating between fetuses with normal or weakened expression of the Fyb antigen when the mother exhibits high titer of anti-Fyb.
Even though no transfusion reaction related symptoms have yet been observed when blood units with weakened expression of Fybwere transfused to Fyb-negative patients, FY DNA typing would have important implications with respect to the good practice of blood transfusion.40 In that respect, the newly described Fyx DNA test should be performed to ascertain that Fyb-negative patients, with a high titer of anti-Fyb, will be transfused by true Fyb-negative blood units and not by Fybweaksamples serologically mistyped as Fyb-negative. Secondly, it is important that RBCs serving as reference for blood bankers are better characterized, both at the phenotypic and genetic levels.
Genotype-phenotype relationship.
The C286T substitution results in an Arg89Cys amino acid change that is predicted to occur in the first cytoplasmic loop of the DARC polypeptide, according to the current seven transmembrane domain topological model.7 Thus, the elucidation of the Fyx-associated substitution represents the first study that highlights the critical role of intracellularly exposed residues in the expression of DARC. The cytoplasmic localization of the Fyx-specific amino acid (Cys89) fits well with the fact that antibodies specific to Fyx RBCs have never been characterized.
Because our data suggest that the overall structure of the membrane-expressed Fyx polypeptide is similar to that of the other allelic forms of DARC (see above), we assume that the presence of a Cysteine residue at position 89 of the Fyxpolypeptide is not associated with an additional disulfide bond that would significantly modify the tertiary structure of DARC.
In agreement with the positive inside rule,42 the three short intracellular loops connecting the seven hydrophobic transmembrane helix of DARC contain several positively charged residues (Arg, Lys) that might be in the vicinity of the polar groups of the phospholipid molecules along the cytoplasmic side of the lipid bilayer. It has been experimentally demonstrated that basic amino acids, through their interaction with negatively charged phospholipids, are critical in the blocking of the translocation of loops across the membrane and thereby should contribute to the control of membrane topology (van Klompenburg et al43 and references herein). Accordingly, we suggest that, by modifying the positive charge of the first intracellular loop, the lack of Arg89, a residue conserved (or replaced by the homologous charged residue Histidine) in the DARC related polypeptides from 11 nonhuman primate species, mouse, and cow18 44 (and our unpublished results), might result in an inefficient insertion of the Fyx polypeptide in the cell membrane and thus account for the very low cell surface expression of DARC in Fyx cells.
The Fyx phenotype and the elusive DARC function.
The precise function of DARC in immunobiology and neurobiology remains uncertain. Because a murine DARC-like protein with conserved chemokine binding properties has been recently characterized,44 it is expected that DARC-deficient mice will be obtained and will help in the understanding of the biological role of DARC. Another approach to determine how much DARC is important in normal and pathological human physiology is to look for a potential correlation between the clinical status and the Duffy phenotypes of a large serie of donors. Hence, the fact that a large majority of blacks do not express DARC on their RBCs without apparent clinical consequence strongly suggested that DARC on RBCs is dispensable.6,45 On the other hand, the demonstration that, in these Fy(a−b−) individuals, the downregulation of DARC was restricted to erythroid cells6,11,21 and thus should represent an adaptative response to resist malaria, reinforced the hypothesis of an important role of DARC in nonerythroid tissues. Conversely, this suggestion became more difficult to support when it was demonstrated that at least 3 apparently healthy Fy(a−b−) Caucasians carry genomic mutations that should results in the lack of DARC in all cells and tissues.20 However, significant, albeit reduced expression of Fya and Fy6 antigens (∼40% as compared with control) were achieved when one of these defective genes that carry a 14-bp deletion resulting in a premature stop signal at codon 118 was transfected in COS-7 cells, but, as expected from structure function/analysis (see above), Fy3 was not expressed and chemokine did not bind to this protein lacking extracellular loops 2 and 3 (our unpublished results). Taken together, these results indicated that, even though a truncated DARC-related protein might be expressed in nonerythroid cells of these rare Caucasian Fy(a−b−) donors, these donors failed to express a functional chemokine receptor in all their tissues without detectable adverse consequences.
Importantly, one finding from the present study, as substantiated by transfectant analysis, is that the substitution identified in theFY*X allele is also predicted to alter DARC expression in all tissues without deleterious consequences for normal physiology. If DARC is involved in transduction of a signal across the membrane upon chemokine binding, by a still undetermined pathway, it is not unlikely that its residual level in homozygous Fyx cells could be sufficient to support normal function. However, it has thus been demonstrated that mutants of the IL-8 RA receptor that poorly bind IL-8 could mediate signal transduction in a way similar to how wild-type receptor does.46
In conclusion, the elucidation of the molecular basis of the Fyx phenotype adds one more degree of complexity to the genetics of the Duffy blood group system, but again raises questions about the importance of DARC in erythroid and nonerythroid tissues. Indeed, if one tries to link up the genetics with biology, it must be postulated that, whatever the precise function of DARC, other structures might operate when it is poorly expressed or absent. Similar compensatory mechanisms have been postulated to account for the observation that inactivation of the water channel AQP-1 gene in individuals with the Colton nul [Co(a−b−)] blood group phenotype is not associated with any apparent clinical consequence.47 Obviously, the challenge now is to develop direct experimental approaches to elucidate the biological role of DARC.
ACKNOWLEDGMENT
The authors are indebted to Dr F. Buffiere, Dr M. Uchikawa, and Dr D. Blanchard for supplying the anti-Fya, anti-Fy3, and anti-Fy6 MoAbs, respectively. We thank Dr P. Bailly for providing the anti-P55 polyclonal antibody and Dr A. Proudfoot for the gift of recombinant IL-8.
Address reprint requests to Yves Colin, PhD, INSERM U76, Institut National de la Transfusion Sanguine, 6 rue Alexandre Cabanel, 75015 Paris, France; e-mail: ycolin@infobiogen.fr.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal