Abstract
Acute promyelocytic leukemia (APL) cells, containing the t(15;17) rearrangement, express the fusion protein, PML/RAR. Clinically, patients respond to all-trans retinoic acid (ATRA) through complete remissions associated with myeloid maturation of leukemic cells. This clinical ATRA response of APL is linked to PML/RAR expression. Unfortunately, these remissions are transient and relapsed APL is often ATRA-resistant. The role PML/RAR plays in the growth and maturation of these APL cells with acquired ATRA resistance has not been fully explored. This study uses an ATRA-resistant NB4 cell line (NB4-R1) to investigate the contribution of PML/RAR expression to ATRA resistance. Targeting of PML/RAR in NB4-R1 cells was undertaken using two approaches: homologous recombination and hammerhead ribozyme-mediated cleavage. Reducing PML/RAR protein in NB4-R1 cells rendered these cells more sensitive to ATRA. These cells were growth-inhibited in ATRA, apoptosis was induced, and there was no apparent signaling of differentiation. Sequence analysis identified a mutation in the ligand binding domain (LBD) of the RAR portion of PML/RAR. Results show that these retinoid-resistant NB4 cells require persistent PML/RAR expression for leukemic cell growth. Taken together, these findings can account for why these cells do not respond to ATRA and how reduction of PML/RAR abrogates the antiapoptotic effect it confers to these leukemic cells.
© 1998 by The American Society of Hematology.
ACUTE PROMYELOCYTIC leukemia (APL) is characterized by the t(15:17) that is specific for the French-American-British (FAB) M3 subset of acute leukemias.1 The t(15;17) yields a fusion product, PML/RARα, which is linked to the induced clinical remissions of APL patients after all-trans retinoic acid (ATRA) differentiation therapy.2-4 These complete remissions are not durable, and relapse is often associated with clinical ATRA resistance.3,4 Whereas variant translocations exist and also involve RARα on chromosome 17, these are much less frequent than t(15;17) APL cases.5,6 The precise role PML/RARα plays in the ATRA response of APL remains to be defined. Studies indicate a dominant-negative interference of this fusion protein on RARα -, PML-, or RXRα -dependent pathways.7-11 It is known that PML/RARα can homodimerize12-14 or heterodimerize with PML10 or RXR.13 It is established that PML/RARα can bind ATRA14 and activate transcription through recognition of retinoic acid response elements (RAREs).15 Transfection experiments indicate that PML/RARα acts as an antiapoptotic factor in non-APL myeloid leukemic cells. When U937 cells were transfected with PML/RARα, growth factor-limited apoptosis was antagonized.11 This finding suggests PML/RARα functions as an antiapoptotic factor in retinoid-sensitive myeloid leukemic cells or perhaps in retinoid-sensitive APL cells. Less is known of the role of PML/RARα in regulating the growth and maturation states of retinoid-resistant APL cells.
The causes of clinical retinoid resistance are under active study. Studies have highlighted several potential retinoid resistance mechanisms. Pharmacokinetic studies in patients undergoing ATRA therapy show that drug plasma concentrations decrease during continuous ATRA therapy.16 Cellular retinoic acid-binding protein (CRABP) is upregulated during ATRA administration,17 which could account for the decreased levels of ATRA measured in plasma. Other studies report that defects in retinoic acid metabolism may account for resistance.18 Some laboratories have highlighted the role of the multidrug resistance gene product in retinoid resistance. P-glycoprotein (Pgp) expression is reported as low in leukemic cells of newly diagnosed APL patients,19 although treatment of APL cells isolated from relapsed ATRA-resistant patients with the Pgp antagonist verapamil allows these cells to mature.18
Because PML/RARα expression is tightly linked to the initial ATRA clinical response in APL,20 this protein has been analyzed in ATRA-resistant NB4 cells.21-23 In one of these lines, a dominant negative mutation in the ligand binding domain of the RARα portion of PML/RARα is reported.24 This mutation leads to altered binding to the ligand. In another ATRA-resistant NB4 line, PML/RARα protein expression is undetected.22 In a different reported line, PML/RARα expression is quite low until cells are treated with ATRA.23 Other reported ATRA-resistant NB4 lines constitutively express PML/RARα protein.23
Using a de novo–derived ATRA-resistant NB4 cell line that we isolated and designated NB4-R1,23 we sought to eliminate or reduce PML/RARα expression and to identify potential PML/RARα mutations through sequence analysis to assess its contribution to ATRA resistance. To accomplish this, PML/RARα DNA was targeted using an homologous recombination vector designed to eliminate this gene product. Clones surviving drug selection displayed a reversal in resistance to the growth-inhibitory effects of ATRA. Unexpectedly, these clones could not be maintained in long-term culture, perhaps due to the growth requirement for persistent PML/RARα expression. An alternative approach taken to evaluate this dependency was to target PML/RARα mRNA using a site-directed hammerhead ribozyme that preferentially cleaves this mRNA four nucleotides downstream from the fusion junction. The expression levels of the catalytic and noncatalytic ribozymes were regulated by the dose of hygromycin used for selection of the episomal vector-based ribozyme-transfected NB4-R1 cells. The engineering of the catalytic ribozyme, APL 1.1, and the noncatalytic, antisense control ribozyme, APL 5.0, was previously reported based on in vitro catalytic properties.25 The current study extends this prior work by demonstrating a dependency on persistent PML/RARα expression for growth of retinoid-resistant APL cells. Whereas myeloid maturation is not triggered by targeting PML/RARα expression in NB4 ATRA-resistant cells, apoptosis is signaled and enhanced by ATRA treatment. Sequence analysis identified a 3-bp deletion in the ligand binding domain (LBD) of the RARα portion of PML/RARα. This deletion eliminated a phenylalanine, but left the reading frame intact. The implications of these findings for retinoid-sensitive and -resistant APL cell growth are discussed.
MATERIALS AND METHODS
Cell lines and culture conditions.
NB4, a human APL cell line bearing the t(15;17), was a gift of Dr M. Lanotte (Paris, France). Two independent clones were isolated from parental NB4 cells: an ATRA-sensitive clone, designated NB4-S1, and a de novo isolated ATRA-resistant clone, designated NB4-R1.23These lines were used in these studies. Cells were cultured in RPMI containing 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mmol/L glutamine, and 10% fetal calf serum. Cells transfected with the knockout plasmid were selected and maintained in RPMI containing 500 μg/mL Geneticin (G-418 Sulfate; GIBCO BRL, Grand Island, NY). Ribozyme-transfected cells were selected and maintained in RPMI containing 65 μg/mL hygromycin. A selection/expression strategy for episomal vector-transfected NB4 cells has been previously reported.26
Genomic DNA library.
Genomic DNA was extracted from NB4-S1 cells using standard techniques and then partially digested with Sau3A I. Genomic DNA ranging from 15 to 20 kb was selected on a sucrose gradient, ligated into λ DASH II (Stratagene, La Jolla, CA) arms purified after a BamHI digest, packaged into empty capsids (Stratagene), and amplified in SRB(P2) bacteria (Stratagene).
Isolation of PML and RARα genomic clones.
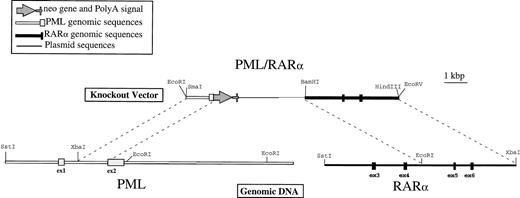
RARα and PML genomic clones were selected using two cDNA-derived radiolabeled probes. The RARα probe was a 500-bp KpnI-Sst I fragment from the 5′ end of the coding sequence corresponding to exons 3, 4, and 5. The PML probe was a 700-bpBgl II-Sst I fragment from the 5′ end of the coding sequence corresponding to exons 1, 2, and 3. For further analysis and subcloning, one representative phage was chosen from each set of positive clones. These two fragments were independently subcloned into Bluescript SK+ (Stratagene) or pUC19 plasmids (American Type Culture Collection, Rockville, MD) for detailed sequencing and mapping analyses. A partial sequence analysis of a coding region from both the PML and RARα selected fragments confirmed that these loci corresponded to the previously published sequences for each gene.7,27 The detailed restriction maps that were derived (Fig 1) were in accord with previously published maps for PML, RARα, and the breakpoint regions in APL.28 29
Physical map of the PML/RAR homologous recombination vector. Genomic DNA was isolated from NB4 cells as described in Materials and Methods and detailed restriction maps were constructed in the breakpoint region 3 (bcr3) for PML and RAR, as shown in this schematic. The knockout vector was constructed using a 1.2-kb 5′ sequence homologous to PML, followed by an in-frame, promotorless, ATGless, neo gene and then by a 4.2-kb 3′ sequence homologous to RAR. The plasmid backbone is Bluescript SK+, and the vector was linearized with EcoRI before transfection into NB4-R1 cells.
Physical map of the PML/RAR homologous recombination vector. Genomic DNA was isolated from NB4 cells as described in Materials and Methods and detailed restriction maps were constructed in the breakpoint region 3 (bcr3) for PML and RAR, as shown in this schematic. The knockout vector was constructed using a 1.2-kb 5′ sequence homologous to PML, followed by an in-frame, promotorless, ATGless, neo gene and then by a 4.2-kb 3′ sequence homologous to RAR. The plasmid backbone is Bluescript SK+, and the vector was linearized with EcoRI before transfection into NB4-R1 cells.
Construction of PML/RARα homologous recombination plasmid.
The plasmid used to knockout the PML/RARα-translocated locus was constructed in Bluescript SK+ (Stratagene). A 5′ region homologous to PML consisted of a 1.2-kb Xba I-Sph I fragment from intron 1 and a portion of the second coding exon (exon 2). This sequence was followed by the neomycin resistance gene,neo, lacking both a promoter and an initiation codon, which was inserted in frame with the AUG of PML. This in frame substitution of the neo coding sequence, confirmed by sequence analysis, was made in the second coding exon to eliminate the maximum amount of the targeted gene product. The 3′ homologous sequence was a 4-kbEcoRI-Xba I fragment containing exons 5 and 6 of RARα. These vectors were linearized with EcoRI before transfection.
Control vectors.
Three control vectors were constructed and used. The first one tested for a functional neo gene in the event of a successful homologous recombination in the PML/RARα locus. The first 86 amino acids of PML (EcoRI to klenow-treated Sph I) were fused to the neo gene30 lacking the 10 amino-terminal amino acids: this PML fragment was ligated to a Bluescript SK+ vector (Stratagene) containing the neo gene and digested with klenow-treated Eag I and EcoRI. The second control vector assessed the incidence of random insertions or recombinations that might occur with the homologous recombination vector that would allow for a functional neo gene. To construct this, the splice acceptor and 36 amino acids from exon 3 of RARα were fused to the above described ATGless neo vector. The third control vector, pKJ-1, was used to control for insertional mutagenesis and for the phenotype of cells maintained in high doses of G418. The pKJ-1 vector (a gift of Dr Jean-Christophe Bories, Paris, France) is a pUC 18 backbone containing the phosphoglycerate kinase (PGK) promoter drivingneo. This was linearized with HindIII before transfection.
Ribozymes and expression vector.
The hammerhead ribozymes used in this study were ribozyme APL 1.1, which is known to catalyze cleavage of PML/RARα mRNA, and APL 5.0, a noncatalytic, antisense control, as described previously.25These ribozymes, contained in self-cleaving cassettes, were cloned into the HindIII/Not I sites of the Eboplpp vector. The episomal Eboplpp vector contains the hygromycin resistance gene for drug selection and is useful to regulate expression of the inserted ribozymes or other genes, as described previously.26
Transfections.
NB4-R1 cells (5 × 106) were washed once and resuspended in phosphate-buffered saline (PBS) (Ca2+/Mg2+ free). Twenty micrograms of plasmid DNA was then added and the cells were electroporated at 0.3 kV and 25 μF.
Protein isolation and Western analysis.
NB4-R1 cells (107) were pelleted, resuspended in 1 mL of lysis buffer (150 mmol/L NaCl; 50 mmol/L Tris, pH 8.0; 1% NP-40; 0.5% deoxycholate; 0.1% sodium dodecyl sulfate [SDS]; 0.5 μg aprotinin, leupeptin, pepstatin, antitrypsin, and chymostatin; and 1 mmol/L phenylmethyl sulfonyl fluoride [PMSF]), and incubated for 30 minutes at 0°C. The samples were centrifuged at 12,000 rpm, and the supernatant was stored on ice. Protein concentration was assessed with the Bradford assay, and 50 μg protein per sample was electrophoresed on a 10% SDS-polyacrylamide gel. The protein was electrotransferred from the gel to supported nitrocellulose (Amersham International, Little Chalfont, UK). The ECL Western blotting protocol and kit (Amersham International) were used for antibody labeling and detection and membranes were exposed to high-speed film (FR Images, Mamaroneck, NY). The RARα (F region) RPα(F) antibody (provided by Dr Pierre Chambon, Strasbourg, France) recognizes the PML/RARα protein. Membranes were stained with Ponceau-S (Sigma, St Louis, MO) to control for equal protein loading.
3-[4,5-Dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide; Thiazolyl blue (MTT) proliferation assay.
On each day of the MTT assay,31 100 μL of cells was taken from each of the culture conditions and placed, in triplicate, in a 96-well plate. Fifty micrograms of MTT was added to each well and this mix was incubated for 4 hours at 37°C. At the end of incubation, 100 μL of 0.04 N HCl in 2-propanol was mixed thoroughly into each well. Plates were read on a Molecular Devices microplate reader (Sunnyvale, CA) at a wavelength of 570 nm, with a background reading at 650 nm subtracted. Triplicate readings for each sample were averaged. Trypan blue dye (Sigma) exclusion was used to assess viability.
Nitroblue tetrazolium (NBT) reduction assay.
For each cell sample, 200 μg NBT solution (Sigma), 105cells, and 100 ng phorbol 12-myristate 13-acetate (PMA; Sigma) were mixed gently. Samples were incubated at 37°C for 15 minutes and then at room temperature for 15 minutes. Subsequently, 0.1 mL of cells from each sample was cytospun, fixed in methanol, and stained with Safranin-O (Fisher Scientific, Pittsburgh, PA). A minimum of 100 cells was scored microscopically as NBT negative or positive (blue).
Terminal deoxynucleotidyl transferase (Tdt) assay.
For fixation, 106 cells were centrifuged, resupended in 1% formaldehyde, incubated on ice for 15 minutes, washed in PBS, and resuspended in 70% ethanol. To perform this assay, cells were centrifuged, rehydrated in PBS, and pelleted. The cell pellet was resuspended in Tdt reaction buffer (40 mmol/L cacodylate buffer; 2.5 mmol/L CoCl2; 7.5 U Tdt enzyme [Boehringer Mannheim, Indianapolis, IN]; and 0.5 nmol/L biotin dUTP [Boehringer Mannheim]) and incubated for 30 minutes at 37°C. After incubation, PBS was added and cells were centrifuged. The pellet was resuspended in 100 μL staining buffer (4× SSC, 0.1% Triton X-100, 5% dry milk, and 25 μg/mL fluoresceinated avidin) and incubated for 30 minutes at room temperature in the dark. A wash solution of PBS with 0.1% Triton X-100 was added and the sample was centrifuged. The cells were washed again and finally resuspended in 0.5 mL propidium iodide stain (5 μg/mL propidium iodide + 0.1% RNAseA), incubated overnight at room temperature, and stored at 4° C until analysis. Samples were analyzed on a FACScan (Becton Dickinson, Franklin Lakes, NJ).
Sequence analysis.
A reverse transcription-polymerase chain reaction (RT-PCR) was performed on DNase (Promega, Madison, WI) -treated total cellular RNA with a 21mer homologous to a sequence in the 3′UTR of RARα. PCR was performed on the resultant cDNA using a set of nested primers. For both sets of inner and outer primers, the 5′ primers were located in PML, 5′ of the fusion junction. The outer 3′ primer was the same as was used for RT-PCR and the inner 3′ primer was just internal to that, also located in the 3′ UTR of RARα. Three independent PCRs were performed and the PCR reactions were cloned into the TOPO TA Cloning kit as per the manufacturer’s instructions (Invitrogen, Carlsbad, CA). One clone from each reaction was sequenced using the ABI Sequencing kit and the ABI 373A Automated Sequencer (Perkin Elmer, Applied Biosystems Division, Foster City, CA). Both strands of each PML/RARα clone were sequenced. The ABI Autoassembler and Prism Sequencing softwares (Perkin Elmer) were used in the sequence analysis.
RESULTS
NB4-R1 cells transfected with the PML/RARα recombination vector reverse ATRA resistance.
To confirm that the neo gene would be functional if there were an effective recombination, NB4-S1 cells (an ATRA-sensitive NB4 clone) were transfected with a control vector that was engineered to contain the expected fusion sequence of PML/neo should the anticipated homologous recombination occur. The PML/neo gene proved functional by allowing transfected NB4-S1 cells to proliferate in doses of G418 that selected against the nontransfected cells.
A second control vector was designed to assess the background incidence of random integration or recombination that would result in an in-frame alignment of the knockout vector with a gene other than PML/RARα. This vector consisted of a splice acceptor fused to a promotorless, ATGless neo. No clones were derived using this vector despite multiple transfections, indicating that precise random integration was rare.
A third vector (pKJ-1) was used to control for phenotypic changes in cells due to insertional mutagenesis or to culture in high doses of G418. This vector was efficiently transfected and integrated into NB4-R1 cells and pools of these transfectants served as controls for these recombination experiments.
Multiple transfections were performed with the NB4-R1 cells using the PML/RARα homologous recombination vector depicted in Fig 1. Rare clones were derived from these transfections. Of the 15 clones isolated after G418 selection, 5 displayed a marked sensitivity to ATRA and the other 10 were similar to the parental NB4-R1 cells, with little to no growth-suppressive response to ATRA treatment.
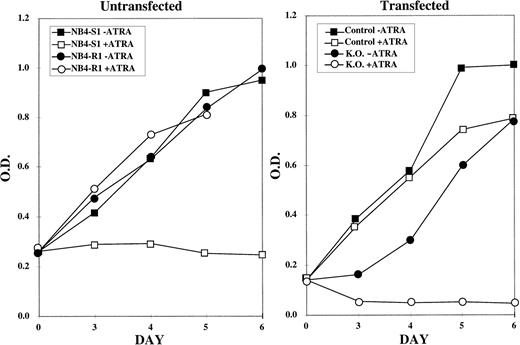
This reversal in ATRA response of the NB4-R1 cells was initially determined using the MTT proliferation assays (Fig 2). The graph on the left of Fig 2depicts the response of NB4-S1 and NB4-R1 cells to 10−5 mol/L ATRA. This is a 10-fold higher dose than is necessary to growth-inhibit and differentiate ATRA-sensitive NB4-S1 cells. This dose was selected to emphasize the ATRA-resistant properties of NB4-R1 cells and to contrast this ATRA response to that observed in transfectants. As shown, NB4-S1 cells are growth-inhibited in the presence of ATRA, whereas NB4-R1 cells are not. The graph on the right of Fig 2 shows that NB4-R1 cells transfected with the control vector (pKJ-1) continue to proliferate in ATRA (but a representative putative knockout clone reversed this resistant phenotype) and became sensitive to the growth-inhibitory effects of ATRA.
MTT proliferation assay performed on control and knockout NB4-R1 cells. The MTT assay was performed on the indicated days (X axis) in the absence (− ATRA) or presence (+ ATRA) of 10−5 mol/L ATRA. The Y axis depicts the optical density (O.D.). The growth curves in the left graph depict untransfected NB4-S1, an ATRA-sensitive NB4 clone, and NB4-R1, the de novo ATRA-resistant clone, from which the knockout subclones were derived. The right graph depicts the growth curves for NB4-R1 cells transfected with the pKJ-1 vector (Control) as a control for insertional mutagenesis and growth in G418 and for one representative knockout subclone (K.O.) that shows a reversal in resistance to growth inhibition by ATRA treatment.
MTT proliferation assay performed on control and knockout NB4-R1 cells. The MTT assay was performed on the indicated days (X axis) in the absence (− ATRA) or presence (+ ATRA) of 10−5 mol/L ATRA. The Y axis depicts the optical density (O.D.). The growth curves in the left graph depict untransfected NB4-S1, an ATRA-sensitive NB4 clone, and NB4-R1, the de novo ATRA-resistant clone, from which the knockout subclones were derived. The right graph depicts the growth curves for NB4-R1 cells transfected with the pKJ-1 vector (Control) as a control for insertional mutagenesis and growth in G418 and for one representative knockout subclone (K.O.) that shows a reversal in resistance to growth inhibition by ATRA treatment.
When this phenotypic change was observed, molecular analysis commenced. Genomic DNA was isolated from nontransfected NB4-S1 and NB4-R1 cells to serve as a negative control. Genomic DNA was also isolated from NB4-R1 cells transfected with the homologous recombination vector, from clones that showed no apparent reversal in phenotype, and from others that did reverse phenotype. Southern analysis was performed on genomic DNA digested with Sst I. When probed with an RARα SalI/Sau I genomic DNA fragment, a unique approximately 4.2-kb band would be present if RARα genomic sequence had been perturbed. This band was identified only in cells that had undergone a reversal in phenotype (data not shown). A neo probe also hybridized to this same band, indicating the homologous recombination vector was present in this approximately 4.2-kb fragment (data not shown). To fully diagnose a recombination and the precise site of recombination, multiple restriction endonuclease digestions were to be coupled with appropriate probes used in Southern analyses. Unexpectedly, transfectants anticipated to have undergone DNA recombination and reversal of retinoid resistance survived only transiently in culture, even in the absence of ATRA. The limited genomic DNA available did not allow for additional analyses. This suggested that targeting PML/RARα was lethal to NB4-R1 cells. It also indicated an alternative strategy was needed to target PML/RARα, as discussed below.
Targeting of PML/RARα mRNA with a site-specific ribozyme.
To circumvent the observed instability of the putative PML/RARα-targeted NB4-R1 cells, another targeting strategy of this fusion gene product was undertaken. The use of a catalytic ribozyme that cleaves PML/RARα mRNA was reported previously.25 It was hypothesized that effective recombination of PML/RARα accounted for the reversal of ATRA resistance in NB4-R1 cells. To learn whether regulated reduction of PML/RARα could be accomplished in NB4-R1 cells, the PML/RARα catalytic ribozyme, designated APL 1.1, and a noncatalytic control, designated APL 5.0, were used. Ribozyme APL 5.0 served as an antisense control, because it would bind to, but not cleave PML/RARα mRNA. The use of episomal vector-based expression of these ribozymes permitted regulation of their expression. Figure 3 depicts schematically the ribozymes as they would hybridize to PML/RARα mRNA, spanning the junction between PML and RARα. As noted, the cleavage site of APL 1.1 is four nucleotides downstream of the junction.
Schematic of the catalytic hammerhead ribozyme, APL 1.1, and the noncatalytic control, APL 5.0, hybridizing to PML/RAR mRNA. The two hybridizing arms (15 and 14 nucleotides [nt], respectively) of APL 1.1 are shown in dark black flanking the catalytic core (22 nt). The noncatalytic control ribozyme, APL 5.0, is identical to APL 1.1 except for one nucleotide substitution and one nucleotide deletion, as indicated. The PML (▧)/RAR (▨) mRNA is depicted, with the cleavage site indicated.
Schematic of the catalytic hammerhead ribozyme, APL 1.1, and the noncatalytic control, APL 5.0, hybridizing to PML/RAR mRNA. The two hybridizing arms (15 and 14 nucleotides [nt], respectively) of APL 1.1 are shown in dark black flanking the catalytic core (22 nt). The noncatalytic control ribozyme, APL 5.0, is identical to APL 1.1 except for one nucleotide substitution and one nucleotide deletion, as indicated. The PML (▧)/RAR (▨) mRNA is depicted, with the cleavage site indicated.
Western analysis of ribozyme transfected NB4-R1 cells.
The ribozyme-transfected NB4-R1 cells were selected and maintained in 65 μg/mL G418 before analyses. To assess cleavage of the PML/RARα mRNA, cells were cultured in 65 or 195 μg/mL hygromycin for 5 days in the absence (−) or presence (+) of 10−6 mol/L ATRA for 5 days (Fig 4). Total cellular protein was extracted and Western analysis was performed. An RARα antibody was used to detect the PML/RARα protein, as described in Materials and Methods. Even at the maintenance dose of 65 μg/mL hygromycin, it is evident from Fig 4 that there is less PML/RARα protein in cells containing the catalytic ribozyme than in those with the control ribozyme. This decrease is augmented when the dose of hygromycin is increased to 195 μg/mL. It is notable that APL 1.1 transfectants have a further decrease in PML/RARα protein expression after exposure to ATRA, at both doses of hygromycin. This is consistent with findings obtained using ATRA-sensitive NB4 cells, which have a decrease in the level of PML/RARα protein after exposure to ATRA.23
Western analysis for PML/RAR expression in ribozyme-transfected NB4-R1 cells. NB4-R1 cells transfected with either the control ribozyme, APL 5.0, or the catalytic ribozyme, APL 1.1, were cultured in 65 or 195 μg/mL hygromycin (hygro) in the absence (−) or presence (+) of 10−6 mol/L ATRA for 5 days. Total cellular protein was extracted and analyzed by Western analysis, using an anti-RAR antibody to detect PML/RAR protein. Relative to APL 5.0 transfectants, this immunoblot indicates that a marked reduction of PML/RAR expression occurs in APL 1.1-transfected NB4-R1 cells, especially after ATRA treatment and selection at the 195 μg/mL hygromycin dosage.
Western analysis for PML/RAR expression in ribozyme-transfected NB4-R1 cells. NB4-R1 cells transfected with either the control ribozyme, APL 5.0, or the catalytic ribozyme, APL 1.1, were cultured in 65 or 195 μg/mL hygromycin (hygro) in the absence (−) or presence (+) of 10−6 mol/L ATRA for 5 days. Total cellular protein was extracted and analyzed by Western analysis, using an anti-RAR antibody to detect PML/RAR protein. Relative to APL 5.0 transfectants, this immunoblot indicates that a marked reduction of PML/RAR expression occurs in APL 1.1-transfected NB4-R1 cells, especially after ATRA treatment and selection at the 195 μg/mL hygromycin dosage.
NB4-R1 cells transfected with a PML/RARα site-directed catalytic ribozyme acquire sensitivity to ATRA.
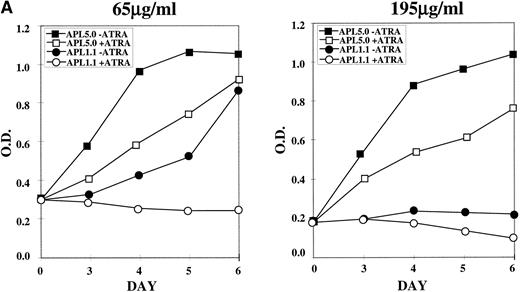
NB4-R1 cells, selected and maintained in 65 μg/mL hygromycin, were cultured in the same or 195 μg/mL hygromycin in the absence (−) or presence (+) of 10−6 mol/L ATRA for 6 days. MTT proliferation assays were performed on days 3 through 6, as shown in Fig 5A. The left graph shows that, in 65 μg/mL hygromycin, cells transfected with APL 5.0, the noncatalytic control, continue to grow, even in ATRA, whereas APL 1.1 transfectants proliferated poorly in the absence of ATRA and did not proliferate in the presence of ATRA. In the graph on the right of Fig 5A, results are displayed for the transfected cells cultured in 195 μg/mL in the absence (−) or presence (+) of ATRA. APL 1.1 transfectants proliferated poorly in the absence or presence of ATRA, whereas cells containing APL 5.0 grew well in the absence of ATRA, with some growth suppression in the presence of ATRA, perhaps due to antisense effects that cooperate with retinoid signals at the higher doses of hygromycin. It is noteworthy that cell cycle analysis of the growth inhibited cells did not indicate that there was arrest at a particular stage in the cell cycle (data not shown).
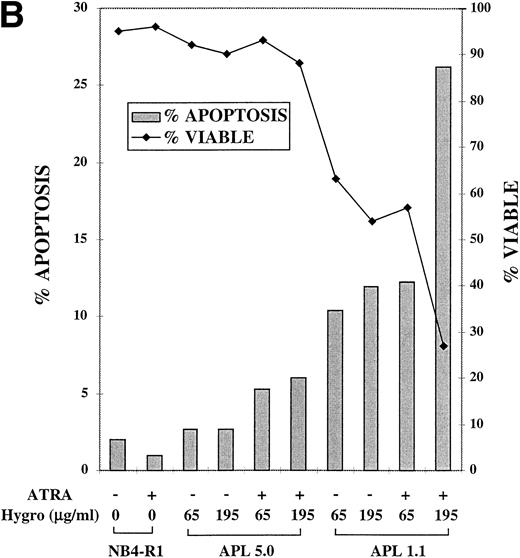
MTT proliferation assay and analysis of apoptosis in ribozyme-transfected NB4-R1 cells. (A) Cells transfected with either the control ribozyme, APL 5.0, or the catalytic ribozyme, APL 1.1, were cultured in 65 or 195 μg/mL hygromycin in the absence (−) or presence (+) of 10−6 mol/L ATRA for 5 days. The MTT assay was performed on days 3 through 6 (X axis). The Y axis depicts the optical density (O.D.). These data are from one experiment representative of four independent analyses. (B) Untransfected NB4-R1 cells and cells transfected with either the control ribozyme, APL 5.0, or the catalytic ribozyme, APL 1.1, were cultured without hygromycin (0 μg/mL) or with 65 or 195 μg/mL hygromycin (hygro) in the absence (−) or presence (+) of 10−6 mol/L ATRA for 4 days. The Tdt assay was used to assses induction of apoptosis (▧) on day 4. Viability was concurrently measured using trypan blue dye exclusion (⧫). These data are from one experiment representative of four independent analyses.
MTT proliferation assay and analysis of apoptosis in ribozyme-transfected NB4-R1 cells. (A) Cells transfected with either the control ribozyme, APL 5.0, or the catalytic ribozyme, APL 1.1, were cultured in 65 or 195 μg/mL hygromycin in the absence (−) or presence (+) of 10−6 mol/L ATRA for 5 days. The MTT assay was performed on days 3 through 6 (X axis). The Y axis depicts the optical density (O.D.). These data are from one experiment representative of four independent analyses. (B) Untransfected NB4-R1 cells and cells transfected with either the control ribozyme, APL 5.0, or the catalytic ribozyme, APL 1.1, were cultured without hygromycin (0 μg/mL) or with 65 or 195 μg/mL hygromycin (hygro) in the absence (−) or presence (+) of 10−6 mol/L ATRA for 4 days. The Tdt assay was used to assses induction of apoptosis (▧) on day 4. Viability was concurrently measured using trypan blue dye exclusion (⧫). These data are from one experiment representative of four independent analyses.
The observed growth inhibition was not associated with induced differentiation, as shown in Table 1. The NBT assay, which measures functional maturation of myeloid cells, indicated in none of the treatment groups was there an appreciable maturation response, as compared with 85% NBT positivity in the ATRA-treated NB4-S1 cells.
The Percentage of NBT-Positive and NB4-R1 Ribozyme-Transfected Cells
| Ribozyme . | Hygromycin (μg/mL) . | ATRA (10−6 mol/L) . | % NBT Positive . |
|---|---|---|---|
| NB4-S1 | 0 | + | 85% |
| NB4-R1 | 0 | + | 1% |
| APL 1.1 | 65 | − | 1% |
| APL 1.1 | 65 | + | 1% |
| APL 1.1 | 195 | − | 1% |
| APL 1.1 | 195 | + | 1% |
| APL 5.0 | 65 | − | 1% |
| APL 5.0 | 65 | + | 1% |
| APL 5.0 | 195 | − | 1% |
| APL 5.0 | 195 | + | 1% |
| Ribozyme . | Hygromycin (μg/mL) . | ATRA (10−6 mol/L) . | % NBT Positive . |
|---|---|---|---|
| NB4-S1 | 0 | + | 85% |
| NB4-R1 | 0 | + | 1% |
| APL 1.1 | 65 | − | 1% |
| APL 1.1 | 65 | + | 1% |
| APL 1.1 | 195 | − | 1% |
| APL 1.1 | 195 | + | 1% |
| APL 5.0 | 65 | − | 1% |
| APL 5.0 | 65 | + | 1% |
| APL 5.0 | 195 | − | 1% |
| APL 5.0 | 195 | + | 1% |
NB4-R1 cells transfected with APL 1.1 or APL 5.0 were cultured as indicated and the NBT assay was performed 5 days after the addition of ATRA (or not) and an increase (or not) in the hygromycin dosage. NB4-S1 cells were used as a positive and parental NB4-R1 cells were a negative control, respectively. This assay was conducted on cells harvested on day 5 from the treatment arms set up for the MTT assays.
A response of NB4-R1 transfectants to the catalytic ribozyme was induction of apoptosis that was enhanced by ATRA treatment, as seen in Fig 5B. For the Tdt assay, cells were cultured in 65 or 195 μg/mL hygromycin in the absence (−) or presence (+) of ATRA (10−6 mol/L) for 4 days. The gray bars in Fig 5B depict the percentage of apoptosis (left Y axis) on day 4. The basal level of apoptosis in nontransfected NB4-R1 cells was 2%, and ATRA treatment did not induce apoptosis (1%). There was a low level of apoptosis measured in cells transfected with APL 5.0 in the absence of ATRA in 65 μg/mL (3%) and 195 μg/mL hygromycin (3%). In ATRA, there was a slight increase in apoptosis at 65 μg/mL hygromycin (5%) and at 195 μg/mL hygromycin (6%). In contrast, NB4-R1 cells transfected with APL 1.1 in the absence of ATRA at 65 μg/mL had an increased level of apoptosis (10%), and when treated with ATRA, apoptosis increased to 12%. When the dose of hygromycin was increased to 195 μg/mL, in the absence of ATRA, there was 12% apoptosis; in the presence of ATRA, this increased to 26%. The viability remained high (>88%) in all of the APL 5.0-transfected cells, but decreased in APL 1.1-expressing cells at 65 μg/mL in the absence (63%) and presence (57%) of ATRA and at 195 μg/mL in the absence (54%) and presence (27%) of ATRA.
DNA sequence analysis identifies a mutation in the ligand binding domain of PML/RARα.
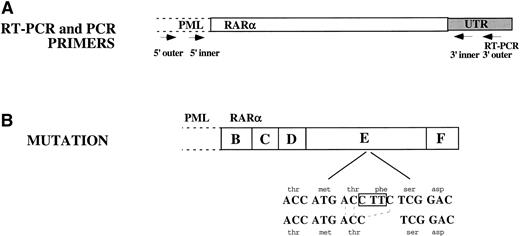
The entire RARα portion of PML/RARα from NB4-R1 cells was sequenced and compared with the published DNA sequence from parental NB4 cells.7 Sequence analysis identified a 3-bp deletion in the LBD of RARα present in each of the clones sequenced. As depicted in Fig 6, the last base pair in codon 778, a threonine (ACC), and the first 2 bp in codon 779, a phenyalanine (TTC), were deleted. The resulting ACC from the 3-bp deletion retained a threonine, but eliminated a phenylalanine. The reading frame was preserved.
Sequence analysis of the RAR portion of PML/RAR. (A) RT-PCR was performed on total cellular RNA from NB4-R1 cells using a primer in the 3′ UTR of RAR as indicated in this figure. Three independent PCR reactions were performed on the cDNA generated from the RT-PCR reaction. Nested primers were used as indicated in the figure: both inner and outer 5′ primers were in PML and both 3′ primers were in the RAR UTR. The 3′ outer primer was the same as was used for the RT-PCR reaction. (B) Sequence analysis showed a 3-nucleotide deletion: C was deleted from codon 778 and TT from codon 779. This eliminated a phenylalanine but retained a threonine. The reading frame was not disrupted.
Sequence analysis of the RAR portion of PML/RAR. (A) RT-PCR was performed on total cellular RNA from NB4-R1 cells using a primer in the 3′ UTR of RAR as indicated in this figure. Three independent PCR reactions were performed on the cDNA generated from the RT-PCR reaction. Nested primers were used as indicated in the figure: both inner and outer 5′ primers were in PML and both 3′ primers were in the RAR UTR. The 3′ outer primer was the same as was used for the RT-PCR reaction. (B) Sequence analysis showed a 3-nucleotide deletion: C was deleted from codon 778 and TT from codon 779. This eliminated a phenylalanine but retained a threonine. The reading frame was not disrupted.
DISCUSSION
Although ATRA treatment of t(15; 17) APL cases induces complete clinical remissions, these remissions are transient. After single-agent ATRA treatment of APL, clinical ATRA resistance often follows.3,4 In the in vivo setting, this ATRA resistance is associated with reduced ATRA levels after continuous ATRA therapy.16 This is associated with defects in retinoic acid metabolism18 and with upregulation of CRABP.17In the in vitro setting, ATRA APL resistance has also been investigated. Retinoid-resistant NB4 cells were derived by several groups, including our laboratory.21-23 In one NB4 retinoid-resistant line, a dominant-negative mutation was found within the ligand binding domain of the RARα region of PML/RARα.24 This results in altered ATRA binding to this receptor. In another ATRA-resistant NB4 line, PML/RARα protein, but not mRNA expression, is undetected.22 In contrast, in the NB4-R1 line examined in this study,23 PML/RARα protein expression is detected both before and after ATRA treatment.
This study was undertaken to explore directly the role of PML/RARα expression in the growth and maturation states of the retinoid-resistant NB4 line, NB4-R1. To explore the biologic effects of PML/RARα in retinoid-resistant NB4-R1 cells, two genetic strategies were taken to target PML/RARα expression. The first strategy was to introduce an homologous recombination vector that would target PML/RARα. In the results displayed within Fig 2, the findings obtained using this targeting vector indicated that a subset of isolated clones displayed an acquired sensitivity to ATRA-mediated growth suppression. Unexpectedly, these clones could not be continuously maintained in culture and could not be genetically characterized in detail. This suggested reduction of PML/RARα expression conferred a major growth disadvantage to these transfectants. Consistent with this view is the finding that two control vectors designed to assess background incidence of random integration failed to elicit this reversal of ATRA resistance. From these experiments, it was speculated that a growth disadvantage existed for these putative PML/RARα-targeted NB4-R1 transfectants and that acquired ATRA sensitivity was linked to reduced PML/RARα expression. Further experiments were undertaken to explore this possibility.
The second strategy taken to target PML/RARα expression involved hammerhead ribozymes engineered to cleave PML/RARα mRNA. Our prior work reported that PML/RARα transcripts are efficiently cleaved by the hammerhead ribozyme, APL 1.1.25 In contrast, a control hammerhead ribozyme, designated APL 5.0, will hybridize to PML/RARα mRNA but will not cleave this transcript. These two hammerhead ribozymes were used in transfection experiments involving the retinoid-resistant APL line, NB4-R1. The results of experiments displayed in Fig 4 indicate that PML/RARα expression is markedly repressed in the APL 1.1 versus APL 5.0 transfectants. When the PML/RARα catalytic but not control ribozymes are introduced into NB4-R1 cells, findings show that NB4-R1 cell growth depends on the selective pressure used to express APL 1.1 hammerhead ribozymes in transfectants. Whereas a reduction of PML/RARα expression was observed at the lower hygromycin dosage for APL 1.1 (see Fig 4), at the higher hygromycin dosage used for this transfectant (195 μg/mL), an even greater reduction of PML/RARα expression was detected. Notably, after ATRA treatment, a further reduction of PML/RARα expression was observed. A tight link was found to exist between this reduced PML/RARα expression and the growth properties of this transfectant, as shown in Fig 5. As expected, the control hammerhead ribozyme, APL 5.0, which can hybridize to PML/RARα but not cleave this transcript, had growth properties similar to parental NB4-R1 cells23(data not shown). For APL 1.1 transfectants maintained at even higher hygromycin dosages, the observed growth suppression was even more prominent, suggesting that a greater reduction of PML/RARα expression was incompatible with the maintenance of leukemic cell growth. This view is supported by the experiments conducted using the homologous recombination vector described in Fig 1.
It is notable that the observed growth suppression of the NB4-R1 cells transfected with the APL 1.1 hammerhead ribozyme was due to reduced viability at the higher dose of hygromycin selection. This reduced viability was at least partly due to the signaling of apoptosis, as shown in Fig 5B. Cell cycle analysis did not indicate arrest in a specific phase of the cell cycle coincident with cell death (data not shown). This observation is consistent with other studies of apoptosis. For example, N-(4-hydroxyphenyl)retinamide (4HPR)-induced apoptosis in HL-60 cells did not block cells in a particular phase of the cell cycle.32 As anticipated by the Western analysis for PML/RARα expression in APL 1.1 transfectants treated with ATRA, an even greater signaling of apoptosis followed ATRA treatment of these transfectants. This suggests that reduced PML/RARα expression in ATRA-resistant APL cells restores at least some of the retinoid sensitivity to growth suppression triggered by ATRA treatment. It is interesting to note that maturation was not triggered in these transfectants having reduced PML/RARα expression, as shown in Table1. This finding argues for an important role for persistent PML/RARα expression to maintain basal leukemic cell growth. These findings are consistent with the view that events in addition to PML/RARα are required to trigger the full retinoid differentiation program in these ATRA-resistant cells.
To explore PML/RARα mutations that may contribute to the phenotype of these ATRA-resistant NB4 cells, the entire RARα portion of PML/RARα isolated from these cells was sequenced. A 3-bp mutation in the ligand binding domain of RARα that eliminated a phenylalanine was identified, as shown in Fig 6. This provides a possible explanation for the ATRA resistance of these NB4-R1 cells and is consistent with other published work of ATRA-resistant myeloid leukemic cells.24,33 34 Although these cells are resistant to ATRA, the mutated PML/RARα appears to continue to act as an antiapoptotic factor, because the elimination or reduction of this product through the targeting strategies reported here abrogates this antiapoptotic effect.
The findings presented in this study indicate that PML/RARα functions as an antiapoptotic translocation product in these ATRA-resistant NB4 cells. That PML/RARα antagonizes an apoptotic program has precedence in prior published work. For instance, induced overexpression of PML/RARα in U937 myeloid leukemic cells leads to an antagonism of growth factor-limited apoptosis.11 This work, highlighting an antiapoptotic role for PML/RARα, was accomplished using a non-APL myeloid leukemic cell line. The current study extends this prior work by performing an analysis of the PML/RARα role in an APL cell context. The current study complements prior work in this field by conducting loss-of-function experiments using two approaches: (1) homologous recombination and (2) hammerhead ribozymes, which target PML/RARα mRNA expression. Both approaches led to similar biologic outcomes, a lethal phenotype, due at least partly to the triggering of apoptosis. That two genetic approaches, each directed towards reducing PML/RARα expression, lead to a similar phenotype provides independent confirmation of these results. The triggering of apoptosis in retinoid-resistant myeloid leukemic cell lines also has precedence. One study reports that apoptosis can be induced in ATRA-resistant NB4 cells when primed with retinoic acid and then triggered with cAMP.35 Prior work demonstrates that treatment with 4HPR triggers apoptosis even in a myeloid leukemic cell line bearing an RARα mutation.32 36 Whether acquired mutations within the LBD of RARα are a general feature of retinoid-resistant promyelocytic cell lines is the subject of work currently in progress.
In summary, the findings reported in this study indicate two strategies to target PML/RARα expression yield similar findings in retinoid-resistant APL cells. This is the requirement for persistent PML/RARα expression to sustain leukemic cell growth. This requirement is linked, at least partly, to the antiapoptotic function of PML/RARα in NB4-R1 APL cells. That reduction of PML/RARα expression is incompatible with leukemic cell growth, even within an established APL cell line, raises the prospect that targeting PML/RARα will have therapeutic potential in retinoid-resistant or -sensitive APL cells. Future work will explore this hypothesis.
ACKNOWLEDGMENT
The authors thank Dr M. Lanotte (INSERM, Paris, France) for the gift of the NB4 cell line from which we derived the NB4-R1 cells. We thank Dr A. Selvakumar (Memorial Sloan-Kettering Cancer Center, New York, NY) for help and advice with the DNA sequencing. We thank Dr A. Goldberg and colleagues at Innovir Laboratories (New York, NY) for helpful consultation and for use of the hammerhead ribozymes 1.1 and 5.0. We thank Dr P. Chambon (INSERM, Strasbourg, France) for release of the anti-RARα antibody used for Western analysis.
Supported by the National Institutes of Health Grants No. RO1 CA 62275-04 (E.D.), NRSA F32 CA61646-01A1 (K.N.-B.), and P01 CA 29502-14A2 (K.N.-B.) and by grants from the Centre National pour la Recherche Scientifique and the Institut Pasteur de Lille.
Address reprint requests to Kathryn Nason-Burchenal, PhD, Memorial Sloan-Kettering Cancer Center, Box 305, 1275 York Ave, New York, NY 10021; e-mail: k-nason-burchenal@ski.mskcc.org.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

![Fig. 3. Schematic of the catalytic hammerhead ribozyme, APL 1.1, and the noncatalytic control, APL 5.0, hybridizing to PML/RAR mRNA. The two hybridizing arms (15 and 14 nucleotides [nt], respectively) of APL 1.1 are shown in dark black flanking the catalytic core (22 nt). The noncatalytic control ribozyme, APL 5.0, is identical to APL 1.1 except for one nucleotide substitution and one nucleotide deletion, as indicated. The PML (▧)/RAR (▨) mRNA is depicted, with the cleavage site indicated.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/5/10.1182_blood.v92.5.1758/4/m_blod41728003x.jpeg?Expires=1769145219&Signature=dDoUitmJvWuW-Zhyv4fKi~vfE9IQzZJkf25mOmlPQKRBCj4nBK0yh8y9b5-XePXKQkfaK0IB3EBBk0cKp6EF8nqXp96xdohYKBOU9AcwHxiER3yai8I0~7q-a88QBXxOTB6Xk3xg9rw6PnzQnzmesbsko824ceHWvpf6OMddUBE~vN~lMlRbs6R2TFIBAGqILEQUB~BKP2G8ZhdGifAzVrDhkhawH48M~K2Gp9FsWU0Vbq4aSa73uHiP4oW80ZkiLmL1Mav9i0vNDfV54MmjBOpRT6E-Q195LVstKpw18nFCl0O8pljiNSApxrpcSl2KLJopH8sRGEqH9oAVlEnXLg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal