Abstract
The 8p11 myeloproliferative syndrome is a rare, aggressive condition associated with reciprocal translocations of chromosome band 8p11, most commonly the t(8;13)(p11;q12). To identify the genes involved in this translocation, we used fluorescence in situ hybridization (FISH) analysis to show that the chromosome 8 breakpoints fell within YAC 899e2 and that the chromosome 13 breakpoints are clustered in a region flanked by YACs 929f11 and 911h8. FISH using chromosome 13 PAC clones indicated that the t(8;13) is not simply a reciprocal translocation but also involves an inversion of 13q11-12. Exon trapping of a PAC that spanned the chromosome 13 translocation breakpoints led to the identification of a gene, ZNF198, that detected rearranged bands when used as a probe against Southern blots of patient DNA. Conceptual translation of the full-length ZNF198 cDNA sequence predicts a protein of 1377 amino acids that shows significant homology to the DXS6673E/KIAA0385 and KIAA0425 proteins. Alignment of these three proteins revealed a novel, conserved Zn-finger–related motif (MYM domain) of the general form CX2C19-22CX3CX13-19CX2CX19-25FCX3CX3F/Y that is repeated five times in each protein. To identify the translocation partner gene on chromosome 8, 5′ and 3′ RACE polymerase chain reactions (PCRs) were performed on patient RNA with several combinations of ZNF198 primers. Clones were identified in which the ZNF198 was fused to exon 9 of the fibroblast growth factor receptor-1 (FGFR1), a gene known to map to 8p11. An identical ZNF198-FGFR1 fusion was detected in the three patients with a t(8;13) for whom RNA was available; reciprocal FGFR1-ZNF198 transcripts were not detected. Rearrangements of both ZNF198 and FGFR1 were found in two further patients by Southern blotting. ZNF198-FGFR1 includes the five MYM domains of ZNF198 and the intracellular tyrosine kinase domain of FGFR1. We hypothesize that this fusion leads to constitutive activation of the FGFR1 tyrosine kinase in a manner analogous to the activation of ABL by BCR in chronic myeloid leukemia.
© 1998 by The American Society of Hematology.
MYELOPROLIFERATIVE disorders (MPDs) are a heterogeneous group of diseases characterized by excess proliferation of cells of the myeloid lineage. The most common MPDs are chronic myeloid leukemia (CML), polycythemia vera (PV), essential thrombocythemia (ET), and myelofibrosis (MF), but several other related disorders with distinct features have been described.1 One such disorder is usually associated with the reciprocal translocation t(8;13)(p11;q12),2,3 but strikingly similar clinical features have also been described in patients with a t(8;9)(p11;q34) or a t(6;8)(q27;p11). These translocations have a common breakpoint at chromosome band 8p11, and hence we have proposed that this disorder be called the 8p11 myeloproliferative syndrome.2
At presentation, the peripheral blood and bone marrow of patients with the 8p11 myeloproliferative syndrome have the appearance of a chronic MPD with prominent eosinophilia. In addition, there is a strikingly high incidence of non-Hodgkin’s lymphoma that may be of either B-cell or, more commonly, T-cell phenotype. The natural history of the disease has similarities to CML with a chronic phase that ultimately transforms into an acute leukemia. Where reported, the same clonal karyotypic abnormality is seen in both the lymphoma and myeloid cells, suggesting a common lymphoid/myeloid stem cell as the target for the original transforming event. Consistent with this hypothesis, the disease appears to be ineradicable by conventional chemotherapy, although some patients have apparently been cured by allogeneic bone marrow transplantation.2
In this report we describe the cloning of the t(8;13) and show that the ZNF198 gene at 13q12 becomes fused to the fibroblast growth factor receptor-1 (FGFR1) gene at 8p11.
MATERIALS AND METHODS
Patient material/details.
Five patients with a t(8;13)(p11;q12) were analyzed. Patients J.B., M.L., and P.R. have been described previously4-6; patients E.C. and J.L. had clinical features typical of the 8p11 myeloproliferative syndrome. Fixed cells, RNA, and DNA were available for analysis for patients J.B., P.R., and E.C.; for patients M.L. and J.L., only DNA was available.
Fluorescence in situ hybridization (FISH).
Yeast containing individual YACs were grown in AHC medium at 30°C for 24 to 72 hours and DNA extracted using a glass-bead method. PAC DNA was isolated from 1.5 mL cultures using a standard miniprep procedure. Probes were labeled with biotin by nick translation, tested on metaphases from phytohemagglutinin-stimulated peripheral blood lymphocytes from a normal individual, and subsequently hybridized to patient metaphases as described.6Hybridization signals were detected using FITC Avidin (Vector, Peterborough, UK). In addition, a prefluorochrome-conjugated full chromosome 8 paint (Biovation, Aberdeen, UK) was included. Chromosomes were counterstained with DAPI/antifade (Biovation). FISH-labeled metaphases were examined using an Olympus Vanox microscope (Olympus, Tokyo, Japan) and images captured on SmartCapture Software (Vysis, Richmond, UK).
Isolation of PAC clones.
Markers mapping to chromosome 13q11-12 were amplified by the polymerase chain reaction (PCR) from either human genomic DNA or YAC DNA and used to probe the gridded human PAC library RPCI1 (obtained from the MRC HGMP Resource Centre, Hinxton, UK). PCR amplification of these markers from multiple overlapping YAC clones indicated an order entirely consistent with that described elsewhere.7 Overlapping clones were identified using probes isolated from the ends of individual PACs by bubble PCR.8
Exon trapping.
Clones were digested with BamHI, BglII, orBamHI + BglII and subcloned into the vector pSPL3B-CAM. Pooled subclones were transfected into COS cells and candidate exons amplified by reverse transcription polymerase chain reaction (RT-PCR) as described.9 Products were cloned into the TA cloning vector pCR2.1 (Invitrogen, Leek, The Netherlands) and sequenced.
RACE-PCR.
RACE-PCR was performed essentially as described.10 Briefly, for 3′ RACE, 1 μg of total RNA extracted from peripheral blood leukocytes of patient E.C. or a normal individual was reverse transcribed using primer RoRidT17(5′-AAGGATCCGTCGACATCGATCCTACGACTCACTATAGGGA-T17-3′. Nested amplification was performed using a series of sense orientation ZNF198 primers in combination with primers Ro(5′-AAGGATCC-GTCGACATCGAT-3′) and Ri(5′-GACATCGATCCTACGACTCA-3′). For 5′ RACE, 1 μg RNA extracted from peripheral blood leukocytes of patient E.C. and a normal individual was reverse transcribed with a series of ZNF198 primers in the antisense orientation. After A-tailing with TdT, second strand cDNA was synthesized using primer RoRidT17 and nested amplification performed using a series of antisense ZNF198 primers in combination with primers Ro and Ri. Products were cloned into pCR2.1 and sequenced. IMAGE clones were obtained from the MRC HGMP Resource Centre, Hinxton, UK.
RT-PCR.
Patient RNA was extracted and reverse transcribed with random hexamer primers.11 Primer combinations used to detect normal ZNF198 and FGFR1 transcripts were DET2 (5′-TAGCTCCTCAATTGCTGCAGGA-3′) plus DET6 (5′-GTACTACAAGGCTGCAAGGTGT-3′) and FGFR6+(5′-ACATCGAGGTGAATGGGAGCAA-3′) plus FGFR9− (5′-GAGGGTCTTC-GGGAAGCTCATA-3′), respectively. Primers to detect ZNF198-FGFR1 and any reciprocal fusion transcripts were DET6 plus FGFR9−and FGFR6+ plus DET2, respectively. All amplifications were performed for 32 cycles with an annealing temperature of 58°C. Other primers used were DET13 (5′-GTTCCTATGCACATGTACAGTC-3′), DET17 (5′-GTATTACCCAAAGGTTC-AAACC-3′), DET22 (5′-TTGTACTGGTTGCCGAACACAG-3′), and DET26 (5′-TCTCAATCCTAAACGTCTGGCA-3′).
Southern and Northern blotting.
DNA extraction and Southern blotting were performed according to standard procedures. ZNF198 and FGFR1 probes were amplified from normal human peripheral blood leukocyte cDNA with primers DET2 plus DET6 and FGFR6+ plus F12-A (5′-TTGGAGGAGAGCTGCTCCTCT-3′), respectively. The same ZNF198 probe was also used to probe a multiple tissue Northern blot (Clontech, Basingstoke, UK).
RESULTS
FISH analysis of chromosome 8.
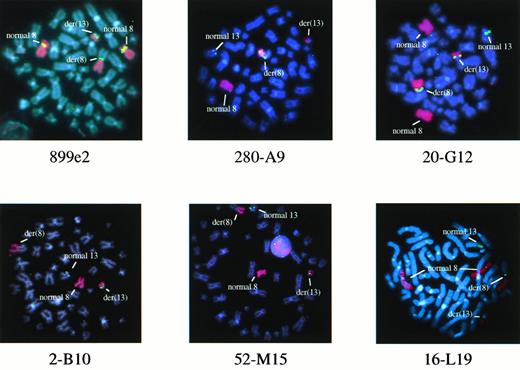
We have previously shown that the chromosome 8 breakpoints in the t(8;13) are telomeric to the MOZ gene and lie between YACs 782H11 and 847B12, a region of approximately 4 centiMorgans.6 In two of two patients tested (patients E.C. and P.R.), YAC 899e2, which maps to this region,12 hybridized to both the der(13) and the der(8) in addition to the normal chromosome 8 (Fig1), suggesting that this clone spanned the breakpoints.
FISH analysis of metaphases from patient E.C. with 899e2 (chromosome 8 YAC clone) or 280-A9, 20-G12, 2-B10, 52-M15, and 16-L19 (chromosome 13 PAC clones). Chromosome 8 sequences are highlighted with a red painting probe and YAC or PAC signals are in green. Patient E.C. had an extra normal chromosome 8 in some cells.
FISH analysis of metaphases from patient E.C. with 899e2 (chromosome 8 YAC clone) or 280-A9, 20-G12, 2-B10, 52-M15, and 16-L19 (chromosome 13 PAC clones). Chromosome 8 sequences are highlighted with a red painting probe and YAC or PAC signals are in green. Patient E.C. had an extra normal chromosome 8 in some cells.
FISH analysis of chromosome 13.
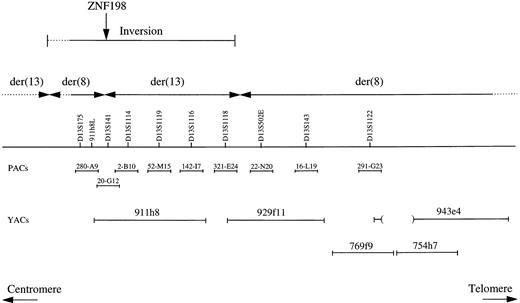
The positions of the YAC clones used for FISH analysis are shown in Fig 2. YAC 943e4 has been reported previously to hybridize to both derivative chromosomes from patients J.B. and M.L.13 However, YACS 769f9 and 754h7 hybridized only to the der(8) in patients P.R. and E.C. (in addition to the normal chromosome 13), indicating that these clones were distal to the breakpoints in these two cases. In contrast, YACs 911h8 and 929f11 hybridized to both derivative chromosomes from patients P.R., E.C., and J.B. This was surprising because these clones do not overlap,7 a fact we confirmed by amplification of internal and end markers. For 911h8, the bulk of the signal was on the der(13), whereas for 929f11 the bulk of the signal was on the der(8) in all three cases. This suggested that the positions of the breakpoints were within or between these clones.
Positions of DNA markers, PAC, and YAC clones within 13q11-12. The results of the FISH analysis are summarized and indicated that a region containing the ZNF198 gene had become inverted at some stage in the formation of the t(8;13).
Positions of DNA markers, PAC, and YAC clones within 13q11-12. The results of the FISH analysis are summarized and indicated that a region containing the ZNF198 gene had become inverted at some stage in the formation of the t(8;13).
To refine the location of the breakpoints, markers that map to the 911h8/929f11 region were used to isolate PAC clones for further FISH analysis on metaphases from patients E.C. and P.R. (Fig 1). All PACs hybridized to 13q11-12 on the normal copies of chromosome 13. In addition, PACs 52-M15 (D13S1119), 142-I7 (D13S1116), and 321-E24 (D13S1118) hybridized to the der(13). PACs 22-N20 (D13S502E), 16-L19 (D13S143), and 291-G23 (D13S1122) hybridized to the der(8), indicating a breakpoint between D13S1118 and D13S502E (Figs 1 and 2B).
Subsequent analysis, however, showed that PAC 20-G12 (D13S141) hybridized to both the der(13) and the der(8), suggesting an additional break within this clone. This was confirmed by hybridization of two overlapping PACs; 280-A9 hybridized to the der(8), whereas 2-B10 hybridized to the der(13). This was the opposite of the expected pattern of hybridization because 280-A9 contained D13S175, a marker proximal to D13S141, and 2-B10 contained D13S1114 and was therefore distal.7 Taken together, these findings suggested that a complex rearrangement had occurred in both patients in which a region between the chromosome 13 centromere and D13S502E had become inverted, presumably before or at the same time as the translocation between chromosomes 8 and 13 (Fig 2B). If this were the case, then the true translocation breakpoint most likely fell within PAC 20-G12.
Identification of the chromosome 13 gene.
Exon trapping and partial sequence analysis of PAC 20-G12 led to the identification of several sequences that matched almost perfectly to multiple human expressed sequence tags (ESTs), some of which corresponded to IMAGE clones (eg, clone nos. 491514, 727984, and 787874). RT-PCR from normal peripheral blood leukocyte cDNA indicated that the ESTs were part of a single gene. Some of these RT-PCR–amplified fragments identified novel bands in Southern blots of patient DNA but not control DNA (see below), suggesting that this gene, subsequently called ZNF198, was disrupted by the t(8;13). The full-length 5050-bp sequence of the ZNF198 cDNA (Genbank: AJ224901) was obtained from the three IMAGE clones and by 5′ and 3′ RACE-PCRs from normal peripheral blood leukocyte cDNA. The sequence of ZNF198 was verified by amplification and direct sequencing of overlapping fragments encompassing the entire gene from normal peripheral blood leukocyte cDNA.
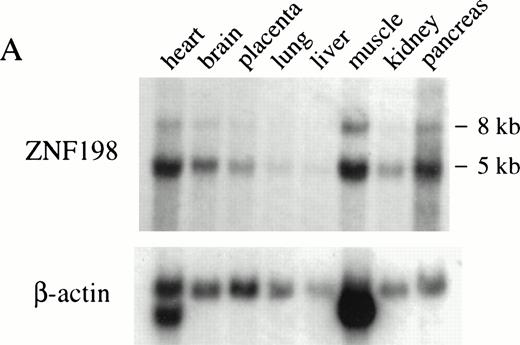
ZNF198 encodes a novel 1377 amino acid protein of predicted molecular weight of 155 kD. The gene is widely expressed, as determined by hybridization to a Northern blot (Fig 3A). A BLAST search revealed that ZNF198 shows significant homology to two genes of unknown function, DXS6673E/KIAA0385 and KIAA0425. Alignment of the predicted amino acid sequences of the three gene products (Fig 4) showed an amino acid identity of 32% within the region shown and the presence of several conserved motifs, notably a novel zinc-finger related structure of the form CX2C19-22CX3CX13-19CX2CX19-25FCX3CX3F/Y that is repeated five times in each protein. We have called this motif a MYM domain because ZNF198 and DXS6673E are implicated in a myeloproliferative syndrome and mental retardation, respectively.
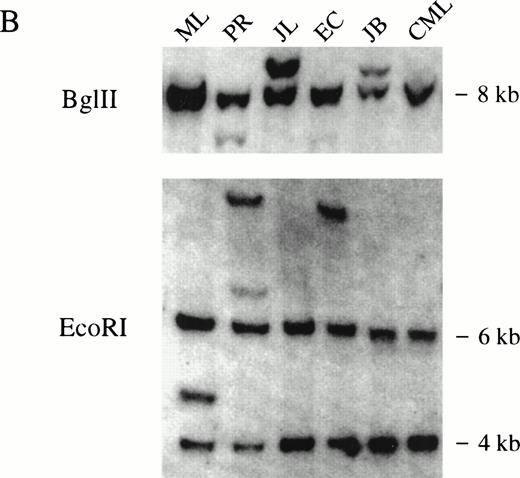
(A) Multiple tissue Northern blot probed with ZNF198 and β-actin. A major ZNF198 transcript of 5.0 kb and a minor transcript of 8.0 kb is seen in all tissues tested. (B) Southern blot ofBglII or EcoRI digested patient and control (CML) DNA probed with ZNF198. Rearrangements are seen for all five patients.
(A) Multiple tissue Northern blot probed with ZNF198 and β-actin. A major ZNF198 transcript of 5.0 kb and a minor transcript of 8.0 kb is seen in all tissues tested. (B) Southern blot ofBglII or EcoRI digested patient and control (CML) DNA probed with ZNF198. Rearrangements are seen for all five patients.
Alignment of the homologous regions of the ZNF198, KIAA0385, and KIAA0425 proteins. The reported sequence of DXS6773E is identical to KIAA0385 apart from a 12–amino acid internal deletion. Amino acids that were identical in all three proteins shown in the consensus. The five MYM domains and the proline-rich domains are highlighted, and the position of the t(8;13) breakpoint in ZNF198 is shown. No significant homology was found between the ZNF198 and KIAA0385 in the N-terminal region.
Alignment of the homologous regions of the ZNF198, KIAA0385, and KIAA0425 proteins. The reported sequence of DXS6773E is identical to KIAA0385 apart from a 12–amino acid internal deletion. Amino acids that were identical in all three proteins shown in the consensus. The five MYM domains and the proline-rich domains are highlighted, and the position of the t(8;13) breakpoint in ZNF198 is shown. No significant homology was found between the ZNF198 and KIAA0385 in the N-terminal region.
Identification of the chromosome 8 gene.
Both 5′ and 3′ RACE-PCR was performed using several combinations of ZNF198 primers and RNA extracted from patient E.C. Amplification products were cloned and sequenced. Most primer combinations resulted in the amplification of ZNF198 sequence only, but 3′ RACE-PCR using primers DET6 and DET13 led to the identification of several clones in which the sequence diverged from ZNF198 after codon 913 (Fig 5A). A database search revealed that the novel sequence immediately downstream of ZNF198 corresponded to exon 9 of FGFR1, a gene known to map to 8p11.14 PCR using primers to FGFR1 exons 8 and 9 confirmed that this gene mapped to YAC 899e2, the clone previously shown to span the t(8;13) chromosome 8 breakpoints.
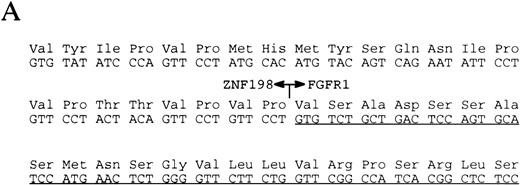
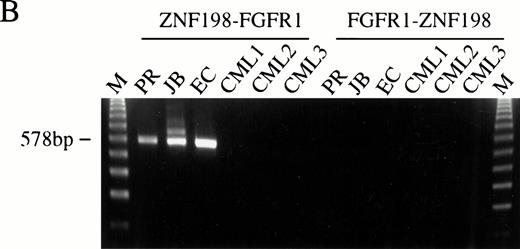
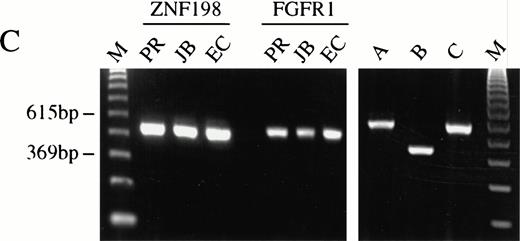
(A) The sequence surrounding the ZNF198-FGFR1 fusion. Proline 913 of ZNF198 is joined to valine 429 of FGFR1.39FGFR1 sequence is underlined. (B) RT-PCR analysis showing the presence of ZNF198-FGFR1 fusion transcripts (578 bp) in patients P.R., J.B., and E.C. but not three patients with CML. Reciprocal FGFR1-ZNF198 transcripts were not detected in any individual. M, 123-bp ladder. (C) (i) Expression of normal ZNF198 (527 bp) and normal FGFR1 (534 bp) in patients P.R., J.B., and E.C. (ii) RT-PCR using (A) primers DET22 + DET26 (564 bp), (B) DET17 + DET26 (395 bp), and (C) DET17 + DET22 + DET26 (564 bp).
(A) The sequence surrounding the ZNF198-FGFR1 fusion. Proline 913 of ZNF198 is joined to valine 429 of FGFR1.39FGFR1 sequence is underlined. (B) RT-PCR analysis showing the presence of ZNF198-FGFR1 fusion transcripts (578 bp) in patients P.R., J.B., and E.C. but not three patients with CML. Reciprocal FGFR1-ZNF198 transcripts were not detected in any individual. M, 123-bp ladder. (C) (i) Expression of normal ZNF198 (527 bp) and normal FGFR1 (534 bp) in patients P.R., J.B., and E.C. (ii) RT-PCR using (A) primers DET22 + DET26 (564 bp), (B) DET17 + DET26 (395 bp), and (C) DET17 + DET22 + DET26 (564 bp).
Confirmation of a ZNF198-FGFR1 fusion in patients with the t(8;13).
RT-PCR analysis using ZNF198 and FGFR1 primers was performed for the three patients (P.R., J.B., and E.C.) for whom RNA was available. An identical size amplification product was obtained for each patient, the sequences of which revealed the same ZNF198-FGFR1 fusion as that found by RACE-PCR. No amplification products were seen in control patients with CML, and reciprocal FGFR1-ZNF198 transcripts were not detected in any patient with several different primer combinations (Fig 5B). Normal ZNF198 and FGFR1 transcripts were detected in all individuals (Fig 5C).
Southern blot analysis using a ZNF198 cDNA probe that spanned the breakpoints detected rearranged bands in genomic DNA extracted from all five patients after digestion with either BglII orEcoRI (Fig 3B). Rearrangements were also found by Southern analysis using an FGFR1 probe encompassing exons 6 to 12 (not shown).
During the preparation of this manuscript, Xiao et al described essentially the same molecular lesion in four t(8;13) patients.15 However, the cDNA sequence and predicted amino acid sequence of ZNF198 as described here (Genbank: AJ224901) are considerably larger than that reported previously (Genbank: AF012126). Specifically, the sequence of Xiao et al predicts a protein that initiates at Met 621 of ZNF198 (Fig 4) and therefore lacks MYM domains 1, 2, and 3 (Fig 6). The reason for this difference is that the sequence described previously contained an insert of 80 bp between Phe 616 and Gln 617 of ZNF198, which introduced an in-frame stop codon. RT-PCR of normal peripheral blood leukocyte cDNA using a sense primer (DET17) to this 80-bp insert in combination with a downstream primer common to both sequences (DET26) resulted in amplification of a product of the expected size. However, semicompetitive PCR using DET17, DET26, and an upstream primer (DET22) resulted in a single visible amplification product of the same size as DET22-DET26 (Fig 5C). Similar results were found with several different primer combinations using normal and leukemic cDNA, indicating that the sequence published previously is probably a minor ZNF198 splice variant. This hypothesis is supported by the fact that our cDNA sequence corresponds to the size of the major transcript detected by Northern blotting and that no bands smaller than 5 kb are visible (Fig3A).
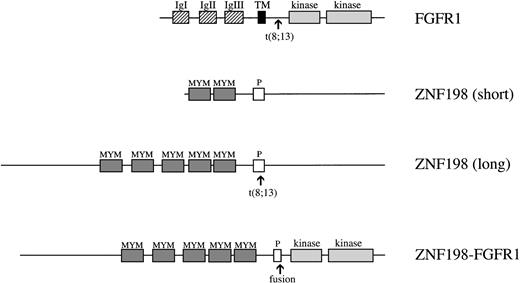
Schematic representations of the normal FGFR1, the shorter form of ZNF198 as described previously,15 the full-length ZNF198 as described here, and the full-length ZNF198-FGFR1 fusion protein. Abbreviations for FGFR1 are: IgI, II, and III, the three extracellular immunoglobulin-like domains; TM, transmembrane domain; kinase, split tyrosine kinase domain. Abbreviations for ZNF198 are: MYM, MYM domains; P, proline-rich domain.
Schematic representations of the normal FGFR1, the shorter form of ZNF198 as described previously,15 the full-length ZNF198 as described here, and the full-length ZNF198-FGFR1 fusion protein. Abbreviations for FGFR1 are: IgI, II, and III, the three extracellular immunoglobulin-like domains; TM, transmembrane domain; kinase, split tyrosine kinase domain. Abbreviations for ZNF198 are: MYM, MYM domains; P, proline-rich domain.
DISCUSSION
We have used a positional cloning strategy to determine the molecular basis of the t(8;13)(p11;q12), the most common cytogenetic abnormality associated with the 8p11 myeloproliferative disorder. We found that a novel gene, ZNF198, on chromosome 13 becomes fused to the FGFR1 on chromosome 8. Using FISH analysis we found that the t(8;13) is not simply a reciprocal translocation but also involves an inversion of a region of 13q11-12 that contains the ZNF198 gene. One possible explanation for this finding is that the orientation of the normal ZNF198 and FGFR1 genes with respect to the centromere are such that a straightforward reciprocal translocation would result in acentric and dicentric derivatives that could not segregate correctly at mitosis. Inversion of ZNF198 would enable the two derivative chromosomes to each carry one centromere. Similar findings have been described for the fusion of TEL to ABL in the t(9;12)16 and AF10 to MLL in the t(10;11).17 Presumably the requirement for a complex rearrangement explains why the t(8;13) is rare and also the fact that apparently widely separated 13q breakpoints have been described in patients with this translocation.2,13 18
Conceptual translation of the ZNF198 cDNA sequence predicts a protein of 1377 amino acids that shows significant homology to the DXS6673E/KIAA0385 and KIAA0425 gene products. The function of these genes is unknown, although DXS6673E is implicated as being responsible for one of the forms of X-linked mental retardation.19Alignment of the three protein sequences reveals a conserved motif of the general form CX2C19-22CX3CX13-19CX2CX19-25FCX3CX3F/Y that is repeated five times in each protein (Fig 4). These MYM domains have not been described before but are clearly reminiscent of the growing family of zinc finger-related motifs.20 Zinc fingers were originally identified as protein domains that mediate specific protein-DNA interactions, but more recently a number of related zinc binding structures have been described that mediate homotypic or heterotypic protein-protein interactions. The MYM domains found in ZNF198 family members more closely resemble the latter type, in particular the RING, LAP/PHD, and LIM domains.20
Fibroblast growth factors (FGFs) are involved in the control of growth, differentiation, and migration of diverse cell types. FGFs, of which there at least 13 types, mediate their effects by binding to four high affinity receptor tyrosine kinases, FGFR1-4, in concert with heparan sulphate proteoglycans.21 In common with other receptor tyrosine kinases, ligand binding induces FGFR dimerization followed by autophosphorylation of several intracellular tyrosine residues. These phosphotyrosines serve as specific docking sites for recruitment of target proteins, such as PLC-γ and effectors of the RAS/MAPK signaling pathway.22
Several lines of evidence have implicated abnormalities of the FGF system in malignancy. FGF4/hst is a frequently identified oncogene in NIH3T3 transformation assays,23 and several FGF family members are activated transcriptionally by integration of Mo-MLTV in murine mammary tumors.24 FGFR2 is constitutively activated by fusion to FRAG1 in rat osteosarcoma cells,25 and FGFR3 is mutated and juxtaposed to the IgH locus in some cases of multiple myeloma.26 Here we have shown that FGFR1 is disrupted by the t(8;13) in the 8p11 myeloproliferative disorder. ZNF198-FGFR1 retains the five MYM domains of ZNF198 and virtually the entire intracellular domain of FGFR1, including the tyrosine kinase domain (Fig 6). It is possible that the MYM domains of ZNF198 mediate dimerization of the fusion protein and thus activation of FGFR1 tyrosine kinase activity in a manner analogous to the coiled-coil domain of BCR in BCR-ABL27 and the helix-loop-helix domain of TEL/ETV6 in TEL-ABL and TEL-PDGFRβ.28,29 In support of this hypothesis, self-association mediated by CCCC zinc fingers has been described for GATA-1 and A20.30 31
Dominant activating mutations of FGFR1 are also found in some individuals with Pfeiffer’s syndrome,32 and activating mutations of FGFR2 and 3 are seen in a number of human craniosynostosis and dwarfism syndromes.33 However, these individuals do not have hematological abnormalities and do not appear to be unduly prone to the development of leukemia. This suggests that FGFR activation in these conditions may be qualitatively or quantitatively different from the activation of FGFR1 by fusion to ZNF198. Alternatively, other cooperating genetic events may be necessary for development of the 8p11 myeloproliferative syndrome, and an increased incidence of hematological abnormalities in patients with Pfeiffer’s syndrome have not been observed due to the rarity of this condition.
Remarkably, tyrosine kinases are emerging as a consistent feature of fusion genes thus far detected in MPDs, ie, BCR-ABL in CML,34,35 TEL-ABL and TEL-JAK2 in Ph-negative/atypical CML,36,37 TEL-PDGRFβ in CMML,38 and now ZNF198-FGFR1 in the 8p11 myeloproliferative syndrome. Although some of these fusions are also found in acute leukemias, it is tempting to speculate that activation of tyrosine kinase signal transduction pathways are a general feature of chronic MPDs.
ACKNOWLEDGMENT
We would like to thank the MRC HGMP Resource Centre (Hinxton, UK) for providing the clones used in this study and M. Fitchett (Oxford, UK) for fixed cells from patient J.B.
Supported by the Leukaemia Research Fund, the Dr Mildred Scheel Stiftung, and the Robert Zikel Memorial Fellowship.
Address reprint requests to Nicholas C.P. Cross, PhD, Department of Haematology, Imperial College School of Medicine, Hammersmith Hospital, Du Cane Road, London W12 ONN, UK; e-mail:ncross@rpms.ac.uk.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal