Abstract
In three Italian patients, two point mutations and a short deletion were found in the intron 7 of factor VII gene, clustered in the donor splice site and located in the first of several repeats. The mutation 9726+5G→A, the most frequent cause of symptomatic factor VII deficiency in Italy, as well as the deletion (9729del4) gave rise in expression studies to abnormally spliced transcripts, which were exclusively produced from the cryptic site in the second repeat. The insertion in the mature mRNA of the first intronic repeat caused (9726+5G→A) a reading frameshift, abolishing most of the factor VII catalytic domain, or produced (9729del4), an altered factor with 11 additional residues, the activity of which was not detectable in the cell medium after mutagenesis and expression studies. Studies of factor VII ectopic mRNA from leukocytes and expression studies indicated that the deleted gene produced 30% of normally spliced transcript. Differently, the 9726+5G→A mutation permitted a very low level (0.2% to 1%) of correct splicing to occur, which could be of great importance to prevent the onset, in the homozygous patients, of most of the life-threatening bleeding symptoms. The 9726+7A→G mutation was found to be a rare and functionally silent polymorphism. These findings, which provide further evidence of the interplay of sequence and position in the 5′ splice site selection, throw light on the heterogeneous molecular bases and clinical phenotypes of FVII deficiency.
© 1998 by The American Society of Hematology.
FACTOR VII (gene symbol, F7; acronym, FVII) is a vitamin K–dependent serine protease glycoprotein circulating as a zymogen in plasma, with a pivotal role in the initiation of blood coagulation.1 While increased levels of FVII are associated with an increased risk of coronary heart disease,2 very low levels of FVII (<2% of normal) cause severe bleeding disorders in patients.3 FVII gene knock-out experiments in mice showed a reduction in the live birth rate and very frequent death in the first day after birth.4 Patterns of mutations have been defined in FVII deficiency,3,5-7 which offer the opportunity to investigate the mechanisms through which the genotypes produce a low level of FVII expression.8-13
FVII gene belongs to a subgroup of coagulation serine proteases, which also includes factor IX, factor X, and protein C, characterized by sequence and structure homology.14,15 Different from the other members of this family, FVII gene16 contains in introns 1A, 1B, 2, 7 and in exon 8, five minisatellites imperfect tandem repeats with monomer element lengths ranging from 14 to 37 bp.17 The intron 7 (IVS7) is characterized by the presence of a variable number (ranging from 5 to 8) of 37-bp repeats,17-19 the first of which contains the IVS7 donor splice site followed by several repeats showing cryptic sites completely homologous in sequence to the functional site. Two mutations, one (9726+5G→A) present in several Italian patients with symptomatic FVII deficiency and another (9726+7A→G) in an asymptomatic subject, have been found in the IVS7 donor splice site.20 However, the functional meaning of both mutations has not been defined.
This gene region, characterized by sequence and repeat variations and thus potentially a “hot spot mutation site,”21 was investigated in one patient with symptomatic FVII deficiency and in two patients selected for reduced FVII level. Mutants were characterized through expression studies in mammalian cells, which provided a sensitive and specific assay for normal and altered splicing products, as well as for mutant FVII.
MATERIALS AND METHODS
Subjects.
Patient 1 was referred at the time of menarca when she showed severe meno-metrorrhagia and was treated with concentrated FVII (Provetin UM; Immuno, Wien, Austria). She experienced two pregnancies without any complication. The FVII activity level was less than 1% as compared with the pooled normal plasma. The enzyme-linked immunoadsorbent assay (ELISA) assay showed an undetectable level of FVII antigen.
Patients 2 and 3 were asymptomatic and were referred for a mildly prolonged prothrombin time (>2 standard deviation from the prothrombin time of the reference plasma), detected in a presurgery screening. FVII:C and FVII:Ag levels were 61% and 58% for patient 2 and 43% and 42% for patient 3.
Genomic studies and reverse transcriptase-polymerase chain reaction (RT-PCR).
DNA and RNA extraction, PCR amplification, restriction and direct sequencing were as previously described.6 Amplification of genomic fragments for cloning was obtained by using the Expand high fidelity PCR system (Boehringer Mannheim, Mannheim, Germany) with the oligos 2-6 (Table 1) in 30 cycles: 20 seconds denaturation at 94°C; 3 seconds annealing at 52°C; 30 seconds extension at 70°C. Total RNA from the buffy-coat fraction of peripheral blood or from 106 baby hamster kidney (BHK) cells was used as template for cDNA synthesis with 40 U of avian myeloblastosis virus RT with 10 nmol of oligo 4 or 5 and the RT reaction mixture (1/10) was amplified with oligos 1-5, 3-4, or 3-5 (Table 1).
Primers
| Primer . | Oligonucleotides Sequence and Numbering . | Exon . |
|---|---|---|
| 1 | 8948AAACCCCAAGGCCGAATTG8966 | 6 |
| 2 | 9568AATGTGACTTCCACACCTCC9587 | 7 |
| 3 | 9641CCCTGATCAACACCATCTGG9663 | 7 |
| 4 | 10609GGGATGATGACCTGCGCCAC10590 | 8 |
| 5 | 10652CAGCGCGATGTCGTGGTT10635 | 8 |
| 6 | 10908CGGCACAGAACATGTACTCC10889 | 8 |
| 7 | 9712TGATCGCGGTGCTGG9726-gtaccactctcccct-10543GCGAGCACGACCTCA10557 | |
| 8 | 10557TGAGGTCGTGCTCGC10543-aggggagagtggtac-9726CCAGCACCGCGATCA9712 | |
| 9 | 9727GTACCACTCTCCCCT9741-gtccgaccgcggtgctgg-10543GCGAGCACGACCTCA10557 | |
| 10 | 10557TGAGGTCGTGCTCGC10543-ccagcaccgcggtcggac-9741AGGGGAGAGTGGTAC9727 |
| Primer . | Oligonucleotides Sequence and Numbering . | Exon . |
|---|---|---|
| 1 | 8948AAACCCCAAGGCCGAATTG8966 | 6 |
| 2 | 9568AATGTGACTTCCACACCTCC9587 | 7 |
| 3 | 9641CCCTGATCAACACCATCTGG9663 | 7 |
| 4 | 10609GGGATGATGACCTGCGCCAC10590 | 8 |
| 5 | 10652CAGCGCGATGTCGTGGTT10635 | 8 |
| 6 | 10908CGGCACAGAACATGTACTCC10889 | 8 |
| 7 | 9712TGATCGCGGTGCTGG9726-gtaccactctcccct-10543GCGAGCACGACCTCA10557 | |
| 8 | 10557TGAGGTCGTGCTCGC10543-aggggagagtggtac-9726CCAGCACCGCGATCA9712 | |
| 9 | 9727GTACCACTCTCCCCT9741-gtccgaccgcggtgctgg-10543GCGAGCACGACCTCA10557 | |
| 10 | 10557TGAGGTCGTGCTCGC10543-ccagcaccgcggtcggac-9741AGGGGAGAGTGGTAC9727 |
Sequence and position of the oligonucleotides used for PCR amplification, reverse transcription, and mutagenesis. The nucleotides inserted by mutagenesis are indicated with lowercase letters.
FVII Expression Levels From Transiently Transfected BHK Cells
| . | FVII Antigen . | FVII Activity . | |
|---|---|---|---|
| Media . | Cell Lysate . | Media . | |
| Wt-FVII | 42.8 | 5.5 | 37 |
| Ins11FVII | 4.8 | 5.0 | 0 |
| . | FVII Antigen . | FVII Activity . | |
|---|---|---|---|
| Media . | Cell Lysate . | Media . | |
| Wt-FVII | 42.8 | 5.5 | 37 |
| Ins11FVII | 4.8 | 5.0 | 0 |
WtFVII, normal FVII; Ins11FVII, mutant FVII with the insertion of 11 residues (GTTLPCPTAVL) at position 208. Levels in cell lysates and culture medium were measured 48 hours after transfection and reported as ng/106 cells.
Cloning.
The genomic region spanning nucleotides 9568-10908 was amplified from DNA of normal subjects (carrying 6 or 7 repeats) and from patients, restricted by Sac I and inserted in the pUC18 vector. Single clones containing the mutation were isolated and subcloned in the eucaryotic expression vector pCDNA3 (Invitrogen Corp, Carlsbad, CA) by using the XbaI and EcoRI restriction sites. Each construct is designated with the name of the mutation inserted. The complete human factor VII cDNA22 was kindly provided by Dr John H. McVey (MRC Clinical Sciencies Centre, London, UK) and cloned in the pCDNA3 by using the XbaI and HindIII restriction sites.
Mutagenesis.
The additional 33-bp (GTACCACTCTCCCCTGTCCGACCGCGGTGCTGG) were inserted at the position 9726 of the FVII-cDNA by using the QuikChange Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to the instruction of the manufacturer. Fifteen base pairs (nt 9727-9741) were inserted using primers 7-8 (Table 1). The mutagenized vector was used as template for a second mutagenesis with oligonucleotides 9-10 (Table 1). For each step, 18 cycles were run as follow: 30 seconds denaturation at 95°C; 1 minute annealing at 45°C; 16 minutes extension at 68°C.
Cell culture, transfection, and expression.
BHK cells were grown in Dulbecco’s modified essential medium supplemented with 10% inactivated fetal calf serum (FCS), 4 mmol/L L-glutamine, 125 U/mL of penicillin, and 125 mg/mL of streptomycin and nonessential amino acids in the 5% CO2 atmosphere at 37°C. A total of 5 × 105 cells were transfected into 10-mm petri-dish using lipofectin (GIBCO-BRL, Gaithersburg, MD). Cells and medium were harvested and studied after 48 hours. Cell lysates were obtained as previously reported.23
Measurement of FVII activity and antigen.
The FVII antigen level was measured by using an enzyme immunoassay (Asserachrom VII:Ag, Diagnostica Stago, Asnieres, France). FVII coagulant activity was determined by a one-stage method using FVII-deficient plasma as a substrate and a commercial human thromboplastin preparation. A standard normal pooled plasma prepared from 50 healthy donors was defined to contain 100% FVII antigen and 100% FVII activity.
RESULTS
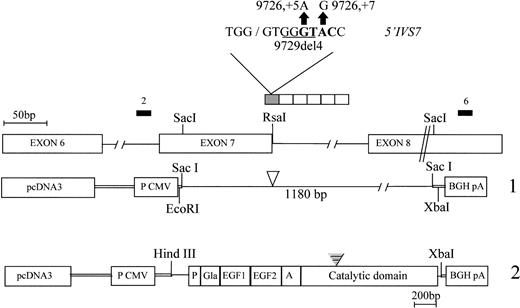
DNA from patients with FVII deficiency was screened by restriction for the presence of FVII gene mutations previously described in the Italian population.7 The suppression of an Rsa I restriction site in the amplified IVS7 (Fig1), which was compatible with two transitions (9726+5G→A, 9726+7A→G) previously reported,20 was detected in patients 1 and 3. The Rsa I restriction produced in patient 2 a shorter band. Sequencing showed in patients 1 and 3 the 9726+5 G to A transition in the homozygous condition and the 9726+7A to G transition in the heterozygous condition, respectively. Differently, a 4-bp deletion (nt 9729-9732, Fig 1) was found in the heterozygous condition in patient 2. The inheritance of this short deletion and of the coagulation phenotype was demonstrated in his granddaughter, characterized by a parallel reduction in FVII activity (47%) and in FVII antigen (42%).
Schematic diagram of the mutated FVII gene region and of the vectors used for the expression of mutant mRNA (1) and protein (2). The sequence of the Rsa I site is reported in bold; 2 and 6, primers. The open and closed triangles indicate the position of the IVS7 mutations and of the additional residues inserted in the FVII cDNA, respectively. P, Gla, EGF1, EGF2 and A, FVII domains.
Schematic diagram of the mutated FVII gene region and of the vectors used for the expression of mutant mRNA (1) and protein (2). The sequence of the Rsa I site is reported in bold; 2 and 6, primers. The open and closed triangles indicate the position of the IVS7 mutations and of the additional residues inserted in the FVII cDNA, respectively. P, Gla, EGF1, EGF2 and A, FVII domains.
The RsaI restriction excluded the presence of these mutations in 100 normal controls. The consensus values (CV) of the wild-type 5′IVS7 splice site, calculated according to Shapiro and Senapathy,24 was 74.7 and a cryptic site with an identical CV is present in each repeat. The 9726+5A and 9729del4 mutations lowered this value to 60.9 and 58.1, respectively. Cryptic sites at positions +4, +19, and +30 are detectable with CV of 63.3, 53.9, and 56.7, respectively. The cryptic site at position +4 was removed by the 9726+5G→A transition and by the 9729del4 mutation.
Because the sequence statistics of Shapiro and Senapathy do not include the position +7, the variation of CV for the 9726+7A→G transition cannot be estimated. However, this mutation makes the regions of the functional donor splice site and of the cryptic site located at +37 completely overlapping for 25 bases. This mutation was found to be associated in patient 3 with the presence of Gln at position 353, a frequent FVII gene polymorphic allele.
mRNA studies.
The availability of fresh white blood cells enabled us to study in patient 2 the FVII gene expression at the mRNA level by RT-PCR of illegitimate transcripts. The size of the cDNA (Fig 2A) was compatible with splicing of exons 7 and 8, whereas the absence of a splicing product 172 bp in size excluded the occurrence of exon 7 skipping. Restriction by BclI showed the expected normal bands (190 bp and 104 bp, Fig 2A) and an additional band larger in size, compatible with the presence in the spliced mRNA of a short intronic sequence.
Studies of ectopic and in vitro–expressed FVII mRNA. (A) RT-PCR amplification of white blood cell mRNA obtained from patient 2 carrying the 9729del4 mutation. PCR products (primers 1-5) were separated by 8% PAGE (left) or by 11% PAGE after Bcl I restriction (right). NC, negative control. (B) RT-PCR amplification of FVII mRNA expressed in cells transfected with constructs containing the three mutations (9729del4, 9726+5A, 9726+7G) or the normal sequence (N). Upper part, ethidium bromide staining of fragments amplified with primers 3 and 4; M, size marker. Lower part, autoradiographs of the same fragments labeled by 32P and restricted by TaqI. Gels were overexposed. (C) Schematic diagrams of FVII cDNAs (exons 6-8) and of fragments amplified from the ectopic mRNA (primers 1-5) or from the mRNA expressed in transfected cells (primers 3-4). The restriction sites used in (A) (Bcl I) and (B) (Taq I) are also indicated together with the sizes of fragments, which are given in bp. Length of abnormally spliced transcripts and of their restricted fragments are reported in parentheses. The 182-bp and 186-bp fragments were obtained from the 9729del4 and 9726+5A constructs, respectively. The 144-bp fragment was obtained by Taq I restriction of the 186-bp amplified fragment. The gray box indicates the repeat inserted in the abnormally spliced transcripts.
Studies of ectopic and in vitro–expressed FVII mRNA. (A) RT-PCR amplification of white blood cell mRNA obtained from patient 2 carrying the 9729del4 mutation. PCR products (primers 1-5) were separated by 8% PAGE (left) or by 11% PAGE after Bcl I restriction (right). NC, negative control. (B) RT-PCR amplification of FVII mRNA expressed in cells transfected with constructs containing the three mutations (9729del4, 9726+5A, 9726+7G) or the normal sequence (N). Upper part, ethidium bromide staining of fragments amplified with primers 3 and 4; M, size marker. Lower part, autoradiographs of the same fragments labeled by 32P and restricted by TaqI. Gels were overexposed. (C) Schematic diagrams of FVII cDNAs (exons 6-8) and of fragments amplified from the ectopic mRNA (primers 1-5) or from the mRNA expressed in transfected cells (primers 3-4). The restriction sites used in (A) (Bcl I) and (B) (Taq I) are also indicated together with the sizes of fragments, which are given in bp. Length of abnormally spliced transcripts and of their restricted fragments are reported in parentheses. The 182-bp and 186-bp fragments were obtained from the 9729del4 and 9726+5A constructs, respectively. The 144-bp fragment was obtained by Taq I restriction of the 186-bp amplified fragment. The gray box indicates the repeat inserted in the abnormally spliced transcripts.
Because the study of illegitimate transcripts was not feasible in patients 1 and 3 and in addition two mutations were present combined with a normal allele (patients 2 and 3), the mechanisms through which the putative gene defects exert their phenotypic effects were investigated in expression studies. The complete IVS7, the 3′ region of exon 7 and the 5′ region of exon 8 were amplified from DNA of each patient and inserted in a mammalian expression vector (Fig1). In addition, the IVS7 from normal controls was also amplified and cloned.
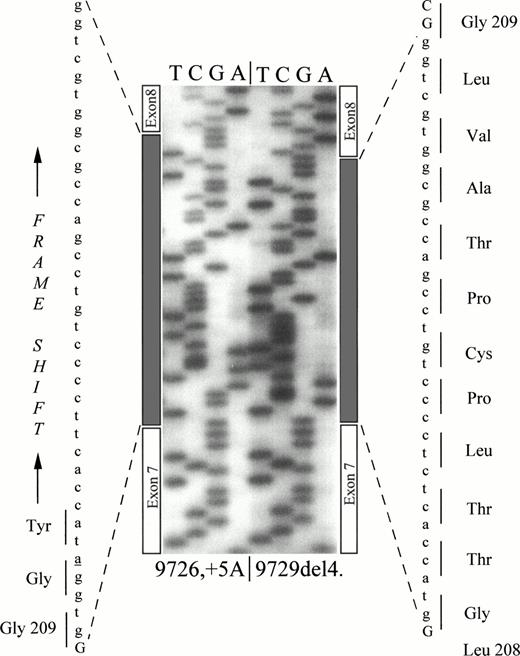
The spliced mRNA was evaluated by RT-PCR of total RNA extracted from transfected cells (Fig 2B). The normal cDNA (149 bp, Fig 2B) was detected in the expression of normal control and of the constructs bearing the 9726+7G or the 9729del4 mutations. A fragment larger in size was detected in the expression of the 9729del4 and 9726+5A mutants. All fragments were characterized by direct sequencing (Fig 3), which showed that both the altered splicing products included the first intronic repeat of IVS7. The abnormal splicing caused the insertion of 37 bp in the mRNA bearing the 9726+5A mutation and of 33 bp in the mRNA transcribed from the 9729del4 mutant. The former insertion predicted a reading frameshift and premature termination, whereas the latter predicted an in-frame insertion of 11 codons (Fig 3).
Sequencing of abnormally spliced cDNA. The gray box indicates the inserted sequences. The 11 additional residues inserted between Leu208 and Gly209 and the frame-shifting after Gly209 are indicated.
Sequencing of abnormally spliced cDNA. The gray box indicates the inserted sequences. The 11 additional residues inserted between Leu208 and Gly209 and the frame-shifting after Gly209 are indicated.
The relative amounts of the normal cDNA and of the cDNA with the 33-bp insertion, evaluated by densitometric scanning of polyacrylamide gels from ectopic mRNA, were similar. In expression experiments the normal band was 30% (mean of three transfections) of the altered one. 32P-labeling of PCR products (Fig 2B) was used to investigate the apparent absence, after ethidium bromide staining, of normally spliced products from the 9726+5A mutant. The densitometric evaluation of the autoradiographic pattern, after three different transfection experiments and serial dilutions (not shown) of RT-PCR products, indicated the presence of a normal cDNA ranging from 0.2% to 1% of the altered cDNA. Restriction with Taq I (Fig 2B and C) produced a fragment 107-bp in size, specific for the normal cDNA.
No altered cDNA including the first repeat was detectable after labeling the RT-PCR products of the 9726+7G mutant (Fig 2B), as well as of the two control IVS7 (6 or 7 repeats).
Protein studies.
The presence of large amounts of normal FVII in the heterozygous patient 2 complicates the evaluation of the altered protein translated from the mRNA with the 33-bp insertion. A vector containing the in-frame insertion of 11 codons in the FVII cDNA was obtained by mutagenesis and the FVII variant was investigated in transient transfection experiments (Table 2). While similar amounts of FVII antigen were detectable in lysate of cells transfected with normal and mutated vectors, a very low level of FVII antigen with undetectable activity was measured in medium of cells transfected with the mutant cDNA.
DISCUSSION
We report the study in the FVII gene of two transitions and a short deletion, localized in the donor splice site of the IVS7 and clustered in a short direct repeat (TGGGTGGGTA), a potential “hot spot” for mutation.21 Among the members of the coagulation serine protease family, this intron provides a peculiar model for the study of splicing mutations because the donor splice site, located in the first of several highly homologous 37-bp repeats, is followed by several cryptic splice sites, some of them with identical CV. The analysis and the estimate of the relative amounts of splicing products were favored by the absence of exon skipping and by the expected negligible differences in reverse transcription and PCR amplification efficiency of fragments similar in size (Fig 2C). Although obtained by transient transfections of BHK cells, which may not reflect relative mRNA levels in the hepatocyte, our studies provided further evidence of the interplay of sequence and position among the several parameters involved in the 5′ splice site selection. In normal, polymorphic, and mutated IVS7, the cryptic sites with lower CV were ignored and only the proximal among the several identical sites with higher score was chosen, which underlines the importance of position of splicing signals for the spliceosome.
Because patient 3 was homozygous for the Gln353 allele of the common polymorphism (Arg353Gln) associated with reduced FVII level,25,26 expression studies were required to define the single role of the 9726+7A→G transition, a position found mutated in other coagulation serine proteases deficiencies.27 After expression and labeling of PCR products, only normally spliced mRNA was detected, thus indicating that the 9726+7A→G mutation is a rare and functionally silent polymorphism and that the reduction of FVII activity in the patient is probably explained by the functional Arg353Gln substitution.
Two of the mutations under study (9726+5G→A, 9729del4), which cause a reduction of the CV, predict a reduction in efficiency of normal splicing and therefore of FVII levels.
The 9726+5G→A mutation is the most frequent mutation responsible for clinically symptomatic FVII deficiency in Italy,20 as demonstrated by the allelic frequency (3%) that we have detected in a small village near Rome and by the seven unrelated homozygous patients so far diagnosed by us in central Italy. The severe impairment of normal splicing and the detection of abnormal spliced transcript including 37 additional bp demonstrated the causative nature of this mutation. The insertion caused a reading frameshift and premature termination of translation at residue 231, thus abolishing most of the C-terminal catalytic domain (residues 168-406). Although the altered mRNA was highly prevalent (approximately 99%), we demonstrated that the mutation permits a minute amount of correct splicing to occur, which predicts the encoding of very low amounts of circulating FVII. Even the very low level of FVII activity at the beginning of blood coagulation may explain the moderate clinical phenotype observed in patient 1. Other Italian patients homozygous for the 9726+5G→A mutation20 were characterized by moderate to severe bleeding symptoms. Differently, in patients with mutations not compatible with residual FVII activity,10,28a severe and life-threatening bleeding condition was observed and in FVII gene knock-out experiments in mice,4 death was very frequent in the first day after birth. Because very low FVII values cannot be properly evaluated with the current assays for FVII:C and FVII:Ag, mRNA studies are of importance to establish the relationship between the residual FVII activity and the clinical phenotype.
Although the 9729del4 produced a donor splicing sequence with a CV lower than that of the 9726+5G→A mutation, it was compatible with the production of sensibly higher amounts of the normally spliced transcript, which defines this short deletion as responsible for a mild FVII deficiency. As expected from the heterozygous condition of the deletion in patient 2, the relative amount of normal transcript was higher in the ectopic mRNA than in the in vitro-expressed mRNA. The deletion of 4 bp caused the in frame insertion of 33 bp, thus producing an altered FVII molecule with 11 additional residues (Fig 3). The inspection of the crystallographic model of FVIIa29 showed that the residues at the site of insertion (Leu208-Gly209) are mostly covered by a chain (residues 219-223) exposed on the surface of the catalytic domain. The insertion of the 11 additional amino acids predicts major rearrangements that led in a theoretical model30 to separate Leu208 from Gly209 by 10 Å and to expose on the surface a new chain. In medium of cells transfected with the mutant cDNA, very low levels of the abnormal protein were detectable by FVII-specific antibodies, and no FVII activity was measured, which is in accordance with the reduction in FVII activity and antigen observed in the plasma of the heterozygous patient and of his granddaughter.
The analysis of the expression of these splicing mutations throws light on the heterogeneous molecular bases of FVII deficiency and elucidates the mechanisms that are able to modulate FVII levels and to produce a spectrum of clinical phenotypes.
ACKNOWLEDGMENT
We thank Dr John H. McVey for providing the FVII-cDNA and Dr Anita Kavlie for helpful suggestions.
Supported by Consiglio Nazionale Delle Ricerche Target Project on Biotechnology and by Ministero Università Ricerca Scientifica Technologica.
Address reprint requests to Francesco Bernardi, BS, Dipartimento di Biochimica e Biologia Molecolare, Universita’ di Ferrara, Via L.Borsari 46, 44100 Ferrara, Italy; e-mail:BER@DNS.UNIFE.IT.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal